Submitted:
28 December 2024
Posted:
30 December 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Sall4 as a Multifaceted Regulator in Embryonic Stem Cells, Development, and Tissue Progenitors
3. Sall4’s Role in Heart Development and Regeneration
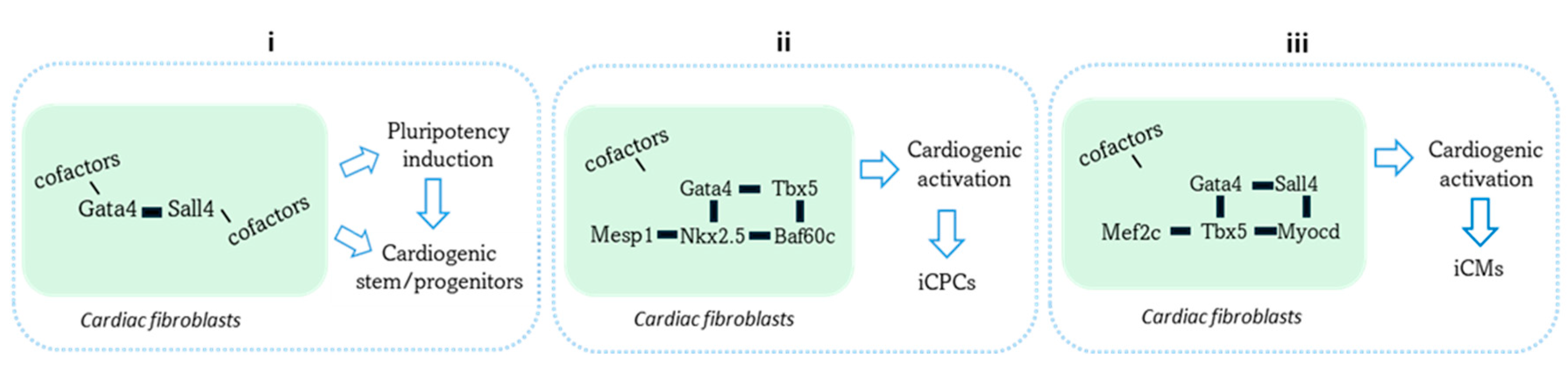
4. Sall4’s Role in Cardiac Cellular Reprogramming and Therapeutics
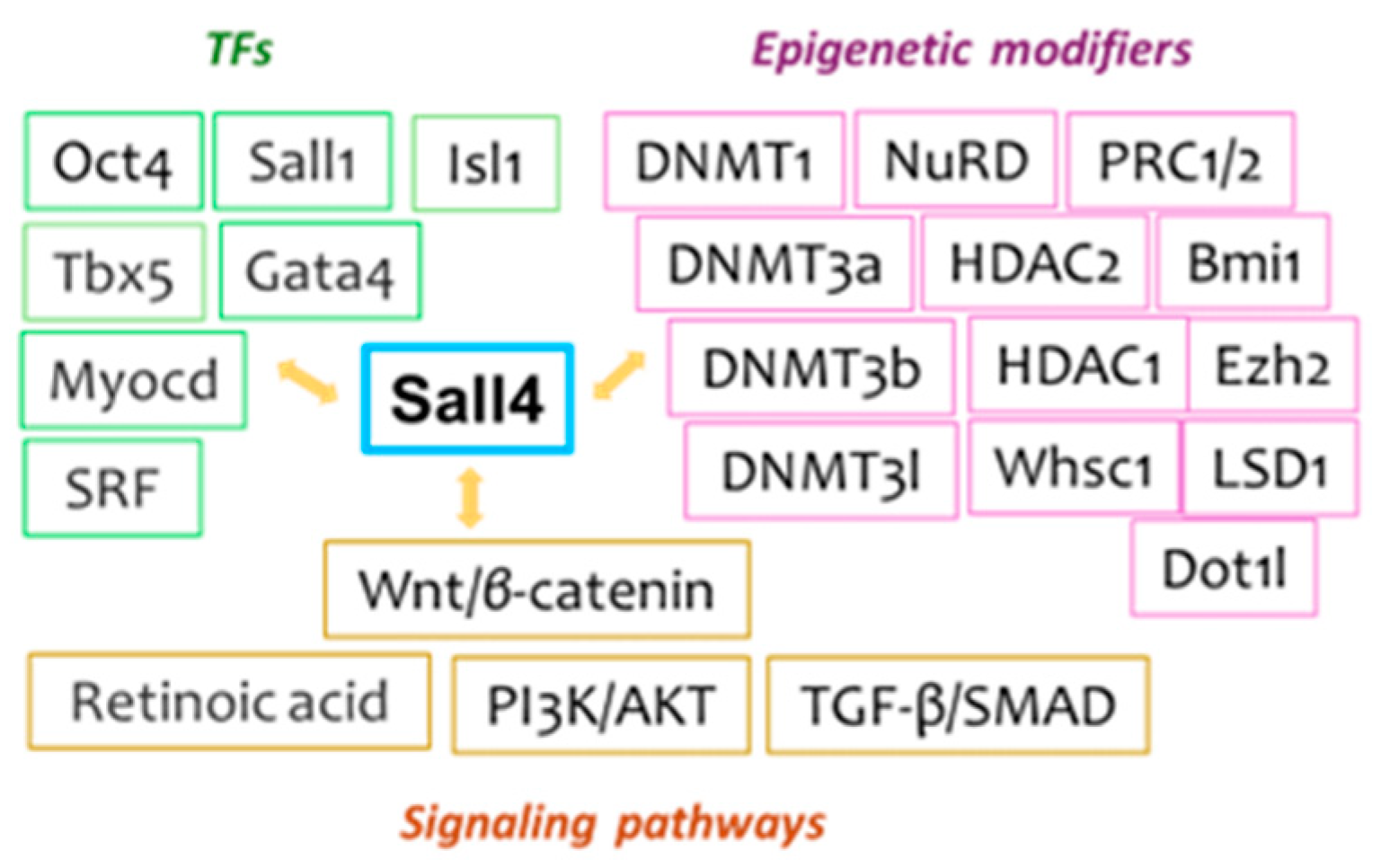
5. Sall4-Mediated Regulatory Networks and Pathways
5.1. Interacting Transcription Factors (TFs)
5.2. Epigenetic Regulation Partners. Epigenetic Modifying Machineries Critically Influence Sall4’s Regulatory Functions, Many of Which Are Vital in Regenerative Cardiac Biology [8,9,10,83,84]. Key Examples Include DNMTs, Histone Modifying Enzymes [85,86,87], and Several Chromatin-Modifying Complexes. Additional Contributors, such as the Set Histone Methyltransferase Whsc1, Further Highlight the Complexity of Sall4's Epigenetic Interactions
5.3. Signaling Pathways. In line with the Complex Transcriptional and Epigenetic Interactions Described Above, a Range of Sall4-Modulated Signaling Pathways Are Involved in Cardiobiology, with Several Important Pathways Outlined Below
5.4. Competitive and Dynamic Regulation of Interacting Pathways
6. Therapeutic Potential for Sall4 in Cardiac Regeneration and Repair
7. Concluding Perspectives
Funding
Informed Consent Statement
Conflicts of Interest
References
- Garry, G.A.; Olson, E.N. Reprogramming of cardiac cell fate as a therapeutic strategy for ischemic heart disease. J Mol Cell Cardiol 2023, 179, 2-6. [CrossRef]
- Hashimoto, H.; Olson, E.N.; Bassel-Duby, R. Therapeutic approaches for cardiac regeneration and repair. Nat Rev Cardiol 2018, 15, 585-600. [CrossRef]
- Kohlhase, J.; Heinrich, M.; Liebers, M.; Frohlich Archangelo, L.; Reardon, W.; Kispert, A. Cloning and expression analysis of SALL4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenet Genome Res 2002, 98, 274-277. [CrossRef]
- Warren, M.; Wang, W.; Spiden, S.; Chen-Murchie, D.; Tannahill, D.; Steel, K.P.; Bradley, A. A Sall4 mutant mouse model useful for studying the role of Sall4 in early embryonic development and organogenesis. Genesis 2007, 45, 51-58. [CrossRef]
- Kohlhase, J.; Heinrich, M.; Schubert, L.; Liebers, M.; Kispert, A.; Laccone, F.; Turnpenny, P.; Winter, R.M.; Reardon, W. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet 2002, 11, 2979-2987. [CrossRef]
- Kohlhase, J.; Chitayat, D.; Kotzot, D.; Ceylaner, S.; Froster, U.G.; Fuchs, S.; Montgomery, T.; Rosler, B. SALL4 mutations in Okihiro syndrome (Duane-radial ray syndrome), acro-renal-ocular syndrome, and related disorders. Hum Mutat 2005, 26, 176-183. [CrossRef]
- Al-Baradie, R.; Yamada, K.; St Hilaire, C.; Chan, W.M.; Andrews, C.; McIntosh, N.; Nakano, M.; Martonyi, E.J.; Raymond, W.R.; Okumura, S.; et al. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet 2002, 71, 1195-1199. [CrossRef]
- Yang, J. SALL4 as a transcriptional and epigenetic regulator in normal and leukemic hematopoiesis. Biomark Res 2018, 6, 1. [CrossRef]
- Tatetsu, H.; Kong, N.R.; Chong, G.; Amabile, G.; Tenen, D.G.; Chai, L. SALL4, the missing link between stem cells, development and cancer. Gene 2016, 584, 111-119. [CrossRef]
- Moein, S.; Tenen, D.G.; Amabile, G.; Chai, L. SALL4: An Intriguing Therapeutic Target in Cancer Treatment. Cells 2022, 11. [CrossRef]
- Lim, C.Y.; Tam, W.L.; Zhang, J.; Ang, H.S.; Jia, H.; Lipovich, L.; Ng, H.H.; Wei, C.L.; Sung, W.K.; Robson, P.; et al. Sall4 regulates distinct transcription circuitries in different blastocyst-derived stem cell lineages. Cell Stem Cell 2008, 3, 543-554. [CrossRef]
- Tahara, N.; Kawakami, H.; Chen, K.Q.; Anderson, A.; Yamashita Peterson, M.; Gong, W.; Shah, P.; Hayashi, S.; Nishinakamura, R.; Nakagawa, Y.; et al. Sall4 regulates neuromesodermal progenitors and their descendants during body elongation in mouse embryos. Development 2019, 146. [CrossRef]
- Pappas, M.P.; Kawakami, H.; Corcoran, D.; Chen, K.Q.; Scott, E.P.; Wong, J.; Gearhart, M.D.; Nishinakamura, R.; Nakagawa, Y.; Kawakami, Y. Sall4 regulates posterior trunk mesoderm development by promoting mesodermal gene expression and repressing neural genes in the mesoderm. Development 2024, 151. [CrossRef]
- Paik, E.J.; Mahony, S.; White, R.M.; Price, E.N.; Dibiase, A.; Dorjsuren, B.; Mosimann, C.; Davidson, A.J.; Gifford, D.; Zon, L.I. A Cdx4-Sall4 regulatory module controls the transition from mesoderm formation to embryonic hematopoiesis. Stem Cell Reports 2013, 1, 425-436. [CrossRef]
- Akiyama, R.; Kawakami, H.; Wong, J.; Oishi, I.; Nishinakamura, R.; Kawakami, Y. Sall4-Gli3 system in early limb progenitors is essential for the development of limb skeletal elements. Proc Natl Acad Sci U S A 2015, 112, 5075-5080. [CrossRef]
- Oikawa, T.; Kamiya, A.; Kakinuma, S.; Zeniya, M.; Nishinakamura, R.; Tajiri, H.; Nakauchi, H. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology 2009, 136, 1000-1011. [CrossRef]
- Yamaguchi, Y.L.; Tanaka, S.S.; Kumagai, M.; Fujimoto, Y.; Terabayashi, T.; Matsui, Y.; Nishinakamura, R. Sall4 is essential for mouse primordial germ cell specification by suppressing somatic cell program genes. Stem Cells 2015, 33, 289-300. [CrossRef]
- Hobbs, R.M.; Fagoonee, S.; Papa, A.; Webster, K.; Altruda, F.; Nishinakamura, R.; Chai, L.; Pandolfi, P.P. Functional antagonism between Sall4 and Plzf defines germline progenitors. Cell Stem Cell 2012, 10, 284-298. [CrossRef]
- Monteon, A.; Hughes, L.; Camberos, V.; Kearns-Jonker, M. Identification of SALL4 Expressing Islet-1+ Cardiovascular Progenitor Cell Clones. Int J Mol Sci 2023, 24. [CrossRef]
- Katano, W.; Mori, S.; Sasaki, S.; Tajika, Y.; Tomita, K.; Takeuchi, J.K.; Koshiba-Takeuchi, K. Sall1 and Sall4 cooperatively interact with Myocd and SRF to promote cardiomyocyte proliferation by regulating CDK and cyclin genes. Development 2023, 150. [CrossRef]
- Gao, C.; Kong, N.R.; Li, A.; Tatetu, H.; Ueno, S.; Yang, Y.; He, J.; Yang, J.; Ma, Y.; Kao, G.S.; et al. SALL4 is a key transcription regulator in normal human hematopoiesis. Transfusion 2013, 53, 1037-1049. [CrossRef]
- Aguila, J.R.; Liao, W.; Yang, J.; Avila, C.; Hagag, N.; Senzel, L.; Ma, Y. SALL4 is a robust stimulator for the expansion of hematopoietic stem cells. Blood 2011, 118, 576-585. [CrossRef]
- Yang, J.; Aguila, J.R.; Alipio, Z.; Lai, R.; Fink, L.M.; Ma, Y. Enhanced self-renewal of hematopoietic stem/progenitor cells mediated by the stem cell gene Sall4. J Hematol Oncol 2011, 4, 38. [CrossRef]
- Abouelnazar, F.A.; Zhang, X.; Wang, M.; Zhang, J.; Yu, D.; Zang, X.; Zhang, J.; Li, Y.; Xu, J.; Yang, Q.; et al. The new advance of SALL4 in cancer: Function, regulation, and implication. J Clin Lab Anal 2023, 37, e24927. [CrossRef]
- Sun, B.; Xu, L.; Bi, W.; Ou, W.B. SALL4 Oncogenic Function in Cancers: Mechanisms and Therapeutic Relevance. Int J Mol Sci 2022, 23. [CrossRef]
- Koshiba-Takeuchi, K.; Takeuchi, J.K.; Arruda, E.P.; Kathiriya, I.S.; Mo, R.; Hui, C.C.; Srivastava, D.; Bruneau, B.G. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet 2006, 38, 175-183. [CrossRef]
- Sakaki-Yumoto, M.; Kobayashi, C.; Sato, A.; Fujimura, S.; Matsumoto, Y.; Takasato, M.; Kodama, T.; Aburatani, H.; Asashima, M.; Yoshida, N.; et al. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development 2006, 133, 3005-3013. [CrossRef]
- Bruneau, B.G.; Nemer, G.; Schmitt, J.P.; Charron, F.; Robitaille, L.; Caron, S.; Conner, D.A.; Gessler, M.; Nemer, M.; Seidman, C.E.; et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 2001, 106, 709-721. [CrossRef]
- Misra, C.; Chang, S.W.; Basu, M.; Huang, N.; Garg, V. Disruption of myocardial Gata4 and Tbx5 results in defects in cardiomyocyte proliferation and atrioventricular septation. Hum Mol Genet 2014, 23, 5025-5035. [CrossRef]
- Wang, B.; Li, L.; Xie, X.; Wang, J.; Yan, J.; Mu, Y.; Ma, X. Genetic variation of SAL-Like 4 (SALL4) in ventricular septal defect. Int J Cardiol 2010, 145, 224-226. [CrossRef]
- Elling, U.; Klasen, C.; Eisenberger, T.; Anlag, K.; Treier, M. Murine inner cell mass-derived lineages depend on Sall4 function. Proc Natl Acad Sci U S A 2006, 103, 16319-16324. [CrossRef]
- Kohlhase, J. SALL4-Related Disorders. In GeneReviews((R)), Adam, M.P., Feldman, J., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Amemiya, A., Eds.; Seattle (WA), 1993.
- Qian, L.; Huang, Y.; Spencer, C.I.; Foley, A.; Vedantham, V.; Liu, L.; Conway, S.J.; Fu, J.D.; Srivastava, D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012, 485, 593-598. [CrossRef]
- Song, K.; Nam, Y.J.; Luo, X.; Qi, X.; Tan, W.; Huang, G.N.; Acharya, A.; Smith, C.L.; Tallquist, M.D.; Neilson, E.G.; et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012, 485, 599-604. [CrossRef]
- Ieda, M.; Fu, J.D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375-386. [CrossRef]
- Tani, H.; Sadahiro, T.; Yamada, Y.; Isomi, M.; Yamakawa, H.; Fujita, R.; Abe, Y.; Akiyama, T.; Nakano, K.; Kuze, Y.; et al. Direct Reprogramming Improves Cardiac Function and Reverses Fibrosis in Chronic Myocardial Infarction. Circulation 2023, 147, 223-238. [CrossRef]
- Mathison, M.; Gersch, R.P.; Nasser, A.; Lilo, S.; Korman, M.; Fourman, M.; Hackett, N.; Shroyer, K.; Yang, J.; Ma, Y.; et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc 2012, 1, e005652. [CrossRef]
- Christoforou, N.; Chakraborty, S.; Kirkton, R.D.; Adler, A.F.; Addis, R.C.; Leong, K.W. Core Transcription Factors, MicroRNAs, and Small Molecules Drive Transdifferentiation of Human Fibroblasts Towards The Cardiac Cell Lineage. Sci Rep 2017, 7, 40285. [CrossRef]
- Ebrahimi, B. Reprogramming barriers and enhancers: strategies to enhance the efficiency and kinetics of induced pluripotency. Cell Regen 2015, 4, 10. [CrossRef]
- Garry, G.A.; Bassel-Duby, R.; Olson, E.N. Direct reprogramming as a route to cardiac repair. Semin Cell Dev Biol 2022, 122, 3-13. [CrossRef]
- yang, l. Sall4 Blocks Cardiac Trans-differentiation but Stimulates Cardiac Stem-like Cell (iPSC) Generation and Improve Post MI Function In Vivo. 2019.
- Gao, H.; Pathan, S.; Dixon, B.; Pugazenthi, A.; Mathison, M.; Mohamed, T.M.A.; Rosengart, T.K.; Yang, J. Sall4 and Gata4 induce cardiac fibroblast transition towards a partially multipotent state with cardiogenic potential. Sci Rep 2024, 14, 24182. [CrossRef]
- Yang, J. Partial Cell Fate Transitions to Promote Cardiac Regeneration. Cells 2024, 13. [CrossRef]
- Zhao, H.; Zhang, Y.; Xu, X.; Sun, Q.; Yang, C.; Wang, H.; Yang, J.; Yang, Y.; Yang, X.; Liu, Y.; et al. Sall4 and Myocd Empower Direct Cardiac Reprogramming From Adult Cardiac Fibroblasts After Injury. Front Cell Dev Biol 2021, 9, 608367. [CrossRef]
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J 1997, 16, 5687-5696. [CrossRef]
- Maitra, M.; Schluterman, M.K.; Nichols, H.A.; Richardson, J.A.; Lo, C.W.; Srivastava, D.; Garg, V. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol 2009, 326, 368-377. [CrossRef]
- Tanimura, N.; Saito, M.; Ebisuya, M.; Nishida, E.; Ishikawa, F. Stemness-related factor Sall4 interacts with transcription factors Oct-3/4 and Sox2 and occupies Oct-Sox elements in mouse embryonic stem cells. J Biol Chem 2013, 288, 5027-5038. [CrossRef]
- Sakamoto, T.; Batmanov, K.; Wan, S.; Guo, Y.; Lai, L.; Vega, R.B.; Kelly, D.P. The nuclear receptor ERR cooperates with the cardiogenic factor GATA4 to orchestrate cardiomyocyte maturation. Nat Commun 2022, 13, 1991. [CrossRef]
- Valimaki, M.J.; Ruskoaho, H.J. Targeting GATA4 for cardiac repair. IUBMB Life 2020, 72, 68-79. [CrossRef]
- Harvey, S.A.; Logan, M.P. sall4 acts downstream of tbx5 and is required for pectoral fin outgrowth. Development 2006, 133, 1165-1173. [CrossRef]
- Cecchetto, A.; Rampazzo, A.; Angelini, A.; Bianco, L.D.; Padalino, M.; Stellin, G.; Daliento, L. From molecular mechanisms of cardiac development to genetic substrate of congenital heart diseases. Future Cardiol 2010, 6, 373-393. [CrossRef]
- Bruneau, B.G. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol 2013, 5, a008292. [CrossRef]
- Karantzali, E.; Lekakis, V.; Ioannou, M.; Hadjimichael, C.; Papamatheakis, J.; Kretsovali, A. Sall1 regulates embryonic stem cell differentiation in association with nanog. J Biol Chem 2011, 286, 1037-1045. [CrossRef]
- Tsubooka, N.; Ichisaka, T.; Okita, K.; Takahashi, K.; Nakagawa, M.; Yamanaka, S. Roles of Sall4 in the generation of pluripotent stem cells from blastocysts and fibroblasts. Genes Cells 2009, 14, 683-694. [CrossRef]
- Yuri, S.; Fujimura, S.; Nimura, K.; Takeda, N.; Toyooka, Y.; Fujimura, Y.; Aburatani, H.; Ura, K.; Koseki, H.; Niwa, H.; et al. Sall4 is essential for stabilization, but not for pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression. Stem Cells 2009, 27, 796-805. [CrossRef]
- Exner, C.R.T.; Kim, A.Y.; Mardjuki, S.M.; Harland, R.M. sall1 and sall4 repress pou5f3 family expression to allow neural patterning, differentiation, and morphogenesis in Xenopus laevis. Dev Biol 2017, 425, 33-43. [CrossRef]
- Bohm, J.; Buck, A.; Borozdin, W.; Mannan, A.U.; Matysiak-Scholze, U.; Adham, I.; Schulz-Schaeffer, W.; Floss, T.; Wurst, W.; Kohlhase, J.; et al. Sall1, sall2, and sall4 are required for neural tube closure in mice. Am J Pathol 2008, 173, 1455-1463. [CrossRef]
- Buganim, Y.; Faddah, D.A.; Cheng, A.W.; Itskovich, E.; Markoulaki, S.; Ganz, K.; Klemm, S.L.; van Oudenaarden, A.; Jaenisch, R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 2012, 150, 1209-1222. [CrossRef]
- Mansour, A.A.; Gafni, O.; Weinberger, L.; Zviran, A.; Ayyash, M.; Rais, Y.; Krupalnik, V.; Zerbib, M.; Amann-Zalcenstein, D.; Maza, I.; et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 2012, 488, 409-413. [CrossRef]
- Furtado, M.B.; Costa, M.W.; Pranoto, E.A.; Salimova, E.; Pinto, A.R.; Lam, N.T.; Park, A.; Snider, P.; Chandran, A.; Harvey, R.P.; et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res 2014, 114, 1422-1434. [CrossRef]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ Res 2015, 116, 237-244. [CrossRef]
- Shu, J.; Zhang, K.; Zhang, M.; Yao, A.; Shao, S.; Du, F.; Yang, C.; Chen, W.; Wu, C.; Yang, W.; et al. GATA family members as inducers for cellular reprogramming to pluripotency. Cell Res 2015, 25, 169-180. [CrossRef]
- Liu, Y.; Asakura, M.; Inoue, H.; Nakamura, T.; Sano, M.; Niu, Z.; Chen, M.; Schwartz, R.J.; Schneider, M.D. Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci U S A 2007, 104, 3859-3864. [CrossRef]
- Belair, D.G.; Lu, G.; Waller, L.E.; Gustin, J.A.; Collins, N.D.; Kolaja, K.L. Thalidomide Inhibits Human iPSC Mesendoderm Differentiation by Modulating CRBN-dependent Degradation of SALL4. Sci Rep 2020, 10, 2864. [CrossRef]
- Afouda, B.A. Towards Understanding the Gene-Specific Roles of GATA Factors in Heart Development: Does GATA4 Lead the Way? Int J Mol Sci 2022, 23. [CrossRef]
- Holtzinger, A.; Rosenfeld, G.E.; Evans, T. Gata4 directs development of cardiac-inducing endoderm from ES cells. Dev Biol 2010, 337, 63-73. [CrossRef]
- Foley, A.C.; Mercola, M. Heart induction by Wnt antagonists depends on the homeodomain transcription factor Hex. Genes Dev 2005, 19, 387-396. [CrossRef]
- Abboud, N.; Morris, T.M.; Hiriart, E.; Yang, H.; Bezerra, H.; Gualazzi, M.G.; Stefanovic, S.; Guenantin, A.C.; Evans, S.M.; Puceat, M. A cohesin-OCT4 complex mediates Sox enhancers to prime an early embryonic lineage. Nat Commun 2015, 6, 6749. [CrossRef]
- Stefanovic, S.; Abboud, N.; Desilets, S.; Nury, D.; Cowan, C.; Puceat, M. Interplay of Oct4 with Sox2 and Sox17: a molecular switch from stem cell pluripotency to specifying a cardiac fate. J Cell Biol 2009, 186, 665-673. [CrossRef]
- Boogerd, C.J.; Evans, S.M. TBX5 and NuRD Divide the Heart. Dev Cell 2016, 36, 242-244. [CrossRef]
- Grunert, M.; Dorn, C.; Rickert-Sperling, S. Cardiac Transcription Factors and Regulatory Networks. Adv Exp Med Biol 2024, 1441, 295-311. [CrossRef]
- Steimle, J.D.; Moskowitz, I.P. TBX5: A Key Regulator of Heart Development. Curr Top Dev Biol 2017, 122, 195-221. [CrossRef]
- Siatra, P.; Vatsellas, G.; Chatzianastasiou, A.; Balafas, E.; Manolakou, T.; Papapetropoulos, A.; Agapaki, A.; Mouchtouri, E.T.; Ruchaya, P.J.; Korovesi, A.G.; et al. Return of the Tbx5; lineage-tracing reveals ventricular cardiomyocyte-like precursors in the injured adult mammalian heart. NPJ Regen Med 2023, 8, 13. [CrossRef]
- Huang, J.; Min Lu, M.; Cheng, L.; Yuan, L.J.; Zhu, X.; Stout, A.L.; Chen, M.; Li, J.; Parmacek, M.S. Myocardin is required for cardiomyocyte survival and maintenance of heart function. Proc Natl Acad Sci U S A 2009, 106, 18734-18739. [CrossRef]
- Gan, P.; Eppert, M.; De La Cruz, N.; Lyons, H.; Shah, A.M.; Veettil, R.T.; Chen, K.; Pradhan, P.; Bezprozvannaya, S.; Xu, L.; et al. Coactivator condensation drives cardiovascular cell lineage specification. Sci Adv 2024, 10, eadk7160. [CrossRef]
- Balza, R.O., Jr.; Misra, R.P. Role of the serum response factor in regulating contractile apparatus gene expression and sarcomeric integrity in cardiomyocytes. J Biol Chem 2006, 281, 6498-6510. [CrossRef]
- Nam, Y.J.; Song, K.; Luo, X.; Daniel, E.; Lambeth, K.; West, K.; Hill, J.A.; DiMaio, J.M.; Baker, L.A.; Bassel-Duby, R.; et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A 2013, 110, 5588-5593. [CrossRef]
- Wada, R.; Muraoka, N.; Inagawa, K.; Yamakawa, H.; Miyamoto, K.; Sadahiro, T.; Umei, T.; Kaneda, R.; Suzuki, T.; Kamiya, K.; et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc Natl Acad Sci U S A 2013, 110, 12667-12672. [CrossRef]
- Christoforou, N.; Chellappan, M.; Adler, A.F.; Kirkton, R.D.; Wu, T.; Addis, R.C.; Bursac, N.; Leong, K.W. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One 2013, 8, e63577. [CrossRef]
- Kwon, C.; Qian, L.; Cheng, P.; Nigam, V.; Arnold, J.; Srivastava, D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol 2009, 11, 951-957. [CrossRef]
- Maven, B.E.J.; Gifford, C.A.; Weilert, M.; Gonzalez-Teran, B.; Huttenhain, R.; Pelonero, A.; Ivey, K.N.; Samse-Knapp, K.; Kwong, W.; Gordon, D.; et al. The multi-lineage transcription factor ISL1 controls cardiomyocyte cell fate through interaction with NKX2.5. Stem Cell Reports 2023, 18, 2138-2153. [CrossRef]
- Xu, H.; Zhou, Q.; Yi, Q.; Tan, B.; Tian, J.; Chen, X.; Wang, Y.; Yu, X.; Zhu, J. Islet-1 synergizes with Gcn5 to promote MSC differentiation into cardiomyocytes. Sci Rep 2020, 10, 1817. [CrossRef]
- Sopic, M.; Robinson, E.L.; Emanueli, C.; Srivastava, P.; Angione, C.; Gaetano, C.; Condorelli, G.; Martelli, F.; Pedrazzini, T.; Devaux, Y.; et al. Integration of epigenetic regulatory mechanisms in heart failure. Basic Res Cardiol 2023, 118, 16. [CrossRef]
- Felisbino, M.B.; McKinsey, T.A. Epigenetics in Cardiac Fibrosis: Emphasis on Inflammation and Fibroblast Activation. JACC Basic Transl Sci 2018, 3, 704-715. [CrossRef]
- Kloet, S.L.; Baymaz, H.I.; Makowski, M.; Groenewold, V.; Jansen, P.W.; Berendsen, M.; Niazi, H.; Kops, G.J.; Vermeulen, M. Towards elucidating the stability, dynamics and architecture of the nucleosome remodeling and deacetylase complex by using quantitative interaction proteomics. FEBS J 2015, 282, 1774-1785. [CrossRef]
- Wang, B.; Li, C.; Ming, J.; Wu, L.; Fang, S.; Huang, Y.; Lin, L.; Liu, H.; Kuang, J.; Zhao, C.; et al. The NuRD complex cooperates with SALL4 to orchestrate reprogramming. Nat Commun 2023, 14, 2846. [CrossRef]
- Xiong, J.; Todorova, D.; Su, N.Y.; Kim, J.; Lee, P.J.; Shen, Z.; Briggs, S.P.; Xu, Y. Stemness factor Sall4 is required for DNA damage response in embryonic stem cells. J Cell Biol 2015, 208, 513-520. [CrossRef]
- Gilsbach, R.; Preissl, S.; Gruning, B.A.; Schnick, T.; Burger, L.; Benes, V.; Wurch, A.; Bonisch, U.; Gunther, S.; Backofen, R.; et al. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun 2014, 5, 5288. [CrossRef]
- Zhong, J.; Agha, G.; Baccarelli, A.A. The Role of DNA Methylation in Cardiovascular Risk and Disease: Methodological Aspects, Study Design, and Data Analysis for Epidemiological Studies. Circ Res 2016, 118, 119-131. [CrossRef]
- Fang, X.; Poulsen, R.; Zhao, L.; Wang, J.; Rivkees, S.A.; Wendler, C.C. Knockdown of DNA methyltransferase 1 reduces DNA methylation and alters expression patterns of cardiac genes in embryonic cardiomyocytes. FEBS Open Bio 2021, 11, 2364-2382. [CrossRef]
- Vujic, A.; Robinson, E.L.; Ito, M.; Haider, S.; Ackers-Johnson, M.; See, K.; Methner, C.; Figg, N.; Brien, P.; Roderick, H.L.; et al. Experimental heart failure modelled by the cardiomyocyte-specific loss of an epigenome modifier, DNMT3B. J Mol Cell Cardiol 2015, 82, 174-183. [CrossRef]
- Madsen, A.; Hoppner, G.; Krause, J.; Hirt, M.N.; Laufer, S.D.; Schweizer, M.; Tan, W.L.W.; Mosqueira, D.; Anene-Nzelu, C.G.; Lim, I.; et al. An Important Role for DNMT3A-Mediated DNA Methylation in Cardiomyocyte Metabolism and Contractility. Circulation 2020, 142, 1562-1578. [CrossRef]
- Wang, W.; Lu, G.; Liu, H.B.; Xiong, Z.; Leung, H.D.; Cao, R.; Pang, A.L.; Su, X.; Law, P.W.N.; Zhao, Z.; et al. Pten Regulates Cardiomyocyte Differentiation by Modulating Non-CG Methylation via Dnmt3. Adv Sci (Weinh) 2021, 8, e2100849. [CrossRef]
- Zhang, D.; Wu, B.; Wang, P.; Wang, Y.; Lu, P.; Nechiporuk, T.; Floss, T.; Greally, J.M.; Zheng, D.; Zhou, B. Non-CpG methylation by DNMT3B facilitates REST binding and gene silencing in developing mouse hearts. Nucleic Acids Res 2017, 45, 3102-3115. [CrossRef]
- Desiderio, A.; Pastorino, M.; Campitelli, M.; Longo, M.; Miele, C.; Napoli, R.; Beguinot, F.; Raciti, G.A. DNA methylation in cardiovascular disease and heart failure: novel prediction models? Clin Epigenetics 2024, 16, 115. [CrossRef]
- Mulder, R.H.; Neumann, A.; Cecil, C.A.M.; Walton, E.; Houtepen, L.C.; Simpkin, A.J.; Rijlaarsdam, J.; Heijmans, B.T.; Gaunt, T.R.; Felix, J.F.; et al. Epigenome-wide change and variation in DNA methylation in childhood: trajectories from birth to late adolescence. Hum Mol Genet 2021, 30, 119-134. [CrossRef]
- Yang, J.; Corsello, T.R.; Ma, Y. Stem cell gene SALL4 suppresses transcription through recruitment of DNA methyltransferases. J Biol Chem 2012, 287, 1996-2005. [CrossRef]
- Montgomery, R.L.; Davis, C.A.; Potthoff, M.J.; Haberland, M.; Fielitz, J.; Qi, X.; Hill, J.A.; Richardson, J.A.; Olson, E.N. Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Genes Dev 2007, 21, 1790-1802. [CrossRef]
- Gillette, T.G. HDAC Inhibition in the Heart: Erasing Hidden Fibrosis. Circulation 2021, 143, 1891-1893. [CrossRef]
- Lu, J.; Qian, S.; Sun, Z. Targeting histone deacetylase in cardiac diseases. Front Physiol 2024, 15, 1405569. [CrossRef]
- Pinnamaneni, J.P.; Singh, V.P.; Kim, M.B.; Ryan, C.T.; Pugazenthi, A.; Sanagasetti, D.; Mathison, M.; Yang, J.; Rosengart, T.K. p63 silencing induces epigenetic modulation to enhance human cardiac fibroblast to cardiomyocyte-like differentiation. Sci Rep 2022, 12, 11416. [CrossRef]
- Singh, V.P.; Pinnamaneni, J.P.; Pugazenthi, A.; Sanagasetti, D.; Mathison, M.; Wang, K.; Yang, J.; Rosengart, T.K. Enhanced Generation of Induced Cardiomyocytes Using a Small-Molecule Cocktail to Overcome Barriers to Cardiac Cellular Reprogramming. J Am Heart Assoc 2020, 9, e015686. [CrossRef]
- Singh, V.P.; Mathison, M.; Patel, V.; Sanagasetti, D.; Gibson, B.W.; Yang, J.; Rosengart, T.K. MiR-590 Promotes Transdifferentiation of Porcine and Human Fibroblasts Toward a Cardiomyocyte-Like Fate by Directly Repressing Specificity Protein 1. J Am Heart Assoc 2016, 5. [CrossRef]
- Lu, J.; Jeong, H.W.; Kong, N.; Yang, Y.; Carroll, J.; Luo, H.R.; Silberstein, L.E.; Yupoma; Chai, L. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One 2009, 4, e5577. [CrossRef]
- Liu, H.; Zhou, R.; Li, S.; Dong, J.; Fang, Y.; Luo, Y.; Su, H.; Lai, B.; Liang, L.; Zhang, D.; et al. Epigenetic repression of Cend1 by lysine-specific demethylase 1 is essential for murine heart development. iScience 2024, 27, 108722. [CrossRef]
- Fei, Q.; Qiu, M.; Fan, G.; Zhang, B.; Wang, Q.; Zhang, S.; Wang, S.; Yang, B.; Zhang, L. Downregulation of Hotair or LSD1 Impaired Heart Regeneration in the Neonatal Mouse. DNA Cell Biol 2021, 40, 1177-1184. [CrossRef]
- Cao, Y.; Dong, Z.; Yang, D.; Ma, X.; Wang, X. LSD1 regulates the expressions of core cardiogenic transcription factors and cardiac genes in oxygen and glucose deprivation injured mice fibroblasts in vitro. Exp Cell Res 2022, 418, 113228. [CrossRef]
- Wang, H.; Cao, N.; Spencer, C.I.; Nie, B.; Ma, T.; Xu, T.; Zhang, Y.; Wang, X.; Srivastava, D.; Ding, S. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep 2014, 6, 951-960. [CrossRef]
- Nicholson, T.B.; Singh, A.K.; Su, H.; Hevi, S.; Wang, J.; Bajko, J.; Li, M.; Valdez, R.; Goetschkes, M.; Capodieci, P.; et al. A hypomorphic lsd1 allele results in heart development defects in mice. PLoS One 2013, 8, e60913. [CrossRef]
- Liu, L.; Souto, J.; Liao, W.; Jiang, Y.; Li, Y.; Nishinakamura, R.; Huang, S.; Rosengart, T.; Yang, V.W.; Schuster, M.; et al. Histone lysine-specific demethylase 1 (LSD1) protein is involved in Sal-like protein 4 (SALL4)-mediated transcriptional repression in hematopoietic stem cells. J Biol Chem 2013, 288, 34719-34728. [CrossRef]
- Guenantin, A.C.; Jebeniani, I.; Leschik, J.; Watrin, E.; Bonne, G.; Vignier, N.; Puceat, M. Targeting the histone demethylase LSD1 prevents cardiomyopathy in a mouse model of laminopathy. J Clin Invest 2021, 131. [CrossRef]
- Yang, Y.; Luan, Y.; Yuan, R.X.; Luan, Y. Histone Methylation Related Therapeutic Challenge in Cardiovascular Diseases. Front Cardiovasc Med 2021, 8, 710053. [CrossRef]
- Pursani, V.; Bhartiya, D.; Tanavde, V.; Bashir, M.; Sampath, P. Transcriptional activator DOT1L putatively regulates human embryonic stem cell differentiation into the cardiac lineage. Stem Cell Res Ther 2018, 9, 97. [CrossRef]
- Cattaneo, P.; Kunderfranco, P.; Greco, C.; Guffanti, A.; Stirparo, G.G.; Rusconi, F.; Rizzi, R.; Di Pasquale, E.; Locatelli, S.L.; Latronico, M.V.; et al. DOT1L-mediated H3K79me2 modification critically regulates gene expression during cardiomyocyte differentiation. Cell Death Differ 2016, 23, 555-564. [CrossRef]
- Cattaneo, P.; Hayes, M.G.B.; Baumgarten, N.; Hecker, D.; Peruzzo, S.; Aslan, G.S.; Kunderfranco, P.; Larcher, V.; Zhang, L.; Contu, R.; et al. DOT1L regulates chamber-specific transcriptional networks during cardiogenesis and mediates postnatal cell cycle withdrawal. Nat Commun 2022, 13, 7444. [CrossRef]
- Nguyen, A.T.; Xiao, B.; Neppl, R.L.; Kallin, E.M.; Li, J.; Chen, T.; Wang, D.Z.; Xiao, X.; Zhang, Y. DOT1L regulates dystrophin expression and is critical for cardiac function. Genes Dev 2011, 25, 263-274. [CrossRef]
- Xu, J.; Wang, J.; Long, F.; Zhong, W.; Su, H.; Su, Z.; Liu, X. Inhibition of the cardiac fibroblast-enriched histone methyltransferase Dot1L prevents cardiac fibrosis and cardiac dysfunction. Cell Biosci 2022, 12, 134. [CrossRef]
- Peng, X.; Feng, G.; Zhang, Y.; Sun, Y. PRC1 Stabilizes Cardiac Contraction by Regulating Cardiac Sarcomere Assembly and Cardiac Conduction System Construction. Int J Mol Sci 2021, 22. [CrossRef]
- Yang, D.; Liu, H.Q.; Yang, Z.; Fan, D.; Tang, Q.Z. BMI1 in the heart: Novel functions beyond tumorigenesis. EBioMedicine 2021, 63, 103193. [CrossRef]
- Yang, J.; Chai, L.; Liu, F.; Fink, L.M.; Lin, P.; Silberstein, L.E.; Amin, H.M.; Ward, D.C.; Ma, Y. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci U S A 2007, 104, 10494-10499. [CrossRef]
- Milanovich, S.; Peterson, J.; Allred, J.; Stelloh, C.; Rajasekaran, K.; Fisher, J.; Duncan, S.A.; Malarkannan, S.; Rao, S. Sall4 overexpression blocks murine hematopoiesis in a dose-dependent manner. Exp Hematol 2015, 43, 53-64 e51-58. [CrossRef]
- Valiente-Alandi, I.; Albo-Castellanos, C.; Herrero, D.; Sanchez, I.; Bernad, A. Bmi1 (+) cardiac progenitor cells contribute to myocardial repair following acute injury. Stem Cell Res Ther 2016, 7, 100. [CrossRef]
- Yang, W.; Wu, Z.; Yang, K.; Han, Y.; Chen, Y.; Zhao, W.; Huang, F.; Jin, Y.; Jin, W. BMI1 promotes cardiac fibrosis in ischemia-induced heart failure via the PTEN-PI3K/Akt-mTOR signaling pathway. Am J Physiol Heart Circ Physiol 2019, 316, H61-H69. [CrossRef]
- Wang, G.; Ye, H.; Wang, X.; Liu, B. Polycomb repressive complex 2 controls cardiac cell fate decision via interacting with RNA: Promiscuously or well-ordered. Front Genet 2022, 13, 1011228. [CrossRef]
- Ren, L.; Deng, H.; Jiang, Y.; Liu, C. Dual-Regulated Mechanism of EZH2 and KDM6A on SALL4 Modulates Tumor Progression via Wnt/beta-Catenin Pathway in Gastric Cancer. Dig Dis Sci 2023, 68, 1292-1305. [CrossRef]
- Robbe, Z.L.; Shi, W.; Wasson, L.K.; Scialdone, A.P.; Wilczewski, C.M.; Sheng, X.; Hepperla, A.J.; Akerberg, B.N.; Pu, W.T.; Cristea, I.M.; et al. CHD4 is recruited by GATA4 and NKX2-5 to repress noncardiac gene programs in the developing heart. Genes Dev 2022, 36, 468-482. [CrossRef]
- Waldron, L.; Steimle, J.D.; Greco, T.M.; Gomez, N.C.; Dorr, K.M.; Kweon, J.; Temple, B.; Yang, X.H.; Wilczewski, C.M.; Davis, I.J.; et al. The Cardiac TBX5 Interactome Reveals a Chromatin Remodeling Network Essential for Cardiac Septation. Dev Cell 2016, 36, 262-275. [CrossRef]
- Wilczewski, C.M.; Hepperla, A.J.; Shimbo, T.; Wasson, L.; Robbe, Z.L.; Davis, I.J.; Wade, P.A.; Conlon, F.L. CHD4 and the NuRD complex directly control cardiac sarcomere formation. Proc Natl Acad Sci U S A 2018, 115, 6727-6732. [CrossRef]
- Goto, N.; Suke, K.; Yonezawa, N.; Nishihara, H.; Handa, T.; Sato, Y.; Kujirai, T.; Kurumizaka, H.; Yamagata, K.; Kimura, H. ISWI chromatin remodeling complexes recruit NSD2 and H3K36me2 in pericentromeric heterochromatin. J Cell Biol 2024, 223. [CrossRef]
- Nimura, K.; Ura, K.; Shiratori, H.; Ikawa, M.; Okabe, M.; Schwartz, R.J.; Kaneda, Y. A histone H3 lysine 36 trimethyltransferase links Nkx2-5 to Wolf-Hirschhorn syndrome. Nature 2009, 460, 287-291. [CrossRef]
- Li, D.; Sun, J.; Zhong, T.P. Wnt Signaling in Heart Development and Regeneration. Curr Cardiol Rep 2022, 24, 1425-1438. [CrossRef]
- Horitani, K.; Shiojima, I. Wnt signaling in cardiac development and heart diseases. In Vitro Cell Dev Biol Anim 2024, 60, 482-488. [CrossRef]
- Naito, A.T.; Shiojima, I.; Akazawa, H.; Hidaka, K.; Morisaki, T.; Kikuchi, A.; Komuro, I. Developmental stage-specific biphasic roles of Wnt/beta-catenin signaling in cardiomyogenesis and hematopoiesis. Proc Natl Acad Sci U S A 2006, 103, 19812-19817. [CrossRef]
- Cohen, E.D.; Wang, Z.; Lepore, J.J.; Lu, M.M.; Taketo, M.M.; Epstein, D.J.; Morrisey, E.E. Wnt/beta-catenin signaling promotes expansion of Isl-1-positive cardiac progenitor cells through regulation of FGF signaling. J Clin Invest 2007, 117, 1794-1804. [CrossRef]
- Lin, L.; Cui, L.; Zhou, W.; Dufort, D.; Zhang, X.; Cai, C.L.; Bu, L.; Yang, L.; Martin, J.; Kemler, R.; et al. Beta-catenin directly regulates Islet1 expression in cardiovascular progenitors and is required for multiple aspects of cardiogenesis. Proc Natl Acad Sci U S A 2007, 104, 9313-9318. [CrossRef]
- Qyang, Y.; Martin-Puig, S.; Chiravuri, M.; Chen, S.; Xu, H.; Bu, L.; Jiang, X.; Lin, L.; Granger, A.; Moretti, A.; et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell 2007, 1, 165-179. [CrossRef]
- Balatskyi, V.V.; Sowka, A.; Dobrzyn, P.; Piven, O.O. WNT/beta-catenin pathway is a key regulator of cardiac function and energetic metabolism. Acta Physiol (Oxf) 2023, 237, e13912. [CrossRef]
- Hahn, J.Y.; Cho, H.J.; Bae, J.W.; Yuk, H.S.; Kim, K.I.; Park, K.W.; Koo, B.K.; Chae, I.H.; Shin, C.S.; Oh, B.H.; et al. Beta-catenin overexpression reduces myocardial infarct size through differential effects on cardiomyocytes and cardiac fibroblasts. J Biol Chem 2006, 281, 30979-30989. [CrossRef]
- Zhou, H.; Dickson, M.E.; Kim, M.S.; Bassel-Duby, R.; Olson, E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc Natl Acad Sci U S A 2015, 112, 11864-11869. [CrossRef]
- Chen, Z.; Yu, X.; Ke, M.; Li, H.; Jiang, Y.; Zhang, P.; Tan, J.; Cao, N.; Yang, H.T. Human embryonic stem cell-derived cardiovascular progenitor cells stimulate cardiomyocyte cell cycle activity via activating the PI3K/Akt pathway. J Mol Cell Cardiol 2024, 197, 5-10. [CrossRef]
- Garay, B.I.; Givens, S.; Abreu, P.; Liu, M.; Yucel, D.; Baik, J.; Stanis, N.; Rothermel, T.M.; Magli, A.; Abrahante, J.E.; et al. Dual inhibition of MAPK and PI3K/AKT pathways enhances maturation of human iPSC-derived cardiomyocytes. Stem Cell Reports 2023, 18, 411. [CrossRef]
- Ghafouri-Fard, S.; Khanbabapour Sasi, A.; Hussen, B.M.; Shoorei, H.; Siddiq, A.; Taheri, M.; Ayatollahi, S.A. Interplay between PI3K/AKT pathway and heart disorders. Mol Biol Rep 2022, 49, 9767-9781. [CrossRef]
- Walkowski, B.; Kleibert, M.; Majka, M.; Wojciechowska, M. Insight into the Role of the PI3K/Akt Pathway in Ischemic Injury and Post-Infarct Left Ventricular Remodeling in Normal and Diabetic Heart. Cells 2022, 11. [CrossRef]
- Qin, W.; Cao, L.; Massey, I.Y. Role of PI3K/Akt signaling pathway in cardiac fibrosis. Mol Cell Biochem 2021, 476, 4045-4059. [CrossRef]
- Tang, Z.; Zhao, P.; Zhang, W.; Zhang, Q.; Zhao, M.; Tan, H. SALL4 activates PI3K/AKT signaling pathway through targeting PTEN, thus facilitating migration, invasion and proliferation of hepatocellular carcinoma cells. Aging (Albany NY) 2022, 14, 10081-10092. [CrossRef]
- Pattanayak, B.; Lameirinhas, A.; Torres-Ruiz, S.; Burgues, O.; Rovira, A.; Martinez, M.T.; Tapia, M.; Zazo, S.; Albanell, J.; Rojo, F.; et al. Role of SALL4 in HER2+ Breast Cancer Progression: Regulating PI3K/AKT Pathway. Int J Mol Sci 2022, 23. [CrossRef]
- Stefanovic, S.; Zaffran, S. Mechanisms of retinoic acid signaling during cardiogenesis. Mech Dev 2017, 143, 9-19. [CrossRef]
- Wang, S.; Moise, A.R. Recent insights on the role and regulation of retinoic acid signaling during epicardial development. Genesis 2019, 57, e23303. [CrossRef]
- Duong, T.B.; Waxman, J.S. Patterning of vertebrate cardiac progenitor fields by retinoic acid signaling. Genesis 2021, 59, e23458. [CrossRef]
- Zawada, D.; Kornherr, J.; Meier, A.B.; Santamaria, G.; Dorn, T.; Nowak-Imialek, M.; Ortmann, D.; Zhang, F.; Lachmann, M.; Dressen, M.; et al. Retinoic acid signaling modulation guides in vitro specification of human heart field-specific progenitor pools. Nat Commun 2023, 14, 1722. [CrossRef]
- Wiesinger, A.; Boink, G.J.J.; Christoffels, V.M.; Devalla, H.D. Retinoic acid signaling in heart development: Application in the differentiation of cardiovascular lineages from human pluripotent stem cells. Stem Cell Reports 2021, 16, 2589-2606. [CrossRef]
- Ghyselinck, N.B.; Duester, G. Retinoic acid signaling pathways. Development 2019, 146. [CrossRef]
- Gely-Pernot, A.; Raverdeau, M.; Teletin, M.; Vernet, N.; Feret, B.; Klopfenstein, M.; Dennefeld, C.; Davidson, I.; Benoit, G.; Mark, M.; et al. Retinoic Acid Receptors Control Spermatogonia Cell-Fate and Induce Expression of the SALL4A Transcription Factor. PLoS Genet 2015, 11, e1005501. [CrossRef]
- Liu, L.; Liu, L.; Leung, L.H.; Cooney, A.J.; Chen, C.; Rosengart, T.K.; Ma, Y.; Yang, J. Knockdown of SALL4 Protein Enhances All-trans Retinoic Acid-induced Cellular Differentiation in Acute Myeloid Leukemia Cells. J Biol Chem 2015, 290, 10599-10609. [CrossRef]
- Zhang, X.; Zhang, P.; Shao, M.; Zang, X.; Zhang, J.; Mao, F.; Qian, H.; Xu, W. SALL4 activates TGF-beta/SMAD signaling pathway to induce EMT and promote gastric cancer metastasis. Cancer Manag Res 2018, 10, 4459-4470. [CrossRef]
- Chen, T.; Tsang, J.Y.S.; Su, X.C.; Li, P.; Sun, W.Q.; Wong, I.L.K.; Choy, K.Y.; Yang, Q.; Tse, G.M.K.; Chan, T.H.; et al. SALL4 promotes tumor progression in breast cancer by targeting EMT. Mol Carcinog 2020, 59, 1209-1226. [CrossRef]
- Saadat, S.; Noureddini, M.; Mahjoubin-Tehran, M.; Nazemi, S.; Shojaie, L.; Aschner, M.; Maleki, B.; Abbasi-Kolli, M.; Rajabi Moghadam, H.; Alani, B.; et al. Pivotal Role of TGF-beta/Smad Signaling in Cardiac Fibrosis: Non-coding RNAs as Effectual Players. Front Cardiovasc Med 2020, 7, 588347. [CrossRef]
- Riching, A.S.; Danis, E.; Zhao, Y.; Cao, Y.; Chi, C.; Bagchi, R.A.; Klein, B.J.; Xu, H.; Kutateladze, T.G.; McKinsey, T.A.; et al. Suppression of canonical TGF-beta signaling enables GATA4 to interact with H3K27me3 demethylase JMJD3 to promote cardiomyogenesis. J Mol Cell Cardiol 2021, 153, 44-59. [CrossRef]
- Stone, N.R.; Gifford, C.A.; Thomas, R.; Pratt, K.J.B.; Samse-Knapp, K.; Mohamed, T.M.A.; Radzinsky, E.M.; Schricker, A.; Ye, L.; Yu, P.; et al. Context-Specific Transcription Factor Functions Regulate Epigenomic and Transcriptional Dynamics during Cardiac Reprogramming. Cell Stem Cell 2019, 25, 87-102 e109. [CrossRef]
- Lalit, P.A.; Salick, M.R.; Nelson, D.O.; Squirrell, J.M.; Shafer, C.M.; Patel, N.G.; Saeed, I.; Schmuck, E.G.; Markandeya, Y.S.; Wong, R.; et al. Lineage Reprogramming of Fibroblasts into Proliferative Induced Cardiac Progenitor Cells by Defined Factors. Cell Stem Cell 2016, 18, 354-367. [CrossRef]
- Wu, J.; Zhao, H.; Tao, Y.; Yang, C.; Yang, Y.; Zhou, B.; Zhao, Y. Improved Factor Combination for In Vivo Reprogramming of Cardiac Myofibroblast to Cardiomyocyte-Like Cell With Dual Recombinase Tracing. Circulation 2023, 148, 1728-1731. [CrossRef]
- Mathison, M.; Singh, V.P.; Sanagasetti, D.; Yang, L.; Pinnamaneni, J.P.; Yang, J.; Rosengart, T.K. Cardiac reprogramming factor Gata4 reduces postinfarct cardiac fibrosis through direct repression of the profibrotic mediator snail. J Thorac Cardiovasc Surg 2017, 154, 1601-1610 e1603. [CrossRef]
- Ma, Y.; Cui, W.; Yang, J.; Qu, J.; Di, C.; Amin, H.M.; Lai, R.; Ritz, J.; Krause, D.S.; Chai, L. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood 2006, 108, 2726-2735. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



