1. Introduction
Many flowers have evolved their morphology to attract pollinators. Generally, they consist of four distinct organs: sepals, petals, stamens, and carpels, whose identities are determined by a combination of floral homeotic genes. This is known as the floral ABCE model, where each class of floral homeotic genes is labeled as A, B, C, or E, and they act together to establish the identity of each organ [
1,
2,
3]. Although the basic structure of flowers, such as the arrangement of organs in a whorled pattern, is remarkably conserved, the morphology and number of each organ can vary widely. A remarkable example of this variation is orchid flowers. Despite having a simple basic structure – three sepals (or outer tepals) in the first whorl, three petals (or inner tepals) in the second whorl, and a single column where male and female organs are combined - approximately 28,000 species exhibit a diverse array of unique shapes and colors, often influenced by coevolution with pollinators [
4]. In particular, many orchid species develop the two lateral petals, and the lip (or labellum) into distinctly different shapes, playing a crucial role in attracting pollinators.
Most of the floral homeotic genes encode MADS-box transcription factors, and they have been identified in ornamental orchids, including
Phalaenopsis [
5,
6,
7],
Oncidium [
8,
9,
10],
Dendrobium [
11],
Orchis [
12],
Erycina [
13], and
Habenaria [
14,
15,
16]. Two main models have been proposed to explain the morphological differences between the lateral petal and the lip. One is the “orchid code”, which suggests that the class B homeotic gene
DEFICIENS (
DEF), functionally differentiated into four genes by gene duplication, alters the morphology of the lateral petal and lip through differential expression in the floral bud [
17,
18,
19,
20,
21]. The other is the perianth code (P-code), which proposes that, in addition to the class B genes,
AGAMOUS-LIKE 6 (
AGL6) forms distinct protein complexes, leading to changes in petal morphology [
22]. In either case, it is clear that the spatiotemporal regulation of DEF-type class B genes and AGL6, along with their protein interactions, results in differences in the identity of the second whorl organs. These models are believed to be conserved in many orchid flowers; however, the morphology of the lip varies greatly among species, suggesting that each species has its own unique lip morphogenesis process, although the mechanisms remain largely unknown.
Habenaria is a large genus within the Orchidaceae family, containing 898 species found in tropical and subtropical regions (Royal Botanic Gardens, Kew,
http://apps.kew.org/wcsp/, accessed on 03 December 2024).
Habenaria radiata (Thunb.) Spreng., syn.
Pecteilis radiata (Thunb.) Raf, is a terrestrial orchid native to wetlands in Japan, China, South Korea, and Far East Russia. The habitats of
Habenaria radiata have been reduced due to environmental destruction and overexploitation, and as a result, this species is listed as Near Threatened on the Red List of Japan (Ministry of the Environment, Red List 2020,
https://ikilog.biodic.go.jp/Rdb/booklist, accessed on 03 December 2024). The leaves of
Habenaria radiata are simple, narrow, and elongated, similar to many other orchids, while its floral organs exhibit a complex shape (
Figure S1). The flower consists of three green sepals, two lateral petals, a lip (labellum), a spur that is a long tubular structure originating from the base of the lip that accumulates nectar, and a column where the male and female reproductive organs are fused (
Figure S1). The lip is the most distinctive part of the floral organs, with a shape resembling a white bird in flight, consisting of a central body part and two lateral wings (
Figure S1). This morphology is believed to be crucial for attracting diurnal butterflies (such as
Parnara guttata,
Polytremis pellucida, and
Pelopidas mathias) and nocturnal hawk moths (such as
Theretra oldenlandiae,
Theretra japonica, and
Theretra nessus) [
23,
24,
25,
26]. However, the mechanisms underlying the formation of the serrations on the edges of the petals, how the lip develops in the bud, and how it unfolds during blooming have not been fully understood.
In this study, we investigated how the complex morphology of the lip of Habenaria radiata is formed and how it expands during blooming. Additionally, we conducted transcriptome analysis of floral buds during the early stages of serration formation and comprehensively obtained gene sequences related to floral organ development. These results provide insights into the mechanism underlying the formation of the complex morphology of orchid petals.
2. Results
2.1. Anthesis and Petal Withering
To examine the lip morphogenesis during anthesis and withering, we took time-lapse photos with fixed-position shooting. Interval shots taken every 10 minutes revealed that the anthesis of a flower - i.e., from sepal opening to petal expansion - takes about 3.5 to 4 hours (
Figure 1A–C, Movie S1). The flowers remained open for more than a week, and we observed that the upper flower began petal withering earlier than the lower flower, despite the lower flower opened earlier (
Figure 1F,G). We found that one of the two pollinia was lost in the upper flower (
Figure 1D,E), but two pollinia remained in the lower flower. This suggests that pollinia removal, likely by pollinator insects, and subsequent pollination stimulates petal withering. Since the lip shape may help attract pollinators such as moths, petals that have completed their role may quickly wither. Petal withering took approximately 8 to 9 hours (
Figure 1F,G, Movie S1). Focusing on the expansion of the lip, the body part opened first, followed by the unfolding of the wing part, which was initially folded inward (
Figure 1H,I). The entire lip expansion process took approximately 2 hours.
2.2. Lip Morphogenesis
To investigate lip development, lips from floral buds of different sizes (1 mm, 2mm, 4mm, 6mm, and 7 mm in bud length,
Figure 2A–E) were dissected and analyzed under a microscope. In the 1 mm floral bud, the lip was flat and round in shape, corresponding to the ‘body’ part (
Figure 2F and S1). The ‘wings’ began to form on the proximal side of the lip (
Figure 2G) and elongated laterally (
Figure 2H). The wings developed serrations on the outer periphery, which deepened during growth, resulting in distinct serrations in the lip (
Figure 2I). Vascular tissue developed in each separated part (
Figure 2J). The spur initiated at the bottom of the buds when the bud length reached 2 mm (
Figure 2B) and developed into a long tubular structure (
Figure 2C–E).
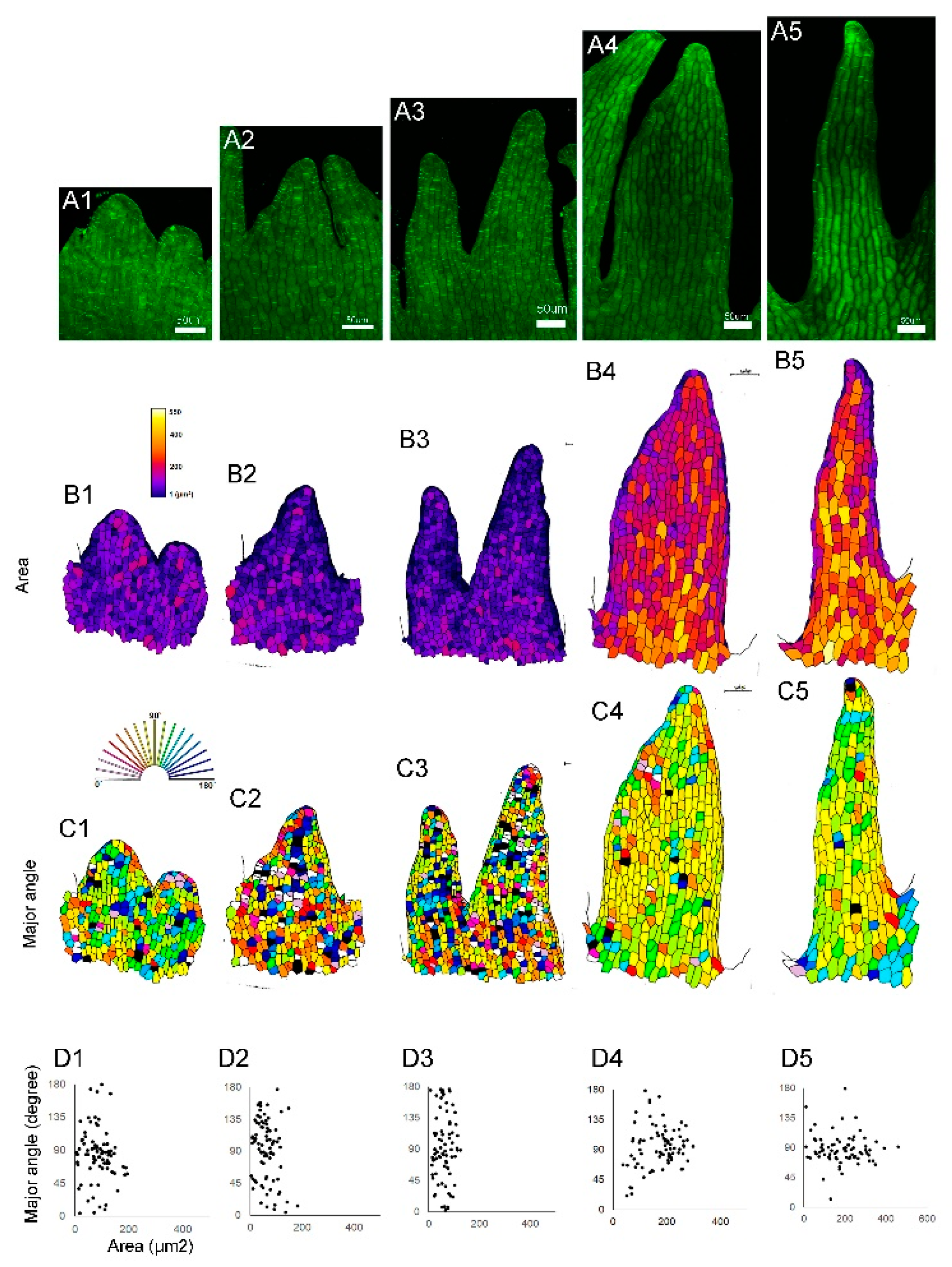
To examine the cellular dynamics during serration development at the lip periphery, lips from different stages were dissected, and the shape of epidermal cells at the serration region was analyzed (
Figure 3A). At early stages, the cell area did not increase; instead, the cell number increased (
Figure 3B
1–B
3). Later, each cell increased in volume (
Figure 3B
4–B
5). The increase in cell volume depends on the directional or polarized cell growth. At early stages, the direction of cell growth was random (
Figure 3C
1–C
3), but later, most cells elongated along the direction of the serration (
Figure 3C
4,C
5). In summary, lip serration begins with the local activation of cell division, followed by cell elongation that increases volume and directs growth, deepening the serration (
Figure 3D
1–D
5).
2.3. Transcriptome Analysis of Floral Buds
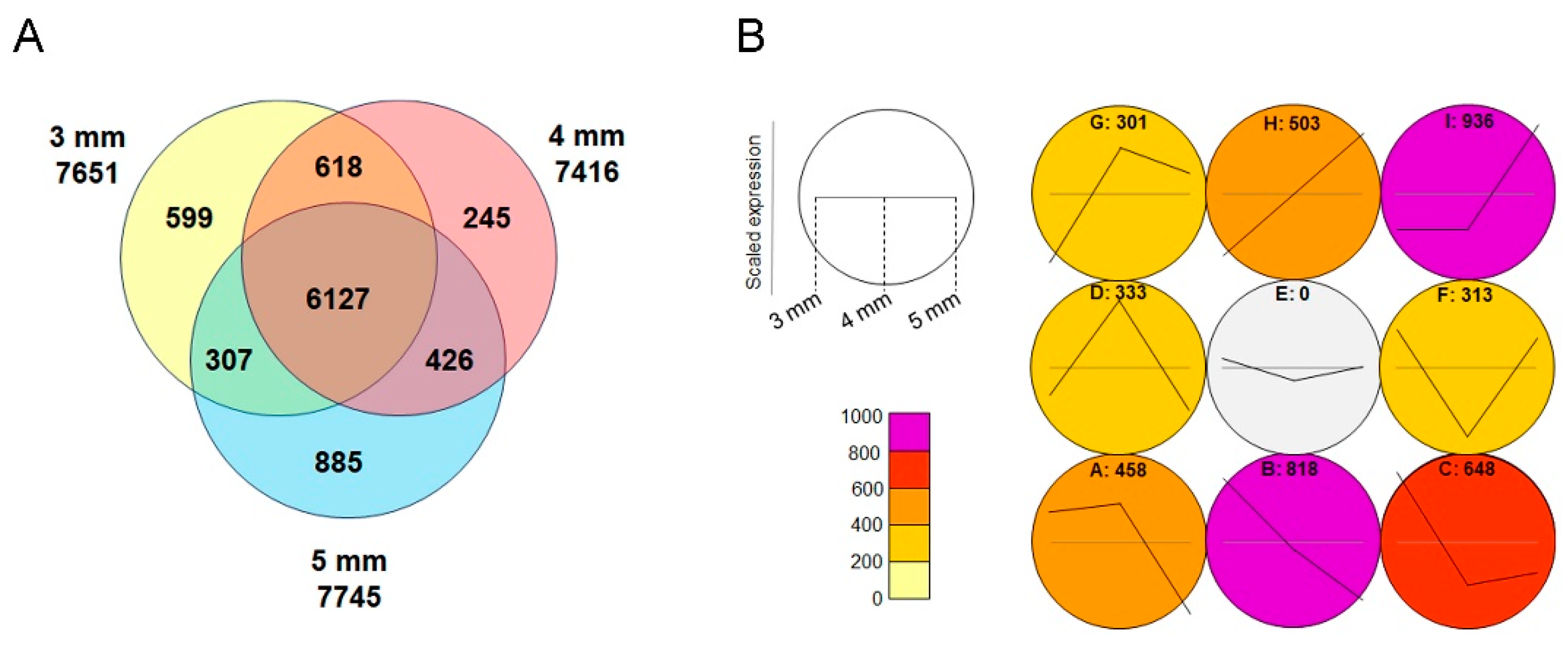
To identify genes involved in floral organ development, particularly those related to the early formation of lip serrations, RNA was extracted from buds measuring 3 mm, 4 mm, and 5 mm in length, as well as from leaves, followed by transcriptome analysis. Approximately 7,500 genes were identified as being expressed more than twice as much in the buds compared to the leaves, and 6,127 of these genes were commonly expressed in all the buds (
Figure 4A). Gene Ontology (GO) analysis of the common genes revealed those related to floral organ formation, mitosis, and meiosis (
Table S1). Among them, 152 genes related to floral organ formation, including floral homeotic genes, were identified (
Table S2). This analysis comprehensively revealed the transcript sequences expressed in Habenaria radiata flowers.
A self-organizing map (SOM) analysis was performed on genes exhibiting expression changes in the 3 mm to 5 mm buds, clustering genes with similar expression patterns (
Figure 4B). Clusters with increasing expression (G, H, I) contained genes related to meiosis (
Table S3), while clusters with decreasing expression (A, B, C) did not (
Table S4), indicating that meiosis of reproductive cells is occurring in the column at this stage.
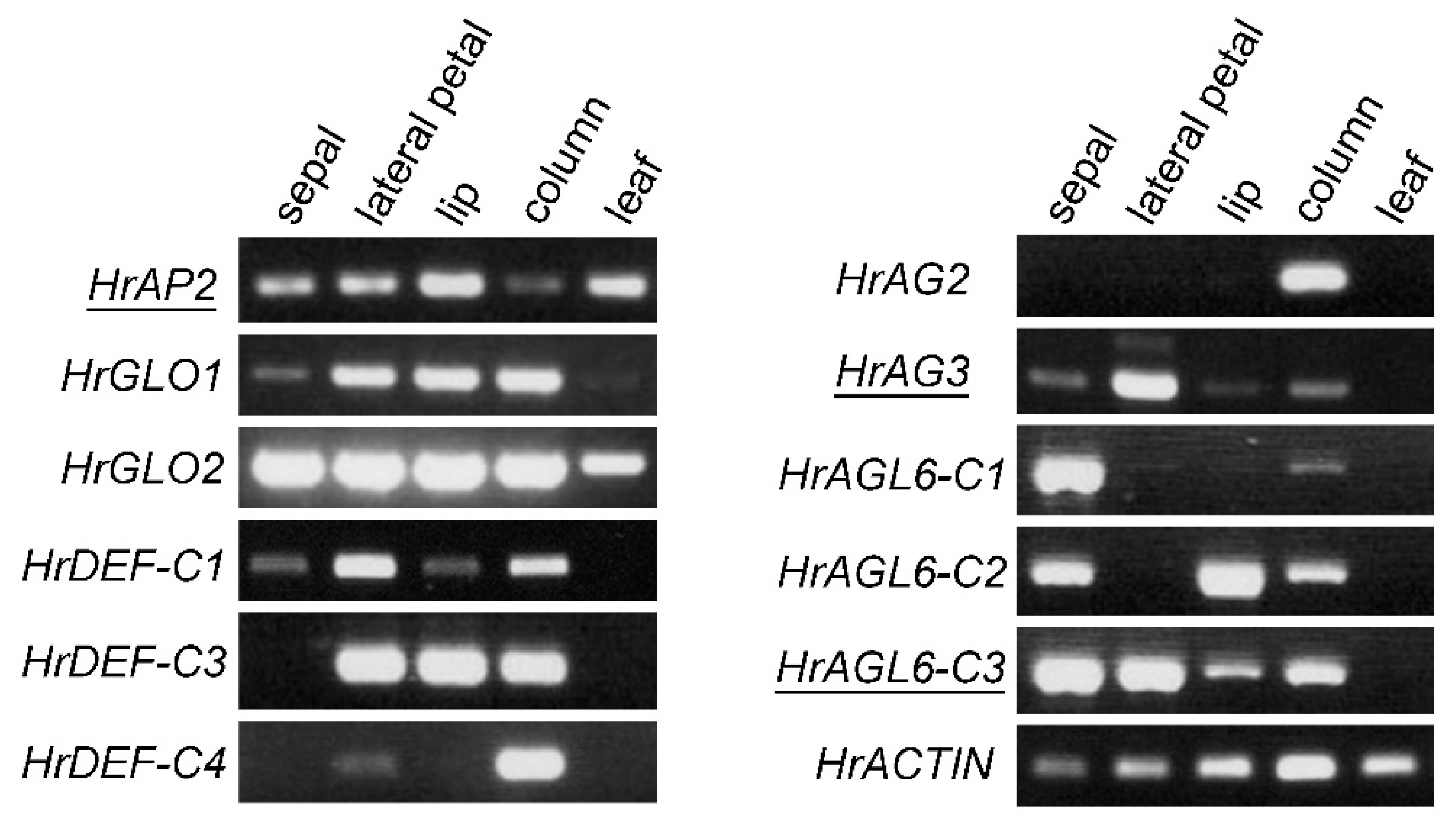
2.4. Expression of Floral Homeotic Genes in Floral Organs
Floral homeotic genes are crucial for floral organ development. We identified the sequences of these genes from the transcriptome data and cloned some of them, which had not been previously reported, using a PCR-based method. We identified Habenaria radiata APETALA2 (HrAP2) as class A gene, GLOBOSA (HrGLO1, HrGLO2) and DEFICIENS (HrDEF-C1, HrDEF-C3, HrDEF-C4) as class B genes, and AGAMOUS (HrAG2, HrAG3) as class C genes. Additionally, we found the AGL6 genes (HrAGL6-C1, HrAGL6-C2, HrAGL6-C3), as AGL6 is involved in petal differentiation in orchids [
22]. Among these, all of the class B genes, HrAG2, HrAGL6-C1, and HrAGL6-C2, have been previously reported [
14,
15,
16].
We examined the expression patterns of these genes in floral organs using RT-PCR (
Figure 5). HrAP2 was expressed in all floral organs, including the leaves. HrGLO1 and HrGLO2 were expressed in all floral organs, whereas the HrDEF genes showed differential expression. Specifically, HrDEF-C3 was expressed in both the petals and the column, while HrDEF-C4 was strongly expressed in the column. HrAG2 was expressed in the column, whereas HrAG3 was strongly expressed in lateral petals but weakly expressed in other floral organs, suggesting that HrAG3 may have a function distinct from class C in flower development. HrAGL6 genes were expressed in the sepals and the column, and of particular interest, they exhibited different expression patterns in the petals. HrAGL6-C1 was either not expressed or weakly expressed in petals, HrAGL6-C2 was expressed in lip, and HrAGL6-C3 was more highly expressed in lateral petals than in the lip. These expression patterns support previous reports [
14,
15,
16] and the two orchid models explaining the differentiation of second whorl organs [
17,
18,
19,
20,
21,
22].
2.5. Transgenic Analysis of Habenaria Radiata Floral Homeotic Genes in a Model Plant
To investigate whether these genes in Habenaria radiata function as floral homeotic genes, we overexpressed them in floral mutants of Arabidopsis thaliana. The constructs were transformed to the heterozygous homeotic mutants, since they produce few or no seeds due to the transformation of floral organs. The transformants were screened on media and genotyped for each mutation. A class A mutant, ap2-3, develops carpels in the first whorl in place of sepals, along with fewer petals and stamens (
Figure 6A). The screening resulted in 14 T1 plants: 3 wild-type, 4 heterozygous, and 6 homozygous. Two homozygous lines produced sepal-like organs in the first whorl and more stamens, and some stamens were petaloid in anther region (
Figure 6B). These results suggest that HrAP2 can weakly replace AP2 in Arabidopsis thaliana, indicating that is has weak activity as class A gene.
We introduced overexpression constructs of HrAG2 or HrAG3 into ag-1 heterozygous plants. The ag-1 developed only sepals and petals, resulting in the double flowers (
Figure 6C). Fourteen T1 plants were obtained: 4 wild-type, 6 heterozygous, and 4 homozygous. In homozygous background, flowers developed stamen-like organs instead of petals (
Figure 6D). In wild-type and heterozygous siblings, flowers developed stamens in place of petals (
Figure 6E). Most of the transformants exhibited hyponastic leaves (
Figure 6E). These results suggests that HrAG2 functions as a class C gene in flower development. In contrast, overexpression of the HrAG3 had no effect on floral organ or leaf development, suggesting that HrAG3 may have a different function in flower development. We also overexpressed the HrDEF-C4 in ap3-5 mutants, but observed no effect on floral organs.
Although they are phylogenetically distant, HrAP2 and HrAG2 partially replaced the floral homeotic genes in Arabidopsis thaliana, suggesting that these genes may also function as floral homeotic genes in Habenaria radiata.
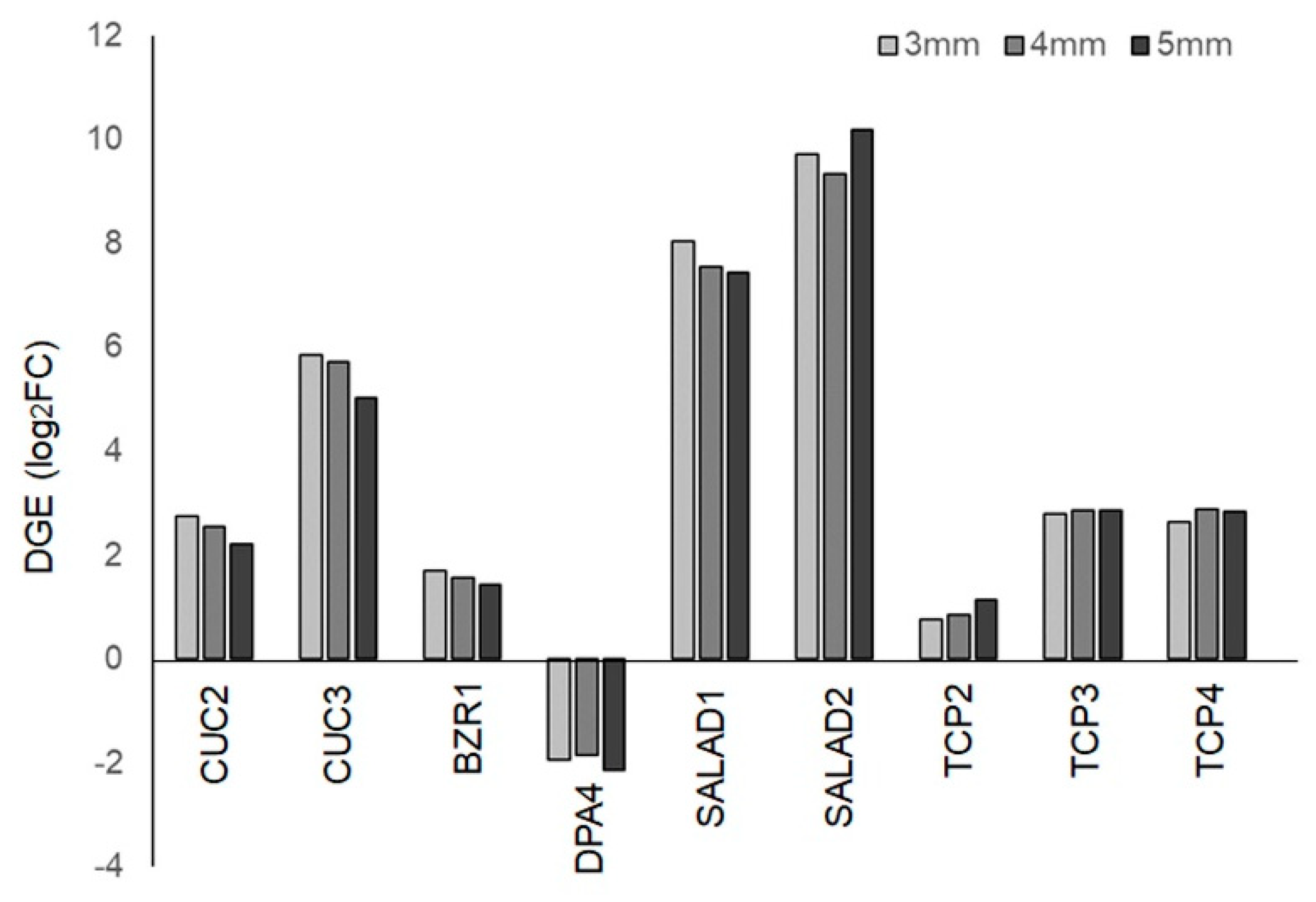
2.6. Digital Expression of Genes Involved in Serration Formation
In Habenaria radiata flowers, the formation of lip serrations is a key process for pollination. Therefore, we focused on genes associated with serration formation in plant organs and examined their digital gene expression (DGE) levels. Homologous genes of CUC2, CUC3, and BZR1, which regulate serration formation in Arabidopsis thaliana leaves [
27,
28,
29], were highly expressed, while the homologous gene of DPA4, a negative regulator of CUC2, showed reduced expression (
Figure 7) [
29,
30]. These factors are suggested to be involved in lip serration formation. On the other hand, homologous genes of SALAD, known negative regulators of CUC2, were highly expressed in floral buds (
Figure 7). The SALAD gene has been shown to modulate the depth of serrations in strawberry leaves by repressing CUC2 expression [
31], suggesting that it may play a role in shaping serrations rather than promoting their growth. Homologous genes of TCP2, TCP3, and TCP4, which are known to regulate serration formation in leaves and petals negatively [
32,
33,
34], showed increased expression in buds compared to leaves (
Figure 7). Since CINCINATA, from the same family in Antirrhinum, is involved in lobe development in petals [
35], these TCP genes are likely involved in floral organ formation in processes other than serration formation.
3. Discussion
We have shown the process of lip formation, anthesis, and petal wilting in the orchid Habenaria radiata. A distinctive feature of this plant is lip serration, and we found that initial cell divisions form protrusions along the lip margin, and subsequently, cells in the serrated region elongate in the same direction, deepening the serrations. While it is known that CUC and TCP genes are involved serration development in the leaves of Arabidopsis thaliana [
27,
28,
29,
30,
31,
33,
34], the serrations on Arabidopsis leaves do not deepen. We found that these genes were expressed in floral buds, but other unknown factors may be involved in deepening the serration at later stage. We found that directional cell growth is important for deepening the serration. Polarized cell growth in plants is regulated by the arrangement of cortical microtubules [
36]; therefore, the relationship between the cytoskeleton and serration growth remains a topic for further investigation.
Plants that form petal serrations are found in several plant families, including Caryophyllaceae, Celastraceae, Cucurbitaceae, Myrtaceae, Saxifragaceae, Tropaeolaceae, and Orchidaceae, suggesting that petal serrations evolved independently across the angiosperms. Notably, species of Cucumis can have extremely long fringes extending several centimeters [
37]. Most species with deep serration on the petal margin are pollinated by nocturnal hawk moths. There are three possible roles for deep serrations in attracting pollinators: the first is a visual effect, where the flower shape becomes more conspicuous, particularly for nocturnal butterflies that need to recognize the flowers. Second, serration increases the surface area for fragrance emission from the petals. Habenaria radiata possesses several varieties with strong fragrance, suggesting that fragrance emission can attract pollinators at night. The third possibility is that serrations provide a scaffold for visiting insects. Hawk moths visiting Habenaria radiata flowers have been observed grasping the lip fringes with their forelegs while inserting their proboscis deep into the spur [
25,
26]. An experiment removing the serrations showed no significant effect on the number of moth visits, but did reduce the time spent on the flower and the seed production [
25]. These findings suggest that the primary role of lip serrations in Habenaria radiata is likely as a scaffold, rather than a visual attractant. The role of serrations as a scaffold has also been investigated in Mitella pauciflora and its pollinator, fungus-gnat, where serrations were found to be important for insects landing [
38].
We previously showed that lip morphology in Habenaria radiata varied across different ecological regions [
26]. It is interesting to investigate whether these regional differences affect pollination and seed production efficiency and whether the morphology of surrounding pollinators, such as proboscis length, varies among habitats. Habenaria radiata is a near-threatened species so successful pollination is crucial for maintaining genetic diversity. Therefore, fundamental knowledge of both Habenaria radiata and its pollinators is essential for ecosystem conservation.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Habenaria radiata “Aoba” was purchased from a gardening store in Nara, Japan. The plants were grown in cultivation soil (Nippi Soil No.1, Nihon Hiryo Co. Ltd., Fujioka, Japan) covered with wet sphagnum, in a glasshouse at Seika campus, Kyoto Prefectural University, Seika, Japan. A GX200 digital camera (RICOH, Japan) was used for interval shooting (time-lapse), and photos were taken every 10 minutes from June 24 to July 7 in 2015. The movies were generated using Image J (version 1.53a) and iMovie (version 10.4).
4.2. Confocal Microscopy
Lips from floral buds at different developmental stages were dissected and fixed in a solution of 10% acetic acid and 90% ethanol for more than 1 hour. After the ethanol series (10 minutes in each concentration: 100%, 70%, 50%, and 30%), the lips were stained in the 50 µg/ml calcofluor solution (Fluorescent Brightener 28, Sigma-Aldrich) for more than 5 minutes. Staining was observed with the TCS SP8 confocal microscopy (Leica, Germany) with 405 nm excitation and detection from 410 to 600 nm. Images were captured using a photomultiplier and a Z-stack with a step size of 0.84.
4.3. Cell Shape Quantification
Cell shape analysis was performed as previously described [
39], with a few modifications. The epidermal cell contours were manually traced from image printouts, which were then scanned with LiDE220 (Cannon) as PNG files with a resolution of 200 dpi. Image J was used for cell shape analysis. The scanned images were converted to 8-bit, and the brightness threshold range was set to ‘Auto’ to segment the cell contours. The “Analyze Particles” function was used to measure the area and fit ellipse to the cells. The cell area and the angle of the major axis of the fitted ellipse were taken for 80 cells from each image. The major axis angle was corrected to ensure that the vertical bisector of an isosceles triangle, aligned with the serrated region, was at 90-degree angle. For result display, the LPX plugin Lpx Measure (measure mode: blobMeasure) (
https://lpixel.net/en/products/lpixel-imagej-plugins/) and pseudo-color from “Lookup
Tables” were used.
4.4. Transcriptome Analysis
Three independent samples were used for RNA extraction and RNA sequencing. Total RNA was isolated from leaves and floral buds (3, 4, or 5 mm in length) using a modified protocol of the RNeasy Plant Mini Kit (QIAGEN, Germany, [
40]). Library preparation, RNA sequencing, sequence assembly and homology searches were performed as described previously [
41]. Total trinity transcripts and putative genes were 771,822 and 447,223, respectively, and median and average contig length were 351 and 657.16, respectively. The data have been deposited with links to BioProject accession number PRJDB19824 in the DDBJ BioProject database. Accession numbers for each dataset are shown in
Table S6. Genes expressed in flowers were selected based on a false discovery rate (FDR) < 0.01, a total read count > 1, and log
2 fold change (log
2FC) > 1 compared to leaves.
4.5. RT-PCR
Total RNA was isolated from leaves, and sepals, lateral petals, lips, and columns from floral buds with 7 mm length, as described above. cDNAs were generated with ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO). PCR was performed using KAPA Taq Extra DNA Polymerase (Kapa Biosystems). Oligonucleotide primers and denature temperature for PCR was listed on
Table S5.
4.6. Transformation of Arabidopsis Thaliana Mutants
HrAP2,
HrAG, and
HrAGL6 genes were cloned into the qCR4-TOPO vector (Invitrogen) and subcloned into the pAN19 vector carrying CaMV 35S promoter and NOS terminator.
35Spro:HrAP2, HrAG2, or HrAG3:NOSter cassette was subcloned into the pBIN30 binary vector and transformed to
Agrobacterium tumefaciens (
Rhizobium radiobacter) strain GV3101_pMP90. The
35S:HrAP2 and
35S:HrAG2 constructs were then transformed into
ap2-3/+ and
ag-1/+ heterozygous plants, respectively, with floral dip method [
42]. T1 plants were selected on MS media containing 10 µg/ml glufosinate and genotyped for the
Arabidopsis AP2 and
AG genes. The
HrAP2,
HrAG3, and
HrAGL6-C3 sequences have been deposited in the GenBank database with accession numbers LC856603, LC856604, and LC856605, respectively.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Morphology of Habenaria radiata; Table S1: Gene ontology analysis of 6,127 genes expressed in floral buds with 3, 4, and 5 mm length; Table S2: 152 genes involved in GO terms related to flower development; Table S3: GO analysis of upregulated genes during flower development (1,080 genes); Table S4: GO analysis of downregulated genes during flower development (1,176 genes); Table S5: Oligonucleotide primers used for RT-PCR; Video S1: Anthesis and withering of Habenaria radiata flower.
Author Contributions
Conceptualization, S.T., Y.N. and T.T.; methodology, S.T., Y.N., T.T, H.T., T.S., and S.K.; software, H.T., T.S. and S.K.; validation, S.T. and S.K.; investigation, S.T. and Y.N.; data curation, S.T. and Y.N.; writing—original draft preparation, S.T. and Y.N.; writing—review and editing, S.T.; visualization, S.T. and Y.N.; supervision, S.T.; project administration, S.T.; funding acquisition, S.T. and S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Numbers JP18K06366 and 21H02513 to ST and SK, respectively.
Data Availability Statement
All data are contained within the article.
Acknowledgments
The authors thank Ms. Kaori Mizuno (Kyoto Sangyo University) for her technical help with library construction and RNA-seq.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Theissen, G.; Saedler, H. Plant biology. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Theissen, G.; Melzer, R.; Rumpler, F. MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 2016, 143, 3259–3271. [Google Scholar] [CrossRef] [PubMed]
- Krizek, B.A.; Fletcher, J.C. Molecular mechanisms of flower development: an armchair guide. Nat Rev Genet 2005, 6, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Tsai, W.C.; Kuoh, C.S.; Chuang, M.H.; Chen, W.H.; Chen, H.H. Four DEF-like MADS box genes displayed distinct floral morphogenetic roles in Phalaenopsis orchid. Plant Cell Physiol 2004, 45, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Tsai, W.C.; Lee, P.F.; Chen, H.I.; Hsiao, Y.Y.; Wei, W.J.; Pan, Z.J.; Chuang, M.H.; Kuoh, C.S.; Chen, W.H.; Chen, H.H. PeMADS6, a GLOBOSA/PISTILLATA-like gene in Phalaenopsis equestris involved in petaloid formation, and correlated with flower longevity and ovary development. Plant Cell Physiol 2005, 46, 1125–1139. [Google Scholar] [CrossRef]
- Tsai, W.C.; Pan, Z.J.; Hsiao, Y.Y.; Jeng, M.F.; Wu, T.F.; Chen, W.H.; Chen, H.H. Interactions of B-class complex proteins involved in tepal development in Phalaenopsis orchid. Plant Cell Physiol 2008, 49, 814–824. [Google Scholar] [CrossRef]
- Hsu, H.F.; Yang, C.H. An orchid (Oncidium Gower Ramsey) AP3-like MADS gene regulates floral formation and initiation. Plant Cell Physiol 2002, 43, 1198–1209. [Google Scholar] [CrossRef]
- Hsu, H.F.; Huang, C.H.; Chou, L.T.; Yang, C.H. Ectopic expression of an orchid (Oncidium Gower Ramsey) AGL6-like gene promotes flowering by activating flowering time genes in Arabidopsis thaliana. Plant Cell Physiol 2003, 44, 783–794. [Google Scholar] [CrossRef]
- Chang, Y.Y.; Kao, N.H.; Li, J.Y.; Hsu, W.H.; Liang, Y.L.; Wu, J.W.; Yang, C.H. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol 2010, 152, 837–853. [Google Scholar] [CrossRef]
- Xu, Y.; Teo, L.L.; Zhou, J.; Kumar, P.P.; Yu, H. Floral organ identity genes in the orchid Dendrobium crumenatum. Plant J 2006, 46, 54–68. [Google Scholar] [CrossRef]
- Aceto, S.; Sica, M.; De Paolo, S.; D'Argenio, V.; Cantiello, P.; Salvatore, F.; Gaudio, L. The analysis of the inflorescence miRNome of the orchid Orchis italica reveals a DEF-like MADS-box gene as a new miRNA target. PLoS One 2014, 9, e97839. [Google Scholar] [CrossRef] [PubMed]
- Dirks-Mulder, A.; Butot, R.; van Schaik, P.; Wijnands, J.W.; van den Berg, R.; Krol, L.; Doebar, S.; van Kooperen, K.; de Boer, H.; Kramer, E.M.; et al. Exploring the evolutionary origin of floral organs of Erycina pusilla, an emerging orchid model system. BMC Evol Biol 2017, 17, 89. [Google Scholar] [CrossRef]
- Kim, S.Y.; Yun, P.Y.; Fukuda, T.; Ochiai, T.; Yokoyama, J.; Kameya, T.; Kanno, A. Expression of a DEFICIENS-like gene correlates with the differentiation between sepal and petal in the orchid, Habenaria radiata (Orchidaceae). Plant Science 2007, 172, 319–326. [Google Scholar] [CrossRef]
- Mitoma, M.; Kanno, A. The Greenish Flower Phenotype of Habenaria radiata (Orchidaceae) Is Caused by a Mutation in the SEPALLATA-Like MADS-Box Gene HrSEP-1. Front Plant Sci 2018, 9, 831. [Google Scholar] [CrossRef]
- Mitoma, M.; Kajino, Y.; Hayashi, R.; Endo, M.; Kubota, S.; Kanno, A. Molecular mechanism underlying pseudopeloria in Habenaria radiata (Orchidaceae). Plant J 2019, 99, 439–451. [Google Scholar] [CrossRef]
- Pan, Z.J.; Cheng, C.C.; Tsai, W.C.; Chung, M.C.; Chen, W.H.; Hu, J.M.; Chen, H.H. The duplicated B-class MADS-box genes display dualistic characters in orchid floral organ identity and growth. Plant Cell Physiol 2011, 52, 1515–1531. [Google Scholar] [CrossRef] [PubMed]
- Mondragon-Palomino, M.; Theissen, G. Conserved differential expression of paralogous DEFICIENS- and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: refining the 'orchid code'. Plant J 2011, 66, 1008–1019. [Google Scholar] [CrossRef] [PubMed]
- Mondragon-Palomino, M. Perspectives on MADS-box expression during orchid flower evolution and development. Front Plant Sci 2013, 4, 377. [Google Scholar] [CrossRef] [PubMed]
- Mondragon-Palomino, M.; Theissen, G. MADS about the evolution of orchid flowers. Trends Plant Sci 2008, 13, 51–59. [Google Scholar] [CrossRef]
- Mondragon-Palomino, M.; Theissen, G. Why are orchid flowers so diverse? Reduction of evolutionary constraints by paralogues of class B floral homeotic genes. Ann Bot 2009, 104, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.F.; Hsu, W.H.; Lee, Y.I.; Mao, W.T.; Yand, J.Y.; Li, J.Y.; Yand, C.H. Model for perianth formation in orchids. Nature Plants 2015, 1, 15046. [Google Scholar] [CrossRef]
- Suetsugu, K.; Tanaka, K. Diurnal butterfly pollination in the orchid Habenaria radiata. Entomol Sci 2014, 17, 443–445. [Google Scholar] [CrossRef]
- Ikeuchi, Y.; Suetsugu, K.; Sumikawa, H. Diurnal skipper Pelodidas mathias (Lepidoptera: Hesperiidae) pollinates Habenaria radiata (Orchidaceae). Entomol News 2015, 125. [Google Scholar]
- Suetsugu, K.; Abe, Y.; Asai, T.; Matsumoto, S.; Hasegawa, M. Specialized petal with conspicuously fringed margin influences reproductive success in Habenaria radiata (Orchidaceae). Ecology 2022, 103, e3781. [Google Scholar] [CrossRef]
- Tachibana, T.; Nishikawa, Y.; Kubo, N.; Takeda, S. Morphological and Genetic Diversities of Habenaria radiata (Orchidaceae) in the Kinki Area, Japan. Int J Mol Sci 2020, 22. [Google Scholar] [CrossRef] [PubMed]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef] [PubMed]
- Bilsborough, G.D.; Runions, A.; Barkoulas, M.; Jenkins, H.W.; Hasson, A.; Galinha, C.; Laufs, P.; Hay, A.; Prusinkiewicz, P.; Tsiantis, M. Model for the regulation of Arabidopsis thaliana leaf margin development. Proc Natl Acad Sci U S A 2011, 108, 3424–3429. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Fang, N.; Lu, T.; Tameshige, T.; Nakata, M.T.; Jiang, Y.; Tan, L.; He, H.; Zhang, X.; Huang, Y.; et al. WOX1 controls leaf serration development via temporally restricting BZR1 and CUC3 expression in Arabidopsis. J Exp Bot 2024. [CrossRef]
- Engelhorn, J.; Reimer, J.J.; Leuz, I.; Gobel, U.; Huettel, B.; Farrona, S.; Turck, F. Development-related PcG target in the apex 4 controls leaf margin architecture in Arabidopsis thaliana. Development 2012, 139, 2566–2575. [Google Scholar] [CrossRef]
- Luo, X.; Guo, L.; Tagliere, E.; Yang, Z.; Liu, Z. Leaf dissection and margin serration are independently regulated by two regulators converging on the CUC2-auxin module in strawberry. Curr Biol 2024, 34, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Ori, N.; Cohen, A.R.; Etzioni, A.; Brand, A.; Yanai, O.; Shleizer, S.; Menda, N.; Amsellem, Z.; Efroni, I.; Pekker, I.; et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 2007, 39, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Mitsuda, N.; Seki, M.; Shinozaki, K.; Ohme-Takagi, M. TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 2010, 22, 3574–3588. [Google Scholar] [CrossRef]
- Koyama, T.; Ohme-Takagi, M.; Sato, F. Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signal Behav 2011, 6, 697–699. [Google Scholar] [CrossRef]
- Crawford, B.C.; Nath, U.; Carpenter, R.; Coen, E.S. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol 2004, 135, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Sun, Z.; Yan, P.; Wang, T.; Zhang, Y. Mechanical regulation of cortical microtubules in plant cells. New Phytol 2023, 239, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- de Boer, H.J.; Schaefer, H.; Thulin, M.; Renner, S.S. Evolution and loss of long-fringed petals: a case study using a dated phylogeny of the snake gourds, Trichosanthes (Cucurbitaceae). BMC Evol Biol 2012, 12, 108. [Google Scholar] [CrossRef] [PubMed]
- Katsuhara, K.R.; Kitamura, S.; Ushimaru, A. Functional significance of petals as landing sites in fungus-gnat pollinated flowers of Mitella Pauciflora (Sexifragaceae). Functional Ecology 2017, 31, 1193–1200. [Google Scholar] [CrossRef]
- Higaki, T.; Takigawa-Imamura, H.; Akita, K.; Kutsuna, N.; Kobayashi, R.; Hasezawa, S.; Miura, T. Exogenous Cellulase Switches Cell Interdigitation to Cell Elongation in an RIC1-dependent Manner in Arabidopsis thaliana Cotyledon Pavement Cells. Plant Cell Physiol 2017, 58, 106–119. [Google Scholar] [CrossRef]
- Brunner, A.M.; Yakovlev, I.A.; Strauss, S.H. Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol 2004, 4, 14. [Google Scholar] [CrossRef]
- Takeda, S.; Yoza, M.; Ueda, S.; Takeuchi, S.; Maeno, A.; Sakamoto, T.; Kimura, S. Exploring the diversity of galls on Artemisia indica induced by Rhopalomyia species through morphological and transcriptome analyses. Plant Direct 2024, 8, e619. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
Figure 1.
Flowering and withering of Habenaria radiata flowers captured by interval shooting (time-lapse). (A-G) Flowering and withering of flowers. The date (dd/mm) and time of capture are shown. Note that upper flower, which bloomed later, withered earlier than lower flower (G), probably due to the loss of pollinium (arrowheads in D and E) and subsequent pollination. (H, I) Side (H) and front (I) views of lip unfolding. The captured time is shown in each panel.
Figure 1.
Flowering and withering of Habenaria radiata flowers captured by interval shooting (time-lapse). (A-G) Flowering and withering of flowers. The date (dd/mm) and time of capture are shown. Note that upper flower, which bloomed later, withered earlier than lower flower (G), probably due to the loss of pollinium (arrowheads in D and E) and subsequent pollination. (H, I) Side (H) and front (I) views of lip unfolding. The captured time is shown in each panel.
Figure 2.
Lip development. (A-E) Floral buds at different stages. Arrowheads in B, C, and D indicate the growing spur. (F-J) Lip inside the floral bud shown in (A-E). The lengths of the floral buds are 1 mm (A, F), 2 mm (B, G), 4 mm (C, H), 6 mm (D, I), and 7 mm (E, J). Scale bars: A, F = 0.5 mm; B, G = 1 mm; C, D, E, H, I, J = 2 mm. .
Figure 2.
Lip development. (A-E) Floral buds at different stages. Arrowheads in B, C, and D indicate the growing spur. (F-J) Lip inside the floral bud shown in (A-E). The lengths of the floral buds are 1 mm (A, F), 2 mm (B, G), 4 mm (C, H), 6 mm (D, I), and 7 mm (E, J). Scale bars: A, F = 0.5 mm; B, G = 1 mm; C, D, E, H, I, J = 2 mm. .
Figure 3.
Cell shape changes during the development of lip serration. (A) Confocal laser microscopy images of petal margin cells. Petals from early (A1) to late (A5) stages were excised, stained, and observed. (B) Distribution of cell area. (C) Elongation direction of each cell. The direction of serration elongation was set as 90 degrees, with the angles relative to this direction shown in different colors. (D) Plot of cell area and elongation direction. Up to time point 3, cell proliferation occurs, and from time point 4 onward, the serrations deepen due to polarized cell elongation. Scale bars: 50 µm. .
Figure 3.
Cell shape changes during the development of lip serration. (A) Confocal laser microscopy images of petal margin cells. Petals from early (A1) to late (A5) stages were excised, stained, and observed. (B) Distribution of cell area. (C) Elongation direction of each cell. The direction of serration elongation was set as 90 degrees, with the angles relative to this direction shown in different colors. (D) Plot of cell area and elongation direction. Up to time point 3, cell proliferation occurs, and from time point 4 onward, the serrations deepen due to polarized cell elongation. Scale bars: 50 µm. .
Figure 4.
Transcriptome analysis of floral buds. (A) Venn diagram showing genes expressed more than twice as much in floral buds of 3 mm, 4 mm, and 5 mm sizes compared to leaves. (B) Self-organizing map (SOM) analysis of the genes expressed in floral buds. The letters represent clusters with similar expression patterns, and the numbers indicate the gene number in each cluster. Clusters G, H, and I show an increase in expression during bud development, while clusters A, B, and C show a decrease in expression.
Figure 4.
Transcriptome analysis of floral buds. (A) Venn diagram showing genes expressed more than twice as much in floral buds of 3 mm, 4 mm, and 5 mm sizes compared to leaves. (B) Self-organizing map (SOM) analysis of the genes expressed in floral buds. The letters represent clusters with similar expression patterns, and the numbers indicate the gene number in each cluster. Clusters G, H, and I show an increase in expression during bud development, while clusters A, B, and C show a decrease in expression.
Figure 5.
RT-PCR of floral homeotic and MADS genes in Habenaria radiata. Underlined genes are reported for the first time in this work. HrACTIN was used as the control.
Figure 5.
RT-PCR of floral homeotic and MADS genes in Habenaria radiata. Underlined genes are reported for the first time in this work. HrACTIN was used as the control.
Figure 6.
Overexpression of HrAP2 and HrAG2 in Arabidopsis thaliana. (A) ap2-3 mutant. (B) 35S:HrAP2 plant in the ap2-3 background. Arrowhead indictes the petaloid stamen. Note that sepal-like organs are generated in the first whorl, and more stamens were produced compared to the ap2-3 mutant. (C) ag-1 flower. (D) 35S:HrAG2 plant in the ag-1 background. Note that stamen-like organs are generated. (E, F) 35S:HrAG2 plants in AG-1 (wild-type sibling) background. (E) Stamens are generated instead of petals in the second whorl. (F) Vegetative phenotype showing hyponastic growth in leaves, resulting in the curled leaves. Scale bars: A, B, C, D, E = 1 mm; F = 5 mm.
Figure 6.
Overexpression of HrAP2 and HrAG2 in Arabidopsis thaliana. (A) ap2-3 mutant. (B) 35S:HrAP2 plant in the ap2-3 background. Arrowhead indictes the petaloid stamen. Note that sepal-like organs are generated in the first whorl, and more stamens were produced compared to the ap2-3 mutant. (C) ag-1 flower. (D) 35S:HrAG2 plant in the ag-1 background. Note that stamen-like organs are generated. (E, F) 35S:HrAG2 plants in AG-1 (wild-type sibling) background. (E) Stamens are generated instead of petals in the second whorl. (F) Vegetative phenotype showing hyponastic growth in leaves, resulting in the curled leaves. Scale bars: A, B, C, D, E = 1 mm; F = 5 mm.
Figure 7.
Digital gene expression (DGE) of genes involved in serration formation in floral buds.
Figure 7.
Digital gene expression (DGE) of genes involved in serration formation in floral buds.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).