Submitted:
23 December 2024
Posted:
24 December 2024
You are already at the latest version
Abstract
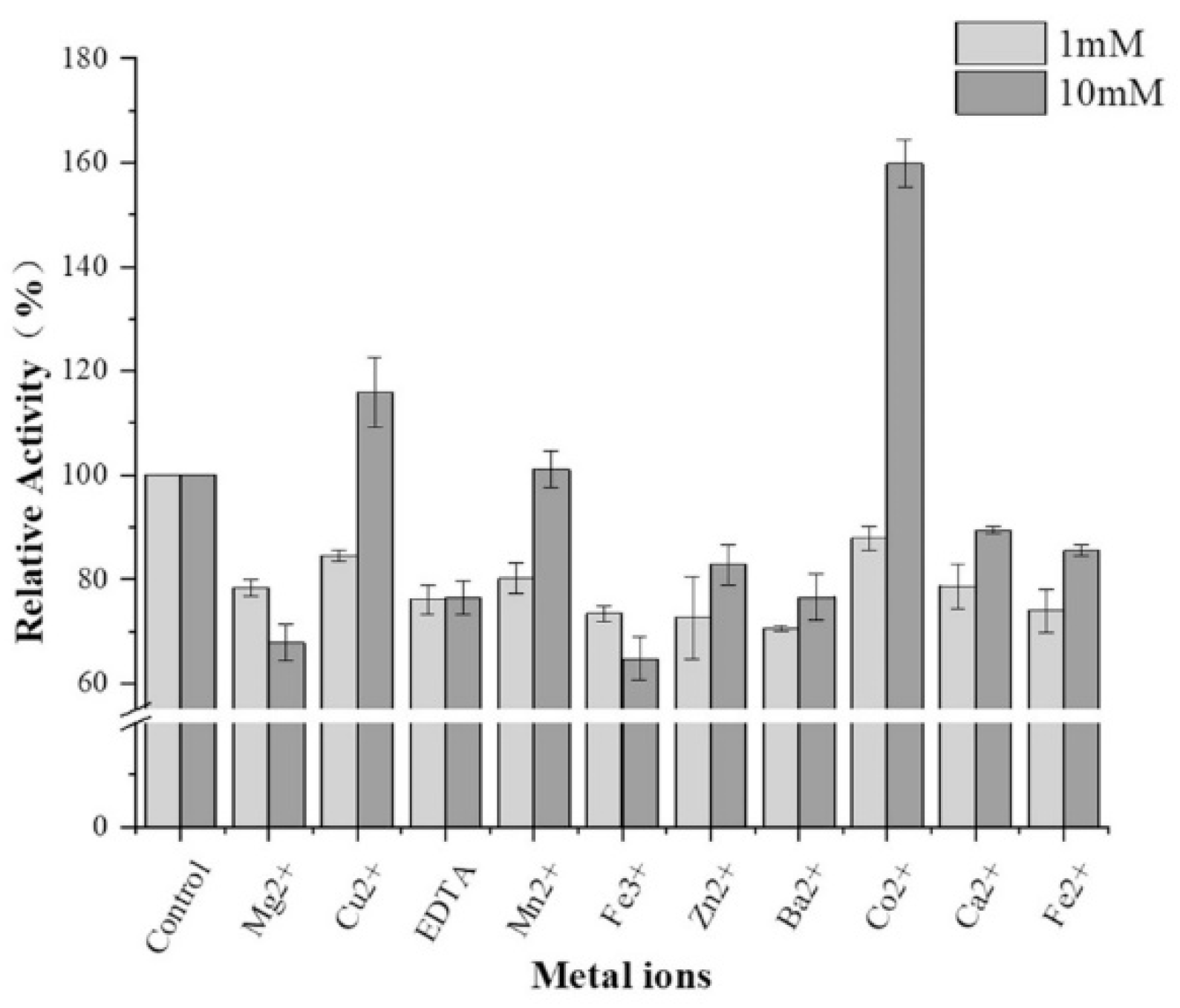
Algin is the most abundant substance in algae. Alginate lyase degrades algin, producing algin monosaccharides, disaccharides, and oligosaccharides, which are widely used in bioenergy, food, medicine, and other fields. In this study, an Exiguobacterium strain isolated from rotten kelp exhibited a robust ability to degrade algae. Sequencing of this strain revealed the presence of three different types of alginate lyase. However, the expression of these three genes in Escherichia coli showed lower alginate lyase activity compared to the original strain. After codon optimization, the gene with the highest activity of the three was successfully expressed in Pichia pastoris to produce recombinant EbAlg664. In 5L high-density fermentation, the activity of the recombinant enzyme reached 1306 U/mg protein, 3.9 times that of the original Exiguobacterium strain. Enzymatic analysis revealed that the optimal temperature and pH range of recombinant EbAlg664 were narrower compared to the original strain. Furthermore, the presence of Cu2+ and Co2+ enhanced enzymatic activity, whereas Mg2+ and Fe3+ inhibited the recombinant alginate lyase. This study provides a theoretical and practical foundation for the industrial-scale production of engineered Pichia pastoris with high alginate lyase activity.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Culture Source and Medium
2.2. Culture and Optimization of Exiguobacterium
2.3. Cloning of Alginate Lyase Gene
2.4. Expression of Alginate Lyase Genes in E. coli
2.5. Heterologous Expression of Alginate Lyase Genes in Pichia Pastoris
2.6. Extraction of Alginate Lyase and Determination of Its Activity
2.7. Effects of pH and Temperature on Alginate Lyase Activity
2.8. Effects of Metal Ions on Alginate Lyase Activity
2.9. Statistical Analyses
3. Results
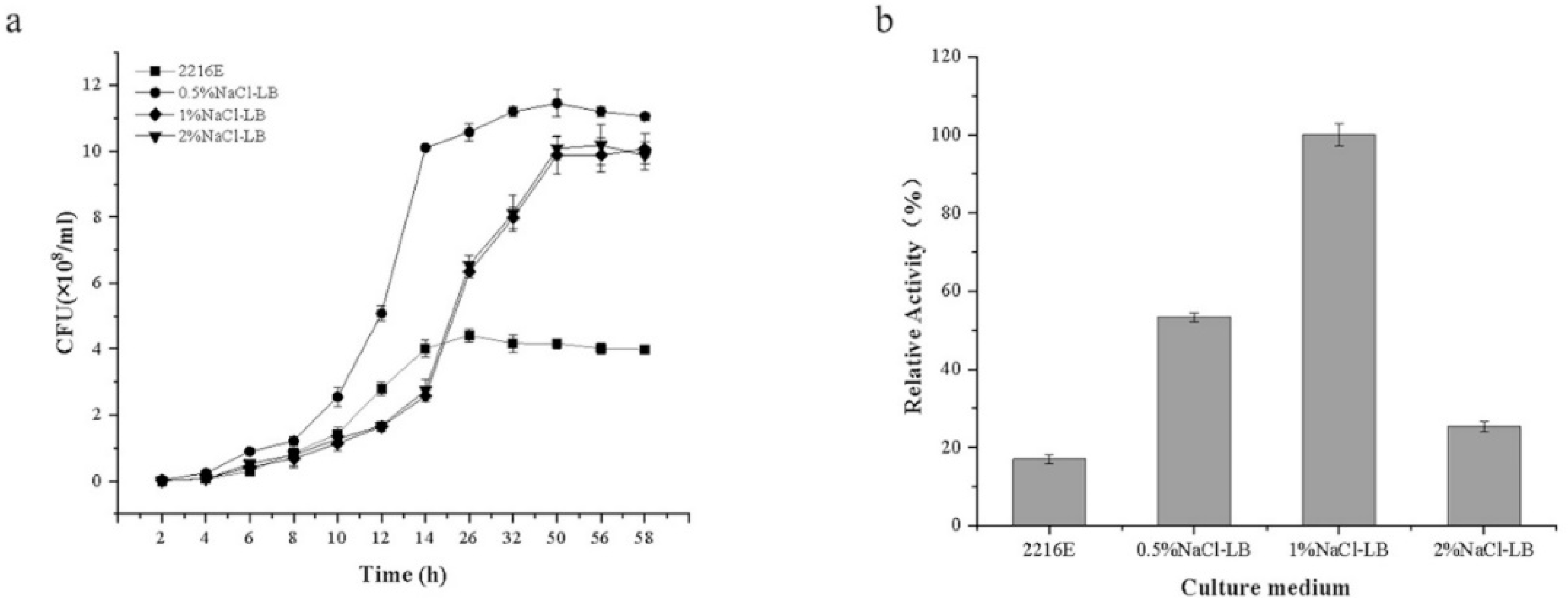
3.1. Media for the Growth and Alginate Lyase Activity of Exiguobacterium
3.2. Gene Sequencing and Protein Analysis of Exiguobacterium
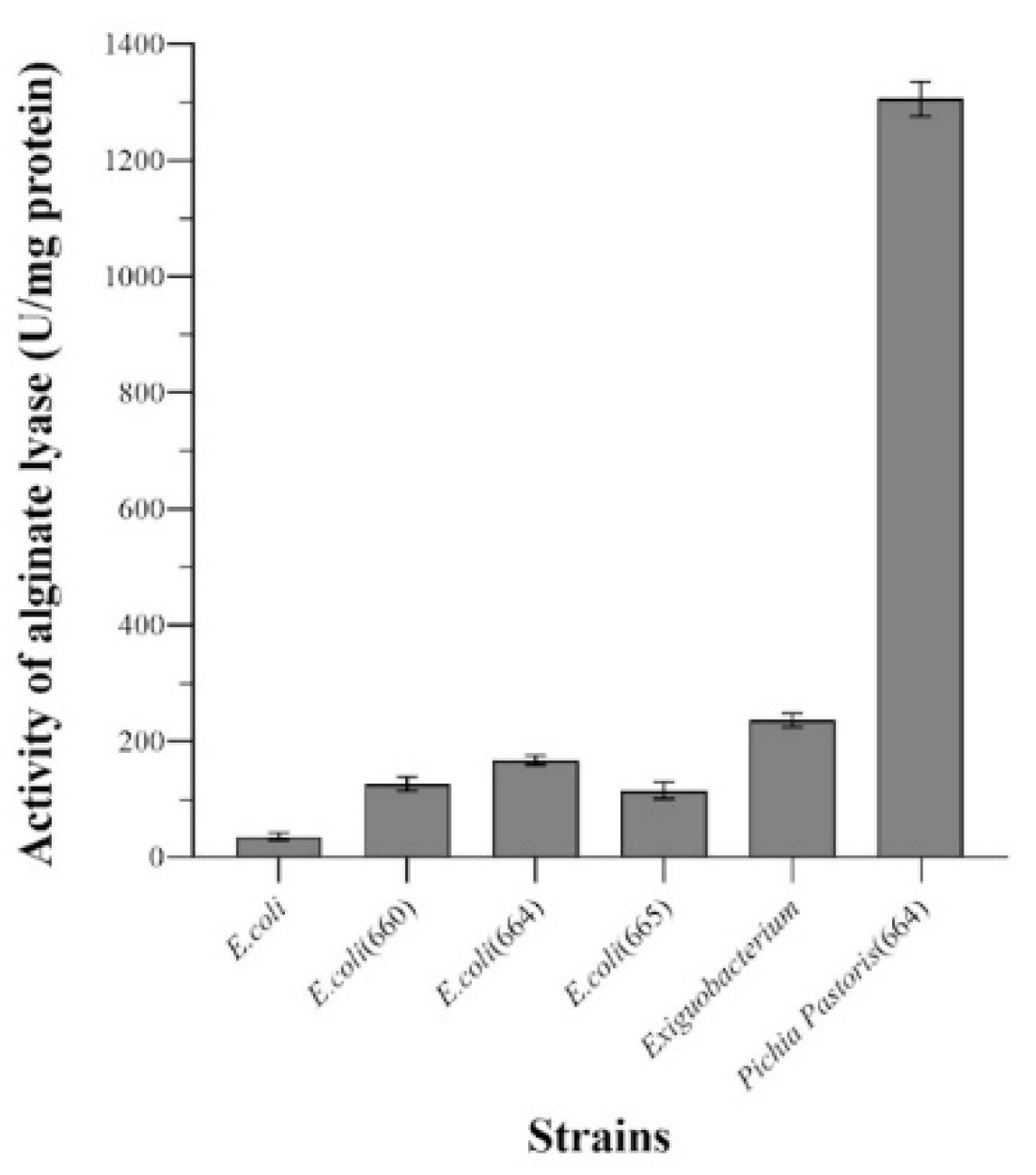
3.3. Expression of Alginate Lyase Gene in E. coli
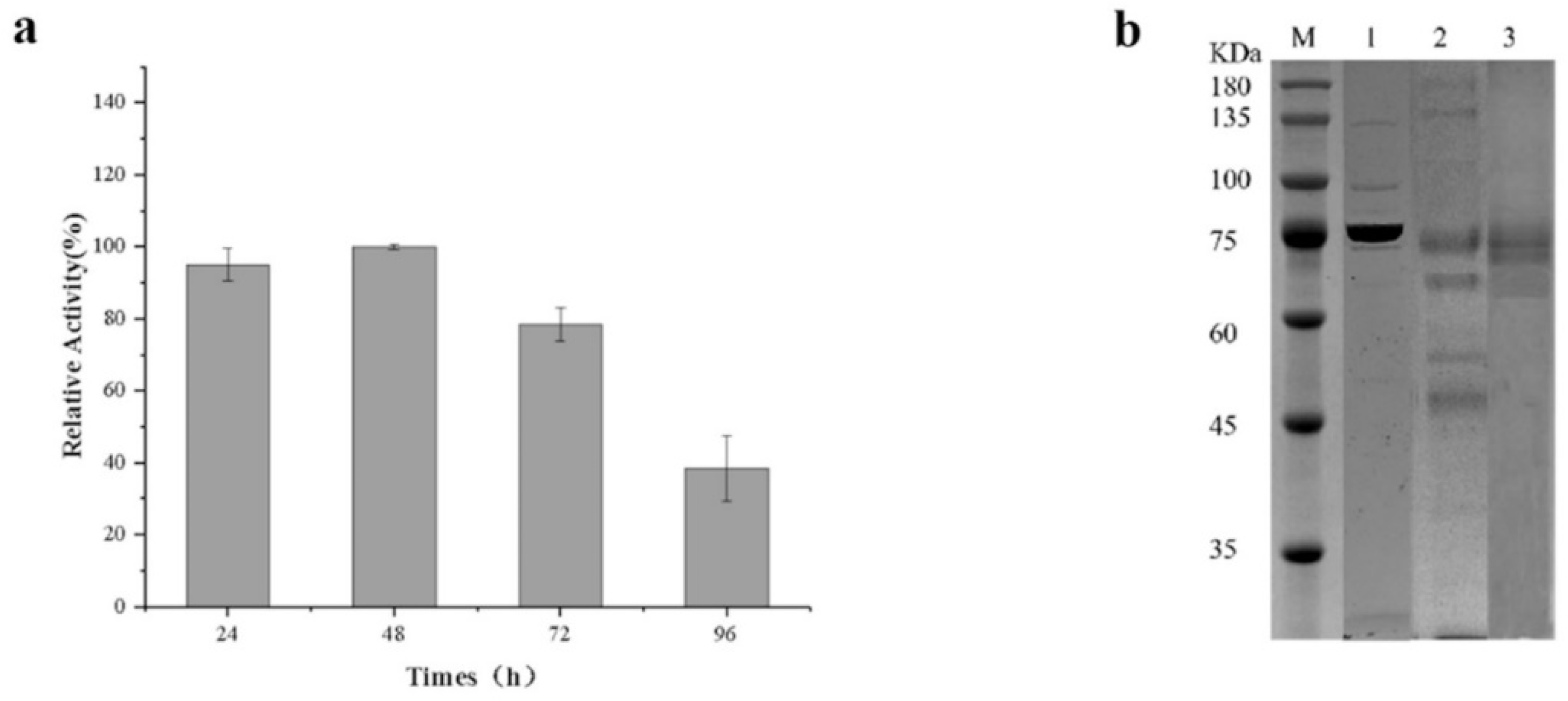
3.4. Expression of Alginate Lyase Gene in Pichia Pastoris
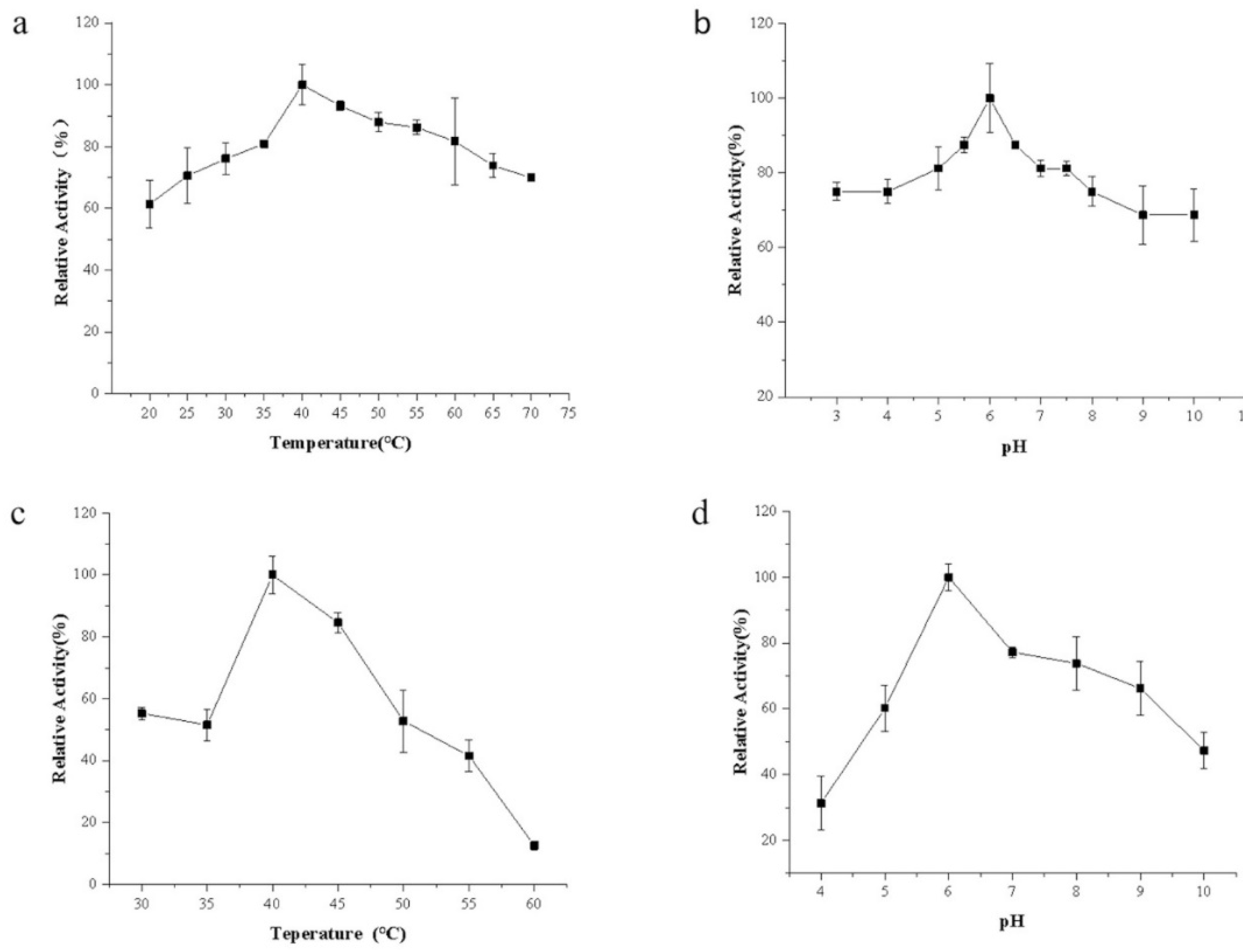
3.5. Enzymatic Properties of Recombinant Alginate Lyase
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- He, Q.; Tong, T.; Yu, C.; Wang, Q. Advances in Algin and Alginate-Hybrid Materials for Drug Delivery and Tissue Engineering. Mar. Drugs. 2022, 21, 14. [Google Scholar] [CrossRef] [PubMed]
- Medipally, S.R.; Yusoff, F.M.; Banerjee, S.; Shariff, M. Microalgae as sustainable renewable energy feedstock for biofuel production. Biomed. Res. Int. 2015, 12, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Rastogi, P.; Kandasubramanian, B. Review of alginate-based hydrogel bioprinting for application in tissue engineering. Biofabrication. 2012, 11, 042001. [Google Scholar] [CrossRef] [PubMed]
- Adamiak, K.; Sionkowska, A. State of Innovation in Alginate-Based Materials. Mar. Drugs. 2023, 21, 353. [Google Scholar] [CrossRef]
- Lu, S.; Na, K.; Wei, J.; Zhang, L.; Guo, X. Alginate oligosaccharides: The structure-function relationships and the directional preparation for application. Carbohydr. Polym. 2022, 284, 119225. [Google Scholar] [CrossRef]
- Vreeland, V. Immunocytochemical localization of the extracellular polysaccharide alginic acid in the brown seaweed, Fucus distichus. J. Histochem. Cytochem. 1972, 20, 358–67. [Google Scholar] [CrossRef]
- Atkins, E.D.; Nieduszynski, I.A.; Mackie, W.; Parker, K.D.; Smolko, E.E. Structural components of alginic acid Ⅱ the crystalline structure of poly-L-mannuronic acid. Results of X-ray diffraction and polarized infrared studies. Biopolymers. 1973, 12, 1865–1878. [Google Scholar] [CrossRef]
- Xu, F.; Wang, P.; Zhang, Y.Z.; Chen, X.L. Diversity of Three-Dimensional Structures and Catalytic Mechanisms of Alginate Lyases. Appl. Environ. Microbiol. 2017, 84, e02040. [Google Scholar] [CrossRef]
- Cheng, D.; Jiang, C.; Xu, J.; Liu, Z.; Mao, X. Characteristics and applications of alginate lyases: A review. Int. J. Biol. Macromol. 2020, 164, 1304–1320. [Google Scholar] [CrossRef]
- Iwasaki, K.; Matsubara, Y. Purification of alginate oligosaccharides with root growth-promoting activity toward lettuce. Biosci. Biotechnol. Biochem. 2000, 64, 1067–1070. [Google Scholar] [CrossRef]
- Xu, X.; Iwamoto, Y.; Kitamura, Y.; Oda, T.; Muramatsu, T. Root growth-promoting activity of unsaturated oligomeric uronates from alginate on carrot and rice plants. Biosci. Biotechnol. Biochem. 2003, 67, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Li, S.Y.; Wang, Z.P.; Wang, L.N.; Peng, J.X.; Wang, Y.N.; Han, Y.T.; et al. Combined enzymatic hydrolysis and selective fermentation for green production of alginate oligosaccharides from Laminaria japonica. Bioresour. Technol. 2019, 32, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Yin, H. Alginate lyase: Review of major sources and classification, properties, structure-function analysis and applications. Bioengineered. 2015, 6, 125–31. [Google Scholar] [CrossRef] [PubMed]
- Schurks, N.; Wingender, J.; Flemming, H.C.; Mayer, C. Monomer composition and sequence of alginates from Pseudomonas aeruginosa. Int. J. Biol. Macromol. 2002, 30, 105–111. [Google Scholar] [CrossRef]
- Long, L.; Hu, Q.; Wang, X.; Li, H.; Li, Z.; Jiang, Z.; et al. A bifunctional exolytic alginate lyase from Microbulbifer sp. ALW1 with salt activation and calcium-dependent catalysis. Enzyme Microb Technol. 2022, 161, 110109. [Google Scholar] [CrossRef]
- Zhou, H.X.; Xu, S.S.; Yin, X.J.; Wang, F.L.; Li, Y. Characterization of a New Bifunctional and Cold-Adapted Polysaccharide Lyase (PL) Family 7 Alginate Lyase from Flavobacterium sp. Mar. Drugs. 2020, 18, 388. [Google Scholar] [CrossRef]
- Yamasaki, M.; Moriwaki, S.; Miyake, O.; Hashimoto, W.; Murata, K.; Mikami, B. Structure and Function of a Hypothetical Pseudomonas aeruginosa Protein PA1167 Classified into Family PL-7: a novel alginate lyase with a beta-sandwich fold. J. Biol. Chem. 2004, 279, 31863–31872. [Google Scholar] [CrossRef]
- Zhu, B.; Ni, F.; Sun, Y.; Ning, L.; Yao, Z. Elucidation of degrading pattern and substrate recognition of a novel bifunctional alginate lyase from Flammeovirga sp. NJ-04 and its use for preparation alginate oligosaccharides. Biotechnol, Biofuels. 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Yang, M.; Yu, Y.; Yang, S.; Shi, X.; Mou, H.; Li, L. Expression and Characterization of a New PolyG-Specific Alginate Lyase From Marine Bacterium Microbulbifer sp. Q7. Front. Microbiol. 2018, 9, 1–13. [Google Scholar] [CrossRef]
- Kasana, R.C.; Pandey, CB. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit. Rev. Biotechnol. 2017, 38, 141–156. [Google Scholar] [CrossRef]
- López, M.C.; Galán, B.; Carmona, M.; Llorens, J.M.N.; Peretó, J.; Porcar, M.; et al. Xerotolerance: A New Property in Exiguobacterium Genus. Microorganisms. 2021. 9, 2455. [CrossRef]
- Okeke, B.C. Bioremoval of hexavalent chromium from water by a salt tolerant bacterium Exiguobacterium sp. GS1. J. Ind. Microbiol. Biotechnol. 2008, 35, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Dhanve, R.S.; Kalyani, D.C.; Phugare S,S. ; Jadhav, J.P. Coordinate action of Exiguobacterial oxidoreductive enzymes in biodegradation of reactive yellow 84A dye. Biodegradation. 2009, 20, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Guo, J.; Yang, Q.; Huang, J. Diluted conventional media improve the microbial cultivability from aquarium seawater. J. Microbiol. 2019, 57, 759–768. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Luan,M. ; Han, N.; Wang, T.; Zhao, X.; Yao, Y. Construction and Analysis of Food-Grade Lactobacillus kefiranofaciens β-Galactosidase Overexpression System. J. Microbiol. Biotechnol. 2021, 31, 550–558. [Google Scholar] [CrossRef]
- He, X.; Han, N.; Wang. Y.P. Cloning, Purification, and Characterization of a Heterodimeric β-Galactosidase from Lactobacillus kefiranofaciens ZW3[J]. J. Microbiol. Biotechnol. 2016, 26, 20–27. [Google Scholar] [CrossRef]
- Chi, F.C.; Kulkarni, S.S.; Zulueta, M.M.; Hung, S.C. Synthesis of Alginate Oligosaccharides Containing L-Guluronic Acids. Chem. Asian. J. 2009.12, 386–390. [CrossRef]
- Wang, S.; Dhulappa, A.; Quadri, S.R.; Jin, P.; Wang, K.; Qiao, H. Reclassification of Some Exiguobacterium Species Based on Genome Analysis. Curr. Microbiol. 2024, 81, 186. [Google Scholar] [CrossRef]
- Mohapatra, B.R. Kinetic and thermodynamic properties of alginate lyase and cellulose co-produced by Exiguobacterium species Alg-S5. Int. J. Biol. Macromol. 2017, 98, 103–110. [Google Scholar] [CrossRef]
- Yue, M.M.; Gong, W.W.; Qiao, Y.; Ding, H. A method for efficient expression of Pseudomonas aeruginosa alginate lyase in Pichia pastoris. Prep. Biochem. Biotechnol. 2016, 46, 165–170. [Google Scholar] [CrossRef]
- Gu, Z.; Niu, F.; Yu, Z.; Bao, Z.; Mukhtar, H.; Yang, P.; et al. High-efficiency expression of alginate lyase in Pichia pastoris facilitated by Vitreoscilla hemoglobin. Int. J. Biol. Macromol. 2024, 282(Pt 2), 137027. [Google Scholar] [CrossRef]
- Liang, Q.; Huang, Y.; Liu, Z.; Xiao, M.; Ren, X.; Liu, T.; et al. A Recombinant Alginate Lyase Algt1 with Potential in Preparing Alginate Oligosaccharides at High-Concentration Substrate. Foods. 2023, 12, 4039. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Liu, Z.; Fu, X.; Zhu, C.; Kong, Q.; Yang, M.; et al. Expression and Characterization of an Alginate Lyase and Its Thermostable Mutant in Pichia pastoris. Mar. Drugs. 2020, 18, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wang, S.; Zhang, Y.; Chen, L. High-Level Expression of a Thermally Stable Alginate Lyase Using Pichia pastoris, Characterization and Application in Producing Brown Alginate Oligosaccharide. Mar. Drugs. 2018, 16, 158–174. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Primer sequences (5’→3’) |
|---|---|
| Ebalg660 F | GGATCCATGAAACGAATCTTACTCGTCCTCG |
| Ebalg660 R | CTCGAGTTAGATGGGCACACTGATCTGTCT |
| Ebalg664 F | GGATCCATGAAACCTTTGTATACACGATTATCG |
| Ebalg664 R | CTCGAGTCATTTTGGTACGTTGATCGTATTT |
| Ebalg665 F | GGATCCATGAGGGAACGAATGGAACATT |
| Ebalg665 R | CTCGAGTCAATATGGAATTCGGATGACG |
| 664(Pichia) F | GAATTCATGAAACCTTTGTATACACGATTATCC |
| 664(Pichia) R | CTCGACTCATTTTGGTACGTTGATCGTATTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




