Introduction
The Vps9 domain demonstrates catalytic activity towards Rab proteins converting them from their inactive state to active state by exchanging GDP for GTP. Specifically, Vps9 domain proteins are commonly known as Rab5 guanine nucleotide exchange factors (GEF). Rab5 is a crucial regulatory component of endocytic pathways through early endosome formation. After the discovery of Rabex-5, the first named Rab5 GEF, it was suggested that any protein that contained the biochemical characteristic of the Vps9 domain may also function as a GEF towards Rab5 [
1]. However, emerging discoveries uncovered the diverse role of Vps9 domain containing proteins linking the endocytic pathway and cell signaling cascades to biological diseases. This functionality stems from the presence of other signaling domains in Vps9 domain containing proteins.
Multidomain proteins have a vast range of functions and roles in biological processes due to the domains acting independent of each other. The domain architecture of Vps9 proteins is a novel topic that continues to reveal its intricate functional and structural plasticity. Emerging discoveries have unveiled unique structural properties such as self-oligomerization, retromer complex formation, and ubiquitin binding. In 2022, the first Vps9 domain GEF and SNX-Bar coat complex (VINE) was discovered [
2], further revealing their involvement in tubular endosomal sorting. From vesicle trafficking to receptor signaling, these proteins exert their influence across a broad spectrum of cellular activities. Moreover, their significance extends beyond normal cellular function, as mounting evidence links dysregulation of Vps9 domain proteins to various human diseases, including neurodegenerative disorders and cancer [
3,
4,
5,
6]. The review also discusses the structural and functional properties of lesser-known Vps9 proteins like VPS9D1. VPS9D1 is unique in its specific GEF activity towards Rab22, rather than Rab5, and plays a role in tubular endosome formation. Additionally, the unknown roles of the coil-coil and MIT domain in VPS9D1 and their specific interactions with other cellular components warrant further investigation. This review aims to shed light on Vps9 proteins, in hopes to provide a better understanding of their multi-functional role in the signaling pathways of biological diseases.
The Vps9 Family Domain Architecture
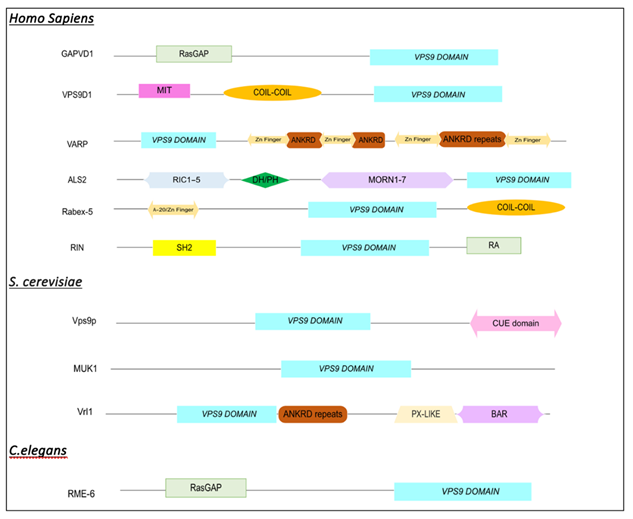
The Vps9 domain family contains 10 proteins that share unique combinations of signaling domains (
Table 1). Vps9 Domain Containing proteins are highly conserved amongst eukaryotes
, Saccharomyces cerevisiae, and Caenorhabditis elegans [
7]. Rabex-5 and its yeast ortholog, Vps9p, both demonstrate ubiquitin activity but contain distinct ubiquitin binding protein (UIB) domains. Rabex-5 contains an A-20 Zinc finger in its N-terminus whereas, Vps9p has a CUE domain in its C-terminus. ANKRD27 contains a conserved four-cysteine motif which is referred to as Zn-fingernails [
8]. This Zn-finger domain is unique to VARP and is not apparent in its yeast homolog, Vrl1
. Another highly structurally diverse Vps9 protein is ALS2. It contains a Pleckstrin homology (PH) domain, regulators of chromosome condensation (RCC1) repeats, a
membrane occupation and recognition nexus (MORN) repeat, a Vps9 domain, and a Rho GEF domain (DH) [
9]
. The highly conserved Ras and Rab interactor family (RIN1, RIN2, RIN3, and RINL) are composed of five domains: Src homology 2 domain (SH2), proline-rich domain, RIN-homology domain, Vps9-domain, and Ras-association domain (RA) [
10]. The RIN family consists of four Vps9-domain containing proteins with the only difference in their structure is the number of proline-rich domains it contains, which symbolizes the number after RIN.
Vps9 Domain Structural Insights
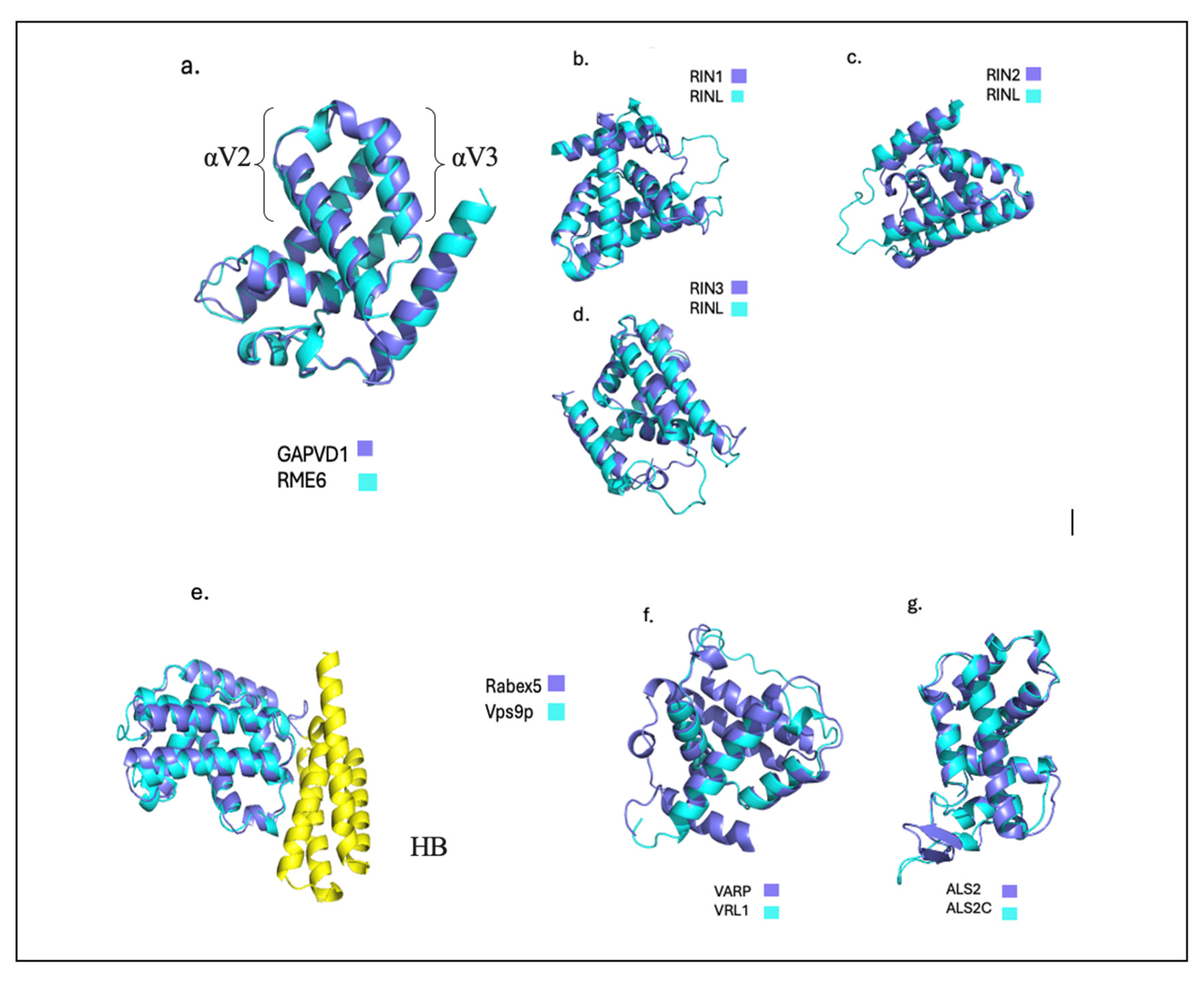
The catalytic core of the Vps9 domain is composed of an N-terminal helical bundle (HB) and a ~140 residue Vps9 domain. The Vps9 domain contains six
α helical folds (αV) and a C-terminal helix. The HB-Vps9 core of Vps9 domain containing proteins is highly specific to Rab proteins. Rabex-5 is the most studied Vps9 domain protein that is also present in yeast as Vps9p. Rabex-5 and its yeast ortholog both function as a guanine nucleotide exchange factor (GEF). The HB-Vps9 domain is commonly termed the catalytic core of Rabex-5 because it has stronger GEF activity towards Rab-5 than its full-length protein [
11]. ALS2CL, is a novel homolog to the C-terminal region of ALS2. Interestingly, ALS2CL demonstrates weaker Rab5 GEF activity when compared to its full-length homolog [
12]. Another notable Vps9 protein is highly conserved Ras and Rab interactor family (RIN). In
Figure 1, previously mentioned proteins and their ortholog pair are displayed to further provide a visual representation. The superimposed structures reveal structural conservation in the helical hairpin formed by
αV2 and αV3. Additionally, VARP and Vrl1 demonstrate the greatest structural variation which seems to be a result of unfolding in the Vps9 domain of Vrl1 (
Figure 1f).
UBIQUITIN Activity of VPS9 Domain Family
Rabex-5
Various signaling domains have been discovered alongside Vps9 domain containing proteins. Specifically, the presence of ubiquitin binding domains (UBD) in Vps9 domain containing proteins suggests an underlying functional interconnection between these two domains. There are three independent instances of acquisition of structurally diverse UBDs: the ZnF in mammalian Rabex5, the CUE domain in yeast, and a UBD in the amoebozoan
Sexangularia sp. [
8]. Rabex-5 forms a ternary complex with Rab5 and its effector, Rabaptin-5, that promotes early endosome formation. The process of this highly dynamic complex formation and Rabex-5 ubiquitination has presented challenges in understanding the functional mechanism. However, advances in modeling techniques and biotechnology have uncovered an emerging role of the ubiquitin binding domain in mediating this Rabex-5: Rabaptin GEF activity [
13,
14]. A recent study used hydrogen deuterium exchange mass spectrometry a method that allows for structural insights in highly dynamic proteins such as Rabex-5 [
13]. It was found that Rabex-5 UBD mutants created destabilization in residues that are part of the Rab5 binding site within the Vps9 domain. This discovery was the first to implicate an additional and potential role of the ubiquitin binding domain modulating Rabex-5 GEF activity. This reinforces the conserved function between ubiquitin binding domains and the Vps9 domain.
GAPVD1
GAPVD1 is a regulator of protein trafficking that acts both as a Rab31 guanine exchange factor (GEF) and a Ras GTPase activating protein (GAP). GAPVD1 is also a substrate for casein kinase 1 delta (CSNK1D) in vitro and in vivo, with 38 phosphorylation sites located in GAPVD1’s unstructured region [
15]. CSNK1D are ubiquitously expressed serine/threonine kinases that regulate multiple cellular processes including endocytosis [
16]. Additionally, GAPVD1 has been linked to EGFR degradation by binding c-Cbl, a ubiquitin E3 ligase [
17]. It was postulated that GAPVD1 degradation of EGFR could be linked to Rab 31 or Rab5 proteins. However, a phosphomimic mutant of GAPVD1 was found to rescue EGF internalization in GAPVD1-/- knockout cell lines [
18] leaving the possibility of phosphorylation-mediated EGF degradation. Phosphorylation may facilitate ubiquitylation of substrates, by regulating the process by which receptors and adaptors recognize substrates [
18]. Thus, the role of GAPVD1 in EGFR degradation may be linked to phosphorylation by CSNK1D or other yet unknown kinases present.
VPS9 Proteins in Biological Diseases
RIN Family
RIN1 has been shown to act as an oncogene in colorectal cancer [
19], non-small cell lung cancer [
20], and gastric adenocarcinoma [
21] but as a tumor suppressor gene in breast cancer [
22]. The mechanisms of the relationship of Rin1 in cancer are still being studied. The most studied mechanistic explanation of RIN1’s tumor suppression activity is through its inactivation of Ras further affecting the RAF/MEK/ERK pathway which is frequently activated in breast tumors [
5]. Another common theme amongst the Vps9 family, specifically the RIN family, is their presence in neurological signaling pathways.
It is postulated that RIN1 acts as an inhibitory modulator of neuronal plasticity through RAS-mediated pathways [
23,
24]. In a recent study conducted, increased RIN1 expression inhibited long-term potentiation in the amygdala by causing EphA4 internalization in rats [
23]. EphA4 regulates neuron proliferation in the hippocampus. Interestingly, a study uncovered similar results with its Vps9 family member, Ras and Rab interactor- like (RINL) [
25].
RINL has the same domain architecture except the RA domain is absent. RINL was found to downregulate EphA8 by forming a ternary complex with ANKS1A [
25]. This suggests an alternative mechanism involved in the RIN1 induced inhibition of neuronal plasticity that is not Ras dependent as the RA domain is absent in RINL.
RIN3 has received attention as a potential key regulator in Alzheimer’s Diseases. Genome wide association studies (GWAS) have presented increasing evidence of the multi-faceted presence of RIN3 in Alzheimer’s pathogenesis [
26]. A case-control study conducted whole-exome sequencing on 93 patients with early-onset Alzheimer’s (EOAD) and discovered a RIN3 missense variant on residue W34N [
6]. Assitionally, through western blot analysis, it was concluded that RIN3 recruits the BIN1 neuronal isoform, BINV1, to early endosomes further preventing Amyloid β processing by APP and BACE1 [
27]. Intrestingly, disruption of retromer complexes and sorting nexins have also been discovered play a role in in the intracellular transport of APP and APP-cleavage secretases (such as BACE1) [
28]. There may be a posssibility that Rin3 and BINV1 forms a retromer complex as this has been seen in other Vps9 protein such as VARP and Vrl1 [
2,
29].
RME-6
RME-6 is the C.elegans homolog of GAPVD1 demonstrating RasGAP and VPS9 domain conservation.
The C.elegans ortholog role acts as a model organism in studying the role of its Vps9 and RasGAP domain in clathirin dependent endocytosis of Alzheimer’s disease.
Knockdown of RME-6 displays a decrease in the endocytosis of the amyloid precursor protein (APP) [
30]
. RME-6 interacts with the PAT1a, the YTSI binding protein, on the tail of amyloid precursor protein that generates
β-amyloid. Considering, RIN3 is also seen in APP production and RIN1 is involved in neuronal growth, its evident the Vps9 domain has an underlying role in the pathogensis of the neuronal system.
ALS2
ALS2 is an intensively studied Vps9 protein linked to autosomal recessive motor neuron diseases such as amyotrophic lateral sclerosis, juvenile-onset primary lateral sclerosis, and spastic paralysis [
31]. ALS2 is also broadly classified as a Rab17/Rab5 GEF, and Rho GEF [
32]. Its multi-functionality stems from its domain architecture that allows it to function in membrane trafficking, axonal growth, endocytosis, and autophagy. ALS2 is committed in a wide range of membrane dynamics. Dysfunctional ALS2 proteins result in motor neuron dysfunction and degeneration that is present in ALS2-linked neurological diseases [
33].
Several in vivo and in vitro studies have discovered the self-oligomerization of ALS2 [
4,
34]. Additionally, studies have found evidence that the self-oligomerization of intrinsically related protein provides stability to ALS related proteins [
35]. ALS2CL and knockout domains of ALS2 have provided evidence that the MORN repeats and Vps9 domain, in its C-terminal half, are both required for Rab5 GEF activity [
34,
36]. The weak Rab5 GEF activity demonstrated by ALS2CL may be explained by a loss of stability in the MORN and Vps9 domain that occurs from an inability to self-oligomerize. ALS2 self-oligomerization serves the possibility to play an important role in protein aggregation and downstream endosome dysfunctions [
35]. To our knowledge, RIN2, VARP, and Rabex-5 are the only other known proteins, in the Vps9 family, that are involved in a complex formation [
10]. The increasing advances of
in-silico computational techniques may provide the tools needed to determine the possibility of other Vps9 proteins forming dimers. The dimerization of Vps9 proteins is a topic that has yet to receive attention and may provide insight into the unanswered questions of a third protein being involved in their Rab GEF activity.
Tubular Endosomal Sorting
VARP and VRL1
VARP (also known as ANKRD27) is a widely expressed Vps9-domain ankyrin repeat protein in most species excluding C. Elegans and D. Melanogaster. VARP and Vrl1 (
Figure 1f) are the only known Vps9 protein that contains two ankyrin repeats (ANKR1 and ANKR2). VARP’s multidomain structure has shown to function as a GEF to Rab proteins (Rab 21, Rab 32, and Rab 38) [
29], the retromer complex [
2], and negative regulation of the VAMP7- mediated snare complex [
37] There is uncertainty in the mechanisms of how Vps9 proteins or other proteins become localized to endosomes and then recycled to plasma membrane and then recycled to endosomes. Recent studies have found that this is made possible through a retromer-dependent pathways [
33,
38].
A recent discovery has linked Vrl1 to form a SNX- BAR coat termed the “VINE” complex [
2]. The VINE complex is the first described SNX-BAR complex with Vps9 domain GEF activity. The mechanism of the VINE complex involves Vin1 recruiting Vrl1 to the endosome membrane through recognition of
an interface between the VPS9 domain and AnkRD. Interestingly, this binding interface is conserved in the Saccharomycetaceae [
2]
family suggesting a vital function in the residues between the AnkRD and Vps9 domain. Additionally, conserved residues in the Vps9 domains of yeast Vrl1, Muk1, and Vps9 have been found to be essential in the production of endosomal PI3P and retromer recruitment [
39].
VPS9D1
VPS9D1 is commonly expressed in mice, yeast, and humans. Our understanding of this protein is still evolving
which suggests a need to further explore its unique structure. Its structure is composed of three helixes in the N-terminus and the Vps9 domain in the C-terminus. In the N-terminus a coil-coil domain is present and that’s function is unknown. The only published study that provides insight into its coil-coil domain was conducted in 1999 [
40] demonstrated its coil-coil domain having similar homology to the B subunit of ATP synthase. This discovery is where its alternative name, ATPBL, stems from. Additionally, VPS9D1 is the first discovered Vps9 domain containing protein that acts as a Rab22 specific GEF
but not Rab5 [
41]. VPS9D1 plays a role in the tubular endosome formation of HeLa Cells.
In a recent study, a conserved residue in the Vps9 domain was mutated to Y628A to further study Vps9d1 specific Rab22 activity [
41]. As expected, the mutant on the Vps9 domain was unable to cause enlargement of Rab22A/B positive endosomes.
Most Rab5 binding proteins have been found to have greater GEF activity with just the Vps9 domain alone. However, the C-terminus of Vps9d1 failed to induce enlargement of Rab22A/B endosomes in vitro [
41]
. This suggests the N-terminus is required for Rab22A/B GEF activity. It may be possible Vps9d1 GEF Rab22 activity requires an additional binding partner to the N-terminus of Vps9d1 to localize it to EEA1.
Further Vps9d1 Structural Discussion
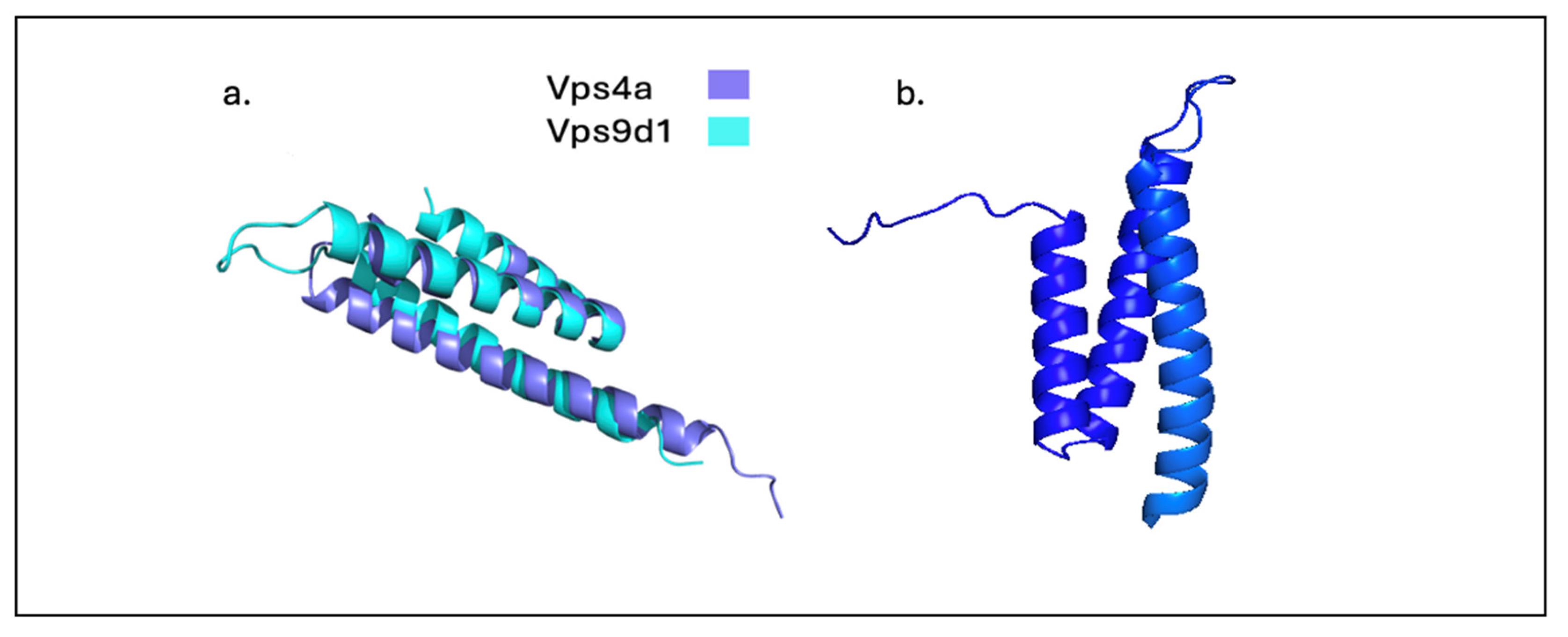
A study on the novel MIT domain briefly mentioned an “uncharacterized protein” containing a Vps9 domain derived from the gene C16orf7 [
42]. To rule out the possibility of structurally related domains producing false positives, the probability of MIT domain presence was computed along with protein-protein profiling. The results demonstrated high MIT presence (probability 72; e-value 4.8× 10−4) in uncharacterized Vps9 domain protein [
42]. Additionally [
43], classified Vps9d1 as a MIT domain containing protein but received no further experimentation due to its inability to interact with ESCRT-III proteins. For the first time, we provide a visual presentation of the MIT domain in VPS9d1 in hopes to shed light on this novel protein (
Figure 2). With the use of AlphaFold3 we superimposed model of the MIT domain of Vps4a, a well characterized protein of the MIT domain family, and its domain resemblance in Vps9d1.
Future Prospects
Emerging discoveries in Vps9 domain proteins have revealed their multifaceted roles beyond their identification as Rab5 GEFs. These proteins are now known to be involved in ubiquitin binding, retromer complex formation, and various cellular processes, including endocytic trafficking and neuronal signaling. The RIN family, particularly RIN3, and RME-6 have been implicated in neurological diseases such as Alzheimer’s, highlighting the clinical significance of these proteins. Future research should focus on elucidating the precise molecular mechanisms by which Vps9 domain proteins regulate these diverse functions. Advances in computational techniques are expected to provide deeper insights into the dynamic properties and potential dimerization of Vps9 proteins. Additionally, the unique roles of lesser-known Vps9 proteins like VPS9D1 and their specific interactions with other cellular components warrant further investigation. Specifically, the presence of a coil-coil and MIT domain in VPS9D1 suggests additional regulatory mechanisms yet to be fully understood. The potential therapeutic implications of targeting Vps9 domain proteins in diseases such as Alzheimer’s and cancer are promising. Understanding the interplay between Vps9 proteins and other cellular pathways could lead to the development of novel therapeutic strategies. As research progresses, the integration of structural biology, computational modeling, and functional studies will be crucial in uncovering the full spectrum of Vps9 protein functions and their impact on human health. Overall, the continued exploration of Vps9 domain proteins holds significant promise for advancing our knowledge of cellular regulation and developing new treatments for a range of diseases.
References
- Esters, H., et al., Vps9, Rabex-5 and DSS4: proteins with weak but distinct nucleotide-exchange activities for Rab proteins. J Mol Biol, 2001. 310(1): p. 141-56. [CrossRef]
- Shortill, S.P., et al., The VINE complex is an endosomal VPS9-domain GEF and SNX-BAR coat. Elife, 2022. 11. [CrossRef]
- Rocca, A., et al., The Predictive and Prognostic Role of RAS-RAF-MEK-ERK Pathway Alterations in Breast Cancer: Revision of the Literature and Comparison with the Analysis of Cancer Genomic Datasets. Cancers (Basel), 2022. 14(21). [CrossRef]
- Sato, K., et al., Altered oligomeric states in pathogenic ALS2 variants associated with juvenile motor neuron diseases cause loss of ALS2-mediated endosomal function. Journal of Biological Chemistry, 2018. 293(44): p. 17135-17153. [CrossRef]
- Milstein, M., et al., RIN1 is a breast tumor suppressor gene. Cancer Res, 2007. 67(24): p. 11510-6.
- Sherva, R., et al., Identification of Novel Candidate Genes for Alzheimer’s Disease by Autozygosity Mapping using Genome Wide SNP Data. Journal of Alzheimer’s Disease, 2011. 23(2): p. 349-359. [CrossRef]
- Crawley-Snowdon, H., et al., Mechanism and evolution of the Zn-fingernail required for interaction of VARP with VPS29. Nat Commun, 2020. 11(1): p. 5031. [CrossRef]
- Herman, E.K., et al., Regulation of early endosomes across eukaryotes: Evolution and functional homology of Vps9 proteins. Traffic, 2018. 19(7): p. 546-563. [CrossRef]
- The Rab5 activator ALS2/alsin acts as a novel Rac1 effector through Rac1-activated endocytosis—PubMed. The Journal of biological chemistry, 06/01/2007. 282(22).
- Saito, K., et al., Purification and analysis of RIN family-novel Rab5 GEFs. Methods Enzymol, 2005. 403: p. 276-83.
- Lauer, J., et al., Auto-regulation of Rab5 GEF activity in Rabex5 by allosteric structural changes, catalytic core dynamics and ubiquitin binding. Elife, 2019. 8.
- Hadano, S., et al., ALS2CL, the novel protein highly homologous to the carboxy-terminal half of ALS2, binds to Rab5 and modulates endosome dynamics. FEBS Lett, 2004. 575(1-3): p. 64-70.
- Lauer, J., et al., Auto-regulation of Rab5 GEF activity in Rabex5 by allosteric structural changes, catalytic core dynamics and ubiquitin binding. eLife, 2019. 8: p. e46302.
- Mattera, R. and J.S. Bonifacino, Ubiquitin binding and conjugation regulate the recruitment of Rabex-5 to early endosomes. Embo j, 2008. 27(19): p. 2484-94. [CrossRef]
- Su, X., C. Kong, and P.D. Stahl, GAPex-5 mediates ubiquitination, trafficking, and degradation of epidermal growth factor receptor. J Biol Chem, 2007. 282(29): p. 21278-84. [CrossRef]
- Guillen, R.X., et al., CRISPR-mediated gene targeting of CK1δ/ε leads to enhanced understanding of their role in endocytosis via phosphoregulation of GAPVD1. Sci Rep, 2020. 10(1): p. 6797. [CrossRef]
- Ibrahim, H., et al., Phosphorylation of GAPVD1 Is Regulated by the PER Complex and Linked to GAPVD1 Degradation. Int J Mol Sci, 2021. 22(7). [CrossRef]
- Chen, Y., et al., Phosphorylation regulates cullin-based ubiquitination in tumorigenesis. Acta Pharm Sin B, 2021. 11(2): p. 309-321. [CrossRef]
- Galvis, A., et al., Inhibition of early endosome fusion by Rab5-binding defective Ras interference 1 mutants. Arch Biochem Biophys, 2009. 482(1-2): p. 83-95. [CrossRef]
- Senda, K., et al., Analysis of RIN1 gene expression in colorectal cancer. Oncol Rep, 2007. 17(5): p. 1171-1175. [CrossRef]
- Wang, Q., et al., Prognostic significance of RIN1 gene expression in human non-small cell lung cancer. Acta Histochem, 2012. 114(5): p. 463-8. [CrossRef]
- Yu, H.F., et al., High RIN1 expression is associated with poor prognosis in patients with gastric adenocarcinoma. Tumour Biol, 2012. 33(5): p. 1557-63. [CrossRef]
- Deininger, K., et al., The Rab5 guanylate exchange factor Rin1 regulates endocytosis of the EphA4 receptor in mature excitatory neurons. Proceedings of the National Academy of Sciences, 2008. 105(34): p. 12539-12544. [CrossRef]
- Han, F., et al., Change of Rin1 and Stathmin in the Animal Model of Traumatic Stresses. Front Behav Neurosci, 2017. 11: p. 62. [CrossRef]
- Kajiho, H., et al., RINL, guanine nucleotide exchange factor Rab5-subfamily, is involved in the EphA8-degradation pathway with odin. PLoS One, 2012. 7(1): p. e30575. [CrossRef]
- Anon, Early-Onset Alzheimer Disease and Candidate Risk Genes Involved in Endolysosomal Transport—PubMed. JAMA neurology, 09/01/2017. 74(9).
- Bhattacharyya, R., et al., The neuronal-specific isoform of BIN1 regulates β-secretase cleavage of APP and Aβ generation in a RIN3-dependent manner. Scientific Reports, 2022. 12(1). [CrossRef]
- Zhang, H., et al., The Retromer Complex and Sorting Nexins in Neurodegenerative Diseases. Frontiers in Aging Neuroscience, 2018. 10. [CrossRef]
- Fukuda, M., Multiple Roles of VARP in Endosomal Trafficking: Rabs, Retromer Components and R-SNARE VAMP7 Meet on VARP. Traffic, 2016/07/01. 17(7). [CrossRef]
- Eggert, S., et al., The Rab5 activator RME-6 is required for amyloid precursor protein endocytosis depending on the YTSI motif. Cellular and Molecular Life Sciences: CMLS, 2020/12. 77(24). [CrossRef]
- Miceli, M., et al., ALS2-Related Motor Neuron Diseases: From Symptoms to Molecules. Biology (Basel), 2022. 11(1). [CrossRef]
- Shimakura, K., et al., The N-terminal intrinsically disordered region mediates intracellular localization and self-oligomerization of ALS2. Biochemical and Biophysical Research Communications, 2021. 569: p. 106-111. [CrossRef]
- Piotrowski, J.T., et al., WASH Knockout T Cells Demonstrate Defective Receptor Trafficking, Proliferation, and Effector Function. Molecular and Cellular Biology, 2013/03. 33(5). [CrossRef]
- Kunita, R., et al., Homo-oligomerization of ALS2 through Its Unique Carboxyl-terminal Regions Is Essential for the ALS2-associated Rab5 Guanine Nucleotide Exchange Activity and Its Regulatory Function on Endosome Trafficking*. Journal of Biological Chemistry, 2004. 279(37): p. 38626-38635. [CrossRef]
- Asakawa, K., H. Handa, and K. Kawakami, Optogenetic modulation of TDP-43 oligomerization accelerates ALS-related pathologies in the spinal motor neurons. Nature Communications, 2020. 11(1): p. 1004. [CrossRef]
- Hadano, S., et al., Molecular and cellular function of ALS2/alsin: Implication of membrane dynamics in neuronal development and degeneration. Neurochemistry International, 2007. 51(2): p. 74-84.
- Kent, M., Helen, et al., Structural Basis of the Intracellular Sorting of the SNARE VAMP7 by the AP3 Adaptor Complex. Developmental Cell, 2012. 22(5): p. 979-988.
- Helfer, E., et al., Endosomal recruitment of the WASH complex: Active sequences and mutations impairing interaction with the retromer. Biology of the Cell, 2013. 105(5): p. 191-207. [CrossRef]
- Bean, B.D.M., et al., Rab5-family guanine nucleotide exchange factors bind retromer and promote its recruitment to endosomes. Molecular Biology of the Cell, 2015. 26(6): p. 1119-1128. [CrossRef]
- Sugimoto, J., T. Hatakeyama, and M. Isobe, Isolation and mapping of a putative b subunit of human ATP synthase (ATP-BL) from human leukocytes. DNA Res, 1999. 6(1): p. 29-35.
- Nakashima, S., T. Matsui, and M. Fukuda, Vps9d1 regulates tubular endosome formation through specific activation of Rab22A. Journal of Cell Science, 2023. 136(6). [CrossRef]
- Rigden, D.J., et al., Ab initio protein modelling reveals novel human MIT domains. FEBS Lett, 2009. 583(5): p. 872-8. [CrossRef]
- Wenzel, D.M., et al., Comprehensive analysis of the human ESCRT-III-MIT domain interactome reveals new cofactors for cytokinetic abscission. Elife, 2022. 11. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).