Submitted:
03 December 2024
Posted:
04 December 2024
You are already at the latest version
Abstract
Keywords:
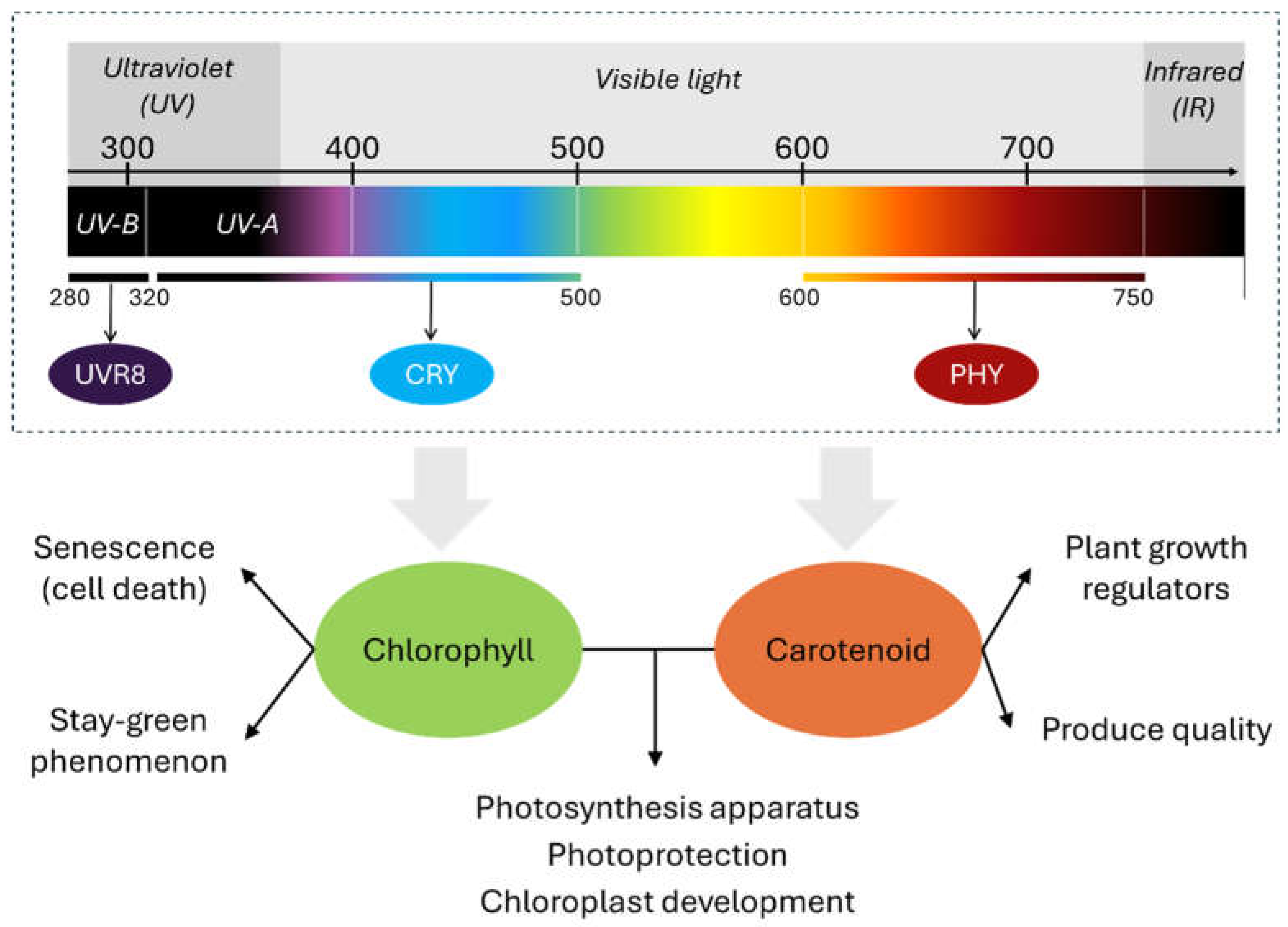
1. Introduction
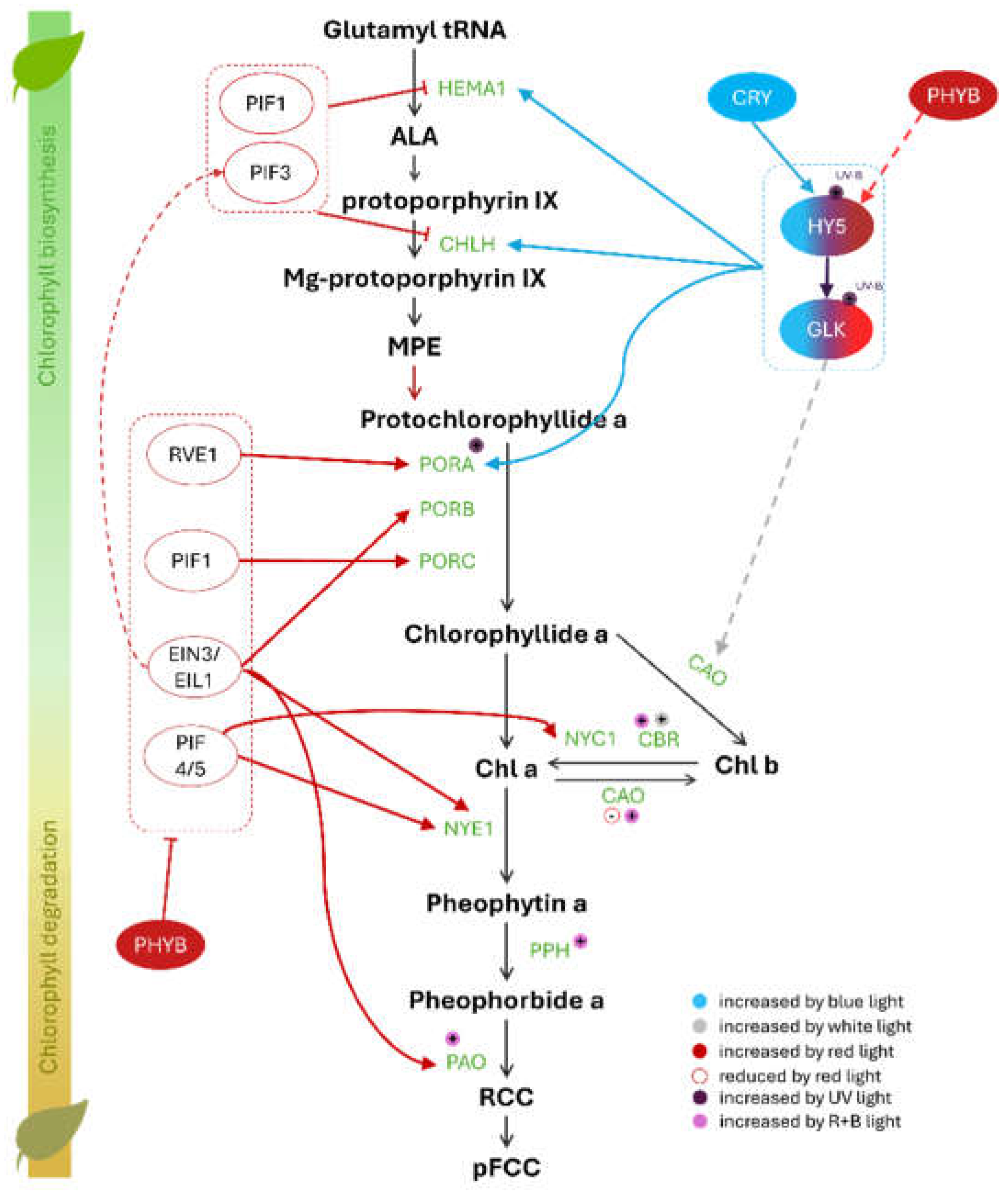
2. Chlorophyll Pathway Under Different Light Spectra
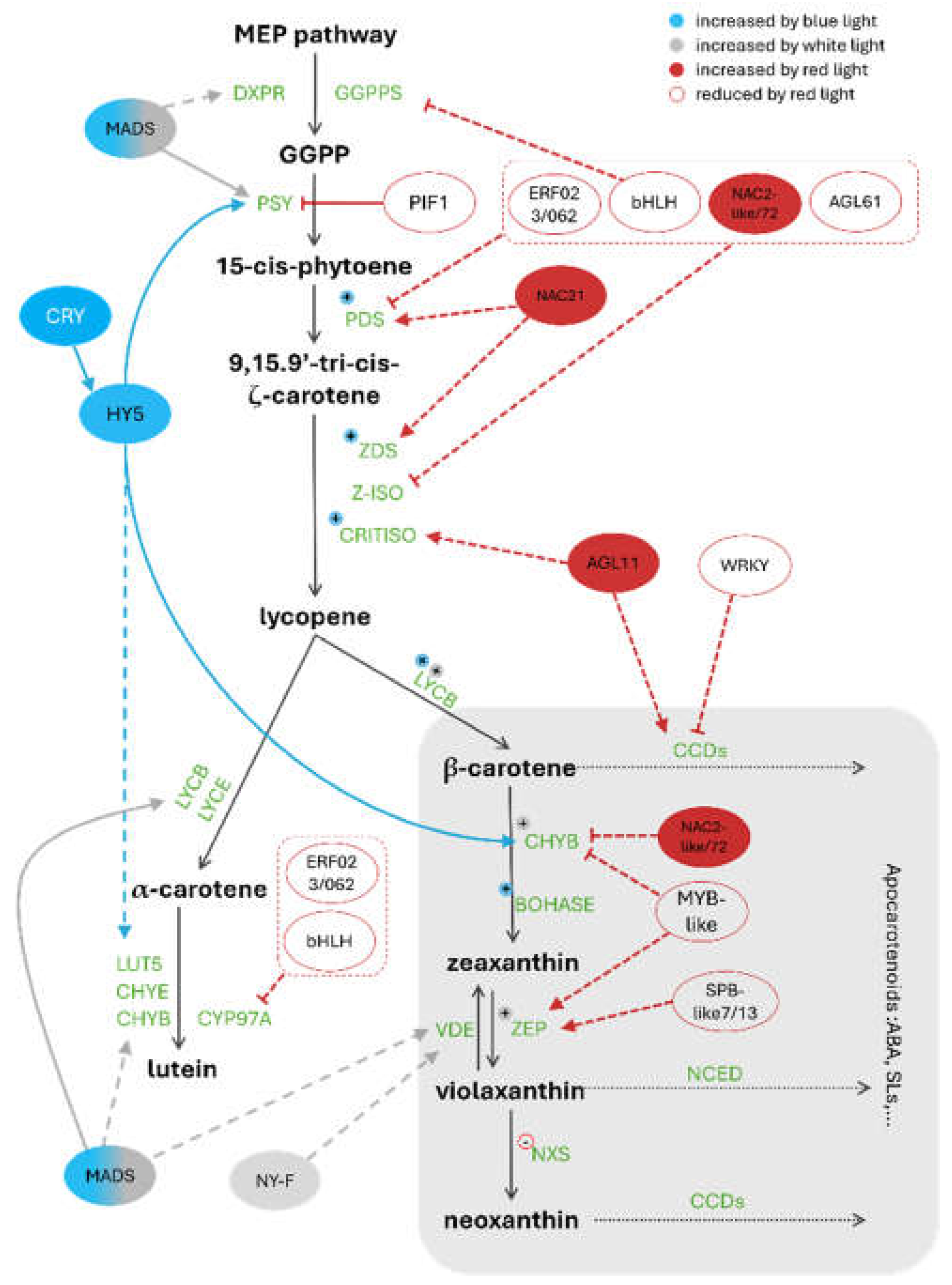
3. Carotenoid Pathway Under Different Light Spectra
4. Conclusions and Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tripathy, B.C.; Pattanayak, G.K. Chlorophyll Biosynthesis in Higher Plants. In; 2012; pp. 63–94.
- Reinbothe, C.; Bakkouri, M. El; Buhr, F.; Muraki, N.; Nomata, J.; Kurisu, G.; Fujita, Y.; Reinbothe, S. Chlorophyll Biosynthesis: Spotlight on Protochlorophyllide Reduction. Trends Plant Sci 2010, 15, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, J.; Qiu, K.; Kuai, B. Phytohormone and Light Regulation of Chlorophyll Degradation. Front Plant Sci 2017, 8. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chlorophyll Metabolism. Curr Opin Plant Biol 2006, 9, 248–255. [Google Scholar] [CrossRef]
- Quian-Ulloa, R.; Stange, C. Carotenoid Biosynthesis and Plastid Development in Plants: The Role of Light. International Journal of Molecular Sciences 2021, 22, 1184. [Google Scholar] [CrossRef] [PubMed]
- Nagarajan, J.; Ramanan, R.N.; Raghunandan, M.E.; Galanakis, C.M.; Krishnamurthy, N.P. Carotenoids. In Nutraceutical and Functional Food Components: Effects of Innovative Processing Techniques; Elsevier Inc., 2017; pp. 259–296. ISBN 9780128052570. [Google Scholar]
- Stanley, L.; Yuan, Y.-W. Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus. Front Plant Sci 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid Metabolism in Plants. Mol Plant 2015, 8, 68–82. [Google Scholar] [CrossRef]
- Kong, S.-G.; Okajima, K. Diverse Photoreceptors and Light Responses in Plants. J Plant Res 2016, 129, 111–114. [Google Scholar] [CrossRef] [PubMed]
- Eng, C.; Wei, Z.; Teo, N.; Shen, L.; Yu, H. Seeing the Lights for Leafy Greens in Indoor Vertical Farming. Trends Food Sci Technol 2020, 106, 48–63. [Google Scholar] [CrossRef]
- Huché-Thélier, L.; Crespel, L.; Gourrierec, J. Le; Morel, P.; Sakr, S.; Leduc, N. Light Signaling and Plant Responses to Blue and UV Radiations—Perspectives for Applications in Horticulture. Environ Exp Bot 2016, 121, 22–38. [Google Scholar] [CrossRef]
- Xiang, N.; Zhao, Y.; Wang, S.; Guo, X. The Modulation of Light Quality on Carotenoids in Maize (Zea Mays L.) Sprouts. Food Chemistry: Molecular Sciences 2022, 5. [Google Scholar] [CrossRef] [PubMed]
- Pizarro, L.; Stange, C. Light-Dependent Regulation of Carotenoid Biosynthesis in Plants. Cienc Investig Agrar 2009, 36. [Google Scholar] [CrossRef]
- Mohanty, B.; Lakshmanan, M.; Lim, S.-H.; Kim, J.K.; Ha, S.-H.; Lee, D.-Y. Light-Specific Transcriptional Regulation of the Accumulation of Carotenoids and Phenolic Compounds in Rice Leaves. Plant Signal Behav 2016, 11, e1184808. [Google Scholar] [CrossRef]
- Llorente, B.; Martinez-garcia, J.F.; Stange, C.; Rodriguez-concepcion, M. Illuminating Colors : Regulation of Carotenoid Biosynthesis and Accumulation by Light. Curr Opin Plant Biol 2017, 37, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Chu, L.; Zhang, Y.; Bian, Y.; Xiao, J.; Xu, D. HY5: A Pivotal Regulator of Light-Dependent Development in Higher Plants. Front Plant Sci 2022, 12. [Google Scholar] [CrossRef]
- Li, Y.; Shi, Y.; Li, M.; Fu, D.; Wu, S.; Li, J.; Gong, Z.; Liu, H.; Yang, S. The CRY2–COP1–HY5–BBX7/8 Module Regulates Blue Light-Dependent Cold Acclimation in Arabidopsis. Plant Cell 2021, 33, 3555–3573. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, R.; Zeng, X.-Y.; Lee, S.; Ye, L.-H.; Tian, S.-L.; Zhang, Y.-J.; Busch, W.; Zhou, W.-B.; Zhu, X.-G.; et al. GLK Transcription Factors Accompany ELONGATED HYPOCOTYL5 to Orchestrate Light-Induced Seedling Development in Arabidopsis. Plant Physiol 2024, 194, 2400–2421. [Google Scholar] [CrossRef] [PubMed]
- McCormac, A.C.; Terry, M.J. Light-signalling Pathways Leading to the Co-ordinated Expression of HEMA1 and Lhcb during Chloroplast Development in Arabidopsis Thaliana. The Plant Journal 2002, 32, 549–559. [Google Scholar] [CrossRef]
- Liu, X.; Yang, M.; Xie, X.; ABM, K.; ATAK, A.; Zhong, C.; Li, D. Effect of Light on Growth and Chlorophyll Development in Kiwifruit Ex Vitro and in Vitro. Sci Hortic 2022, 291, 110599. [Google Scholar] [CrossRef]
- Stephenson, P.G.; Fankhauser, C.; Terry, M.J. PIF3 Is a Repressor of Chloroplast Development. Proceedings of the National Academy of Sciences 2009, 106, 7654–7659. [Google Scholar] [CrossRef]
- Liu, X.; Li, Y.; Zhong, S. Interplay between Light and Plant Hormones in the Control of Arabidopsis Seedling Chlorophyll Biosynthesis. Front Plant Sci 2017, 8. [Google Scholar] [CrossRef]
- Jiang, Z.; Xu, G.; Jing, Y.; Tang, W.; Lin, R. Phytochrome B and REVEILLE1/2-Mediated Signalling Controls Seed Dormancy and Germination in Arabidopsis. Nat Commun 2016, 7, 12377. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Shen, X.; Liu, R.; Xue, C.; Wei, N.; Deng, X.W.; Zhong, S. The Red Light Receptor Phytochrome B Directly Enhances Substrate-E3 Ligase Interactions to Attenuate Ethylene Responses. Dev Cell 2016, 39, 597–610. [Google Scholar] [CrossRef]
- Zhong, S.; Zhao, M.; Shi, T.; Shi, H.; An, F.; Zhao, Q.; Guo, H. EIN3/EIL1 Cooperate with PIF1 to Prevent Photo-Oxidation and to Promote Greening of Arabidopsis Seedlings. Proceedings of the National Academy of Sciences 2009, 106, 21431–21436. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.-Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-Interacting Transcription Factors PIF4 and PIF5 Induce Leaf Senescence in Arabidopsis. Nat Commun 2014, 5, 4636. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; van Iersel, M.W. Photosynthetic Physiology of Blue, Green, and Red Light: Light Intensity Effects and Underlying Mechanisms. Front Plant Sci 2021, 12. [Google Scholar] [CrossRef]
- Smith, H.L.; McAusland, L.; Murchie, E.H. Don’t Ignore the Green Light: Exploring Diverse Roles in Plant Processes. J Exp Bot 2017, 68, 2099–2110. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cheng, X.; Cao, J.; Zhu, W.; Wan, X.; Liu, L. GOLDEN 2-LIKE Transcription Factors Regulate Chlorophyll Biosynthesis and Flavonoid Accumulation in Response to UV-B in Tea Plants. Hortic Plant J 2023, 9, 1055–1066. [Google Scholar] [CrossRef]
- Huang, X.; Hu, L.; Kong, W.; Yang, C.; Xi, W. Red Light-Transmittance Bagging Promotes Carotenoid Accumulation of Grapefruit during Ripening. Commun Biol 2022, 5, 1–10. [Google Scholar] [CrossRef]
- Toledo-Ortiz, G.; Huq, E.; Rodríguez-Concepción, M. Direct Regulation of Phytoene Synthase Gene Expression and Carotenoid Biosynthesis by Phytochrome-Interacting Factors. Proceedings of the National Academy of Sciences 2010, 107, 11626–11631. [Google Scholar] [CrossRef]
- XIE, B. xing; WEI, J. jing; ZHANG, Y. ting; SONG, S. wei; SU, W.; SUN, G. wen; HAO, Y. wei; LIU, H. cheng Supplemental Blue and Red Light Promote Lycopene Synthesis in Tomato Fruits. J Integr Agric 2019, 18, 590–598. [Google Scholar] [CrossRef]
- Fu, C.; Han, Y.; Kuang, J.; Chen, J.; Lu, W. Papaya CpEIN3a and CpNAC2 Co-Operatively Regulate Carotenoid Biosynthesis-Related Genes CpPDS2/4, CpLCY-e and CpCHY-b During Fruit Ripening. Plant Cell Physiol 2017, 58, 2155–2165. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wu, M.; Cao, L.; Yuan, W.; Dong, M.; Wang, X.; Chen, W.; Shang, F. Characterization of OfWRKY3, a Transcription Factor That Positively Regulates the Carotenoid Cleavage Dioxygenase Gene OfCCD4 in Osmanthus Fragrans. Plant Mol Biol 2016, 91, 485–496. [Google Scholar] [CrossRef]
- Quian-Ulloa, R.; Stange, C. Carotenoid Biosynthesis and Plastid Development in Plants: The Role of Light. Int J Mol Sci 2021, 22, 1184. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Ye, J.; Zhu, K.; Zhang, Y.; Zhang, M.; Xu, Q.; Deng, X. A Fruit Ripening-Associated Transcription Factor CsMADS5 Positively Regulates Carotenoid Biosynthesis in Citrus. J Exp Bot 2021, 72, 3028–3043. [Google Scholar] [CrossRef]
- Zhu, K.; Chen, H.; Mei, X.; Lu, S.; Xie, H.; Liu, J.; Chai, L.; Xu, Q.; Wurtzel, E.T.; Ye, J.; et al. Transcription Factor CsMADS3 Coordinately Regulates Chlorophyll and Carotenoid Pools in Citrus Hesperidium. Plant Physiol 2023, 193, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Ngamwonglumlert, L.; Devahastin, S.; Chiewchan, N.; Raghavan, V. Plant Carotenoids Evolution during Cultivation, Postharvest Storage, and Food Processing: A Review. Compr Rev Food Sci Food Saf 2020, 19, 1561–1604. [Google Scholar] [CrossRef]
- Wang, T.; Liu, S.; Tian, S.; Ma, T.; Wang, W. Light Regulates Chlorophyll Biosynthesis via ELIP1 during the Storage of Chinese Cabbage. Sci Rep 2022, 12, 11098. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



