2.1. Generation and Characterization of CD19 Knock-Out Lymphoma Cell Lines
To test our hypothesis that CD19 is important for CXCR4 signaling and overall survival of WM cells, we generated CD19 knock-out (KO) BCWM.1 cells by CRISPR-Cas9 methods (
Figure S1). In parallel, we also generated IgM KO BCWM.1 cells to investigate the influence of BCR on the survival of these cells. We adopted a two-step strategy that supports CRISPR-Cas9 engineering both through lentivirus (BSL2) and murine ecotropic γ-retrovirus (BSL1) based delivery of the targeting sgRNA (see methods). Supporting our hypothesis, cells transduced with the CD19 targeting sgRNA accompanying a GFP reporter were outcompeted by the untransduced cells and lost from the mixture between 7-15 days post transfection and prior to single cell sorting (
Figure S1A). We, therefore first sorted all GFP positive transduced cells in bulk, grow them for few days and then sorted as single cell for generating clones. The average numbers of growing clones were significantly lower for CD19 KO cells compared to WT cells (
Figure S1B). In case of IgM KO, the effect was even stronger (
Figure S1B). Upon collecting the growing CD19 KO BCWM.1 clones, we first confirmed the loss of CD19 expression in these cells by flow cytometry (
Figure S1C) and determined the mutation in the CD19 locus by sequencing (
Figure S1D). Compared to WT BCWM.1 cells, CD19 KO cells did not show any differences in the surface expression of IgM-BCR and CXCR4 (
Figure S1E) as well as in IgM secretion (
Figure S1F), suggesting no autoregulation and compromised receptor expressions in this model. Following the same method, we also generated CD19 KO of GCB-DLBCL derived DHL6 cells and characterized them (
Figure S1G-H).
2.2. CD19 Is Required for Growth and CXCL12 Induced Migration of Lymphoma Cells
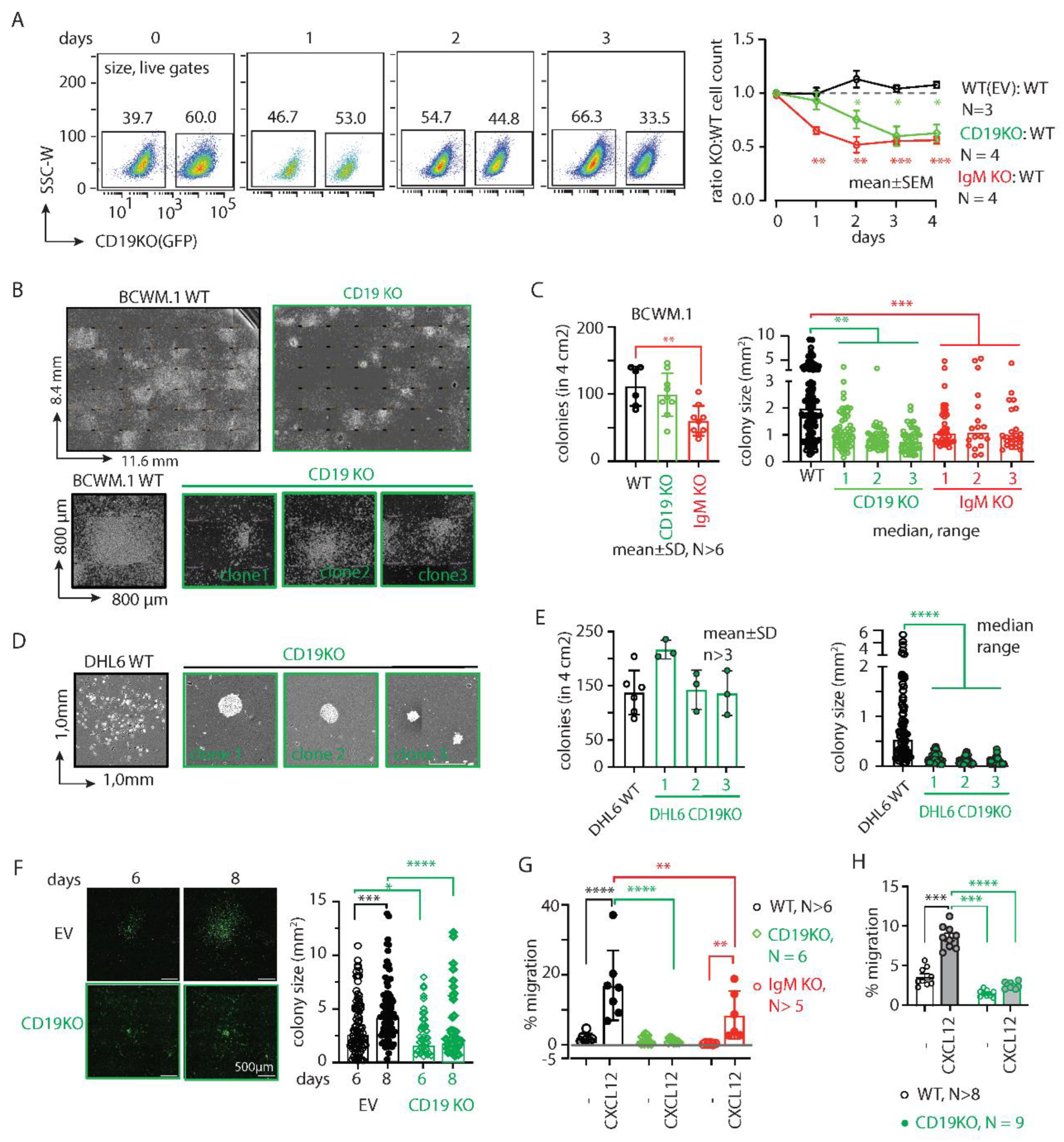
To investigate the survival competence of CD19 KO BCWM.1 cells in competition with the WT cells, we cocultured them at 1:1 ratio and measured cell growth by counting of the cell number by flow cytometry (
Figure S1I). Expectedly, the CD19 KO clones were slow growing and were ~40% lost (surviving fractions on day2-4: 75.5±16.9; 60.1±18.3 and 62.8±15.9) in competition to WT counterpart within 3-4 days of coculturing (
Figure 1A). IgM KO cells, on the other hand, were more rapidly declined to <60% (day2-4: 52.1±14.9; 55.5±6.7; 56.3±6.5) within 2 days (
Figure 1A), indicative to the stronger BCR signaling dependence of lymphoma cells. Overall, this data suggests that both CD19 and IgM play crucial role in survival and overall growth of BCWM.1 cells.
Next, we assessed the colony forming ability of these cells by culturing them in methylcellulose-based matrix and monitored the colony formation (
Figure 1B-C). Compared to WT BCWM.1 cells, CD19 KO clones were grown significantly slower and produced smaller sized colonies (
Figure 1B). While the number of colonies produced by the CD19 KO clones (mean±SD, 100±32) was unchanged compared to WT (103±29), significantly reduced numbers of colonies were produced by the IgM KO (59±22) BCWM.1 clones (
Figure 1C). This suggests a specific role of CD19 in cell proliferation and growth. In contrast, IgM KO cells failed to begin the colony formation due to severe survival disadvantage and produced significantly reduced numbers of colonies (
Figure 1C). Like WM, the loss of CD19 in GCB-DLBCL derived DHL6 cells also caused reduced colony size without changes in the number of colonies (
Figure 1D-E). Notably, we failed to generate IgM KO DHL6 cells suggesting an indispensable role of BCR signaling for survival of DLBCL cells [
12,
24]. Intriguingly, the CD19 KO colonies appeared condensed compared to WT colonies for both cell types, most prominently for DHL6 cells, suggesting a role of CD19 in intra-colony cell mobilization, spreading and subsequent growth of colony size.
We, therefore performed live cell imaging to monitor colony formation and spreading of BCWM.1 cells over time (
Figure 1F). Like CFC assay we plated CD19 KO BCWM.1 cells and empty vector (EV) transduced GFP positive control cells in methylcellulose-based matrix and monitored the cell growth every 24 hours by real-time imaging (see method). Between 6-8 days, the colonies became visible and spread around the proliferation center. As depicted in
Figure 1F, CD19 KO BCWM.1 cells failed to grow and spread resulting in smaller sized and condensed colonies compared EV transduced cells.
Next, we analyzed the CXCR4 response in CD19 KO cells by testing CXCL12 induced migration. Both BCWM.1 and DHL6 failed to migrate in response to CXCL12 upon loss of CD19 (
Figure 1G-H). IgM KO BCWM.1 cells, on the other hand, exhibited only partial reduction in CXCL12 induced migration suggesting differential role of CD19 and BCR on CXCR4 signaling (
Figure 1G). Notably, the rate of specific migration by WT DHL6 cells (no cytokine control, 3.3±1.5%; 60nM CXCL12, 9.7±1.5%) was much lower than BCWM.1 cells (no cytokine control, 2.4±2.0%; 60nM CXCL12, 19±8%) and below 10% which could not be improved by increasing the incubation period or dose of CXCL12 in the migration assay (
Figure 1H). This is in line with the general CXCR4 non-responsiveness of the DLBCL cells[
25]. Together, these data suggest a specific role of CD19 in cell growth, colony spreading and CXCL12 induced migration.
2.3. CD19 mAbs Increase Survival and CXCL12 Induced Migration of WM Cells
Knowing the essential role of CD19 in survival and CXCL12 induced migration of both BCWM.1 and DHL6 lymphoma cells, we tested whether treatment with anti-CD19 monoclonal antibodies (mAb) have any effect on these cells. To this end, we generated a humanized anti-CD19 monoclonal antibody (mAbo) by cloning the variable sequences from an anti-CD19 hybridoma, and we compared its effects with those of commercially available humanized anti-CD19 mAbs (mAb1 and mAb2) used in therapeutic and preclinical studies (see Methods). Notably, most therapeutic and preclinical mAbs from commercial sources are available only in limited quantities for research use, with minimal information and no options for experimental modification. Therefore, we sought to produce mAbo in HEK293T cells and purified it using HiTrap Protein G affinity chromatography (
Figure S2A-C). Upon determining the amount and purity by SDS-PAGE and ELISA (
Figure S2B-C), we tested the binding specificity of mAbo on human peripheral B cells and BCWM.1 cells compared to non-B cells and CD19 KO BCWM.1 cells, respectively (
Figure S2D). In both cases, mAbo specifically detected CD19 positive B cells.
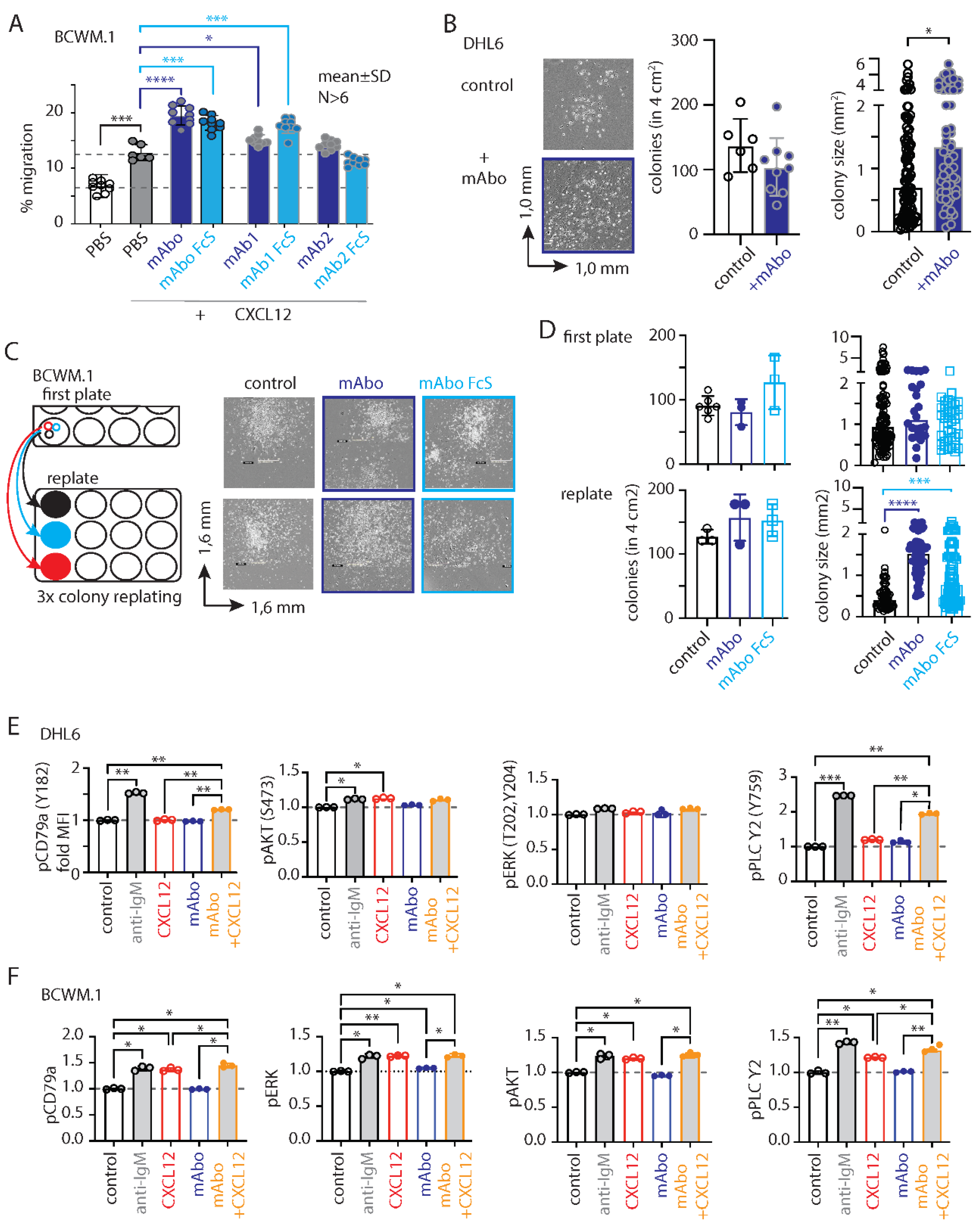
Next, we performed migration assays on BCWM.1 cells in the presence of mAbo. As depicted in
Figure 2A, mAbo treatment significantly enhanced the migration of these cells towards CXCL12, suggesting a role for activated CD19 signaling crosstalk with CXCR4 signaling. As controls, we used two other commercial humanized CD19 clones, mAb1 and mAb2. The mAb1 clone effectively increased CXCL12-induced migration similar to our mAbo. In contrast, the mAb2 clone minimally affected the migration, suggesting differential activation responses through CD19 antibody clones (
Figure 2A). Additionally, we tested the Fc silent (FcS) mutant versions of all these humanized mAbs to avoid aberrant Fc receptor (FcR) mediated recruitment. In all cases, the FcS versions behaved similarly to the unmutated mAbs, thereby eliminating interference through FcR on the B cell surface. When tested on CD19 KO BCWM.1 cells, none of these mAbs altered CXCL12-induced migration (
Figure S2E). In parallel to migration, we assessed the effect of mAbo on colony formation (
Figure 2B-D). While DHL6 colony sizes significantly increased upon the addition of mAbo (
Figure 2B), BCWM.1 colonies only exhibited a trend toward increase after 10 days (
Figure 2C). Therefore, we replated the colonies to allow them to grow on a new surface, where BCWM.1 colonies were found to significantly increase in size in response to mAbo treatment (
Figure 2C-D). Notably, mAbo treatment did not affect the number of colonies, suggesting no impact on colony seeding or initiation. Instead, it led to enhanced proliferation and spreading, resulting in larger colonies.
As shown in
Figure 1H, DHL6 cells exhibited minimal migration in response to CXCL12. We, therefore, analyzed the CXCR4 and BCR proximal phosphorylation signaling using an intracellular phospho-flow cytometry assay (
Figure 2E-F and
Figure S2F). To optimize assay conditions, we tested phosphorylation signals for pCD79a (Y182), pERK (T202, Y204), pAKT (S473) and pPLC-γ2 (Y759) in DHL6 cells following anti-IgM or CXCL12 treatment for 5 and 10 minutes (
Figure S2F). While BCR stimulation with anti-IgM readily increased phosphorylation of all tested markers, CXCL12 treatment selectively induced pAKT and pPLC-γ2 in DHL6 cells (
Figure 2E and
Figure S2F). Notably, DHL6 cells were only minimally responsive to CXCL12 induced migration as compared to BCWM.1 (
Figure 1G-H). Interestingly, the combination of mAbo and CXCL12 treatments resulted in a greater increase in pPLC-γ2 and pCD79a, suggesting a synergistic CXCR4 signal amplification through CD19. Similar results were observed for BCWM.1 cells (
Figure 2F). In contrast to DHL6 cells, CXCL12 treatment alone induced phosphorylation of all tested markers in BCWM.1 cells [
26], reaching levels comparable to those induced by anti-IgM treatment. This suggests differential effects of BCR and CXCR4 signaling between WM and DLBCL.
2.4. Variable Efficiencies of CD19 mAbs in Inducing ADCC
The clinical success of anti-CD19 mAbs relies on B cell–depleting cytolytic activity, specifically through antibody dependent cell mediated cytotoxicity (ADCC) in presence of activated natural killer (NK) cells[
27]. We therefore evaluated the efficacy of the CD19 mAbs in ADCC in presence of IL-2 stimulated NK cells isolated from the human peripheral blood (
Figure 3 and
Figure S3). We optimized a FACS based quantitative analysis to determine the absolute live cell count and distinguish cell types based on endogenous markers post ADCC assay (
Figure S3A-E)). Briefly, cell numbers were normalized to the number of live lymphoma cells obtained from the 4 hours control experimental ADCC coculture of lymphoma and rhIL-2 activated NK cells without any mAb (no mAb) addition (
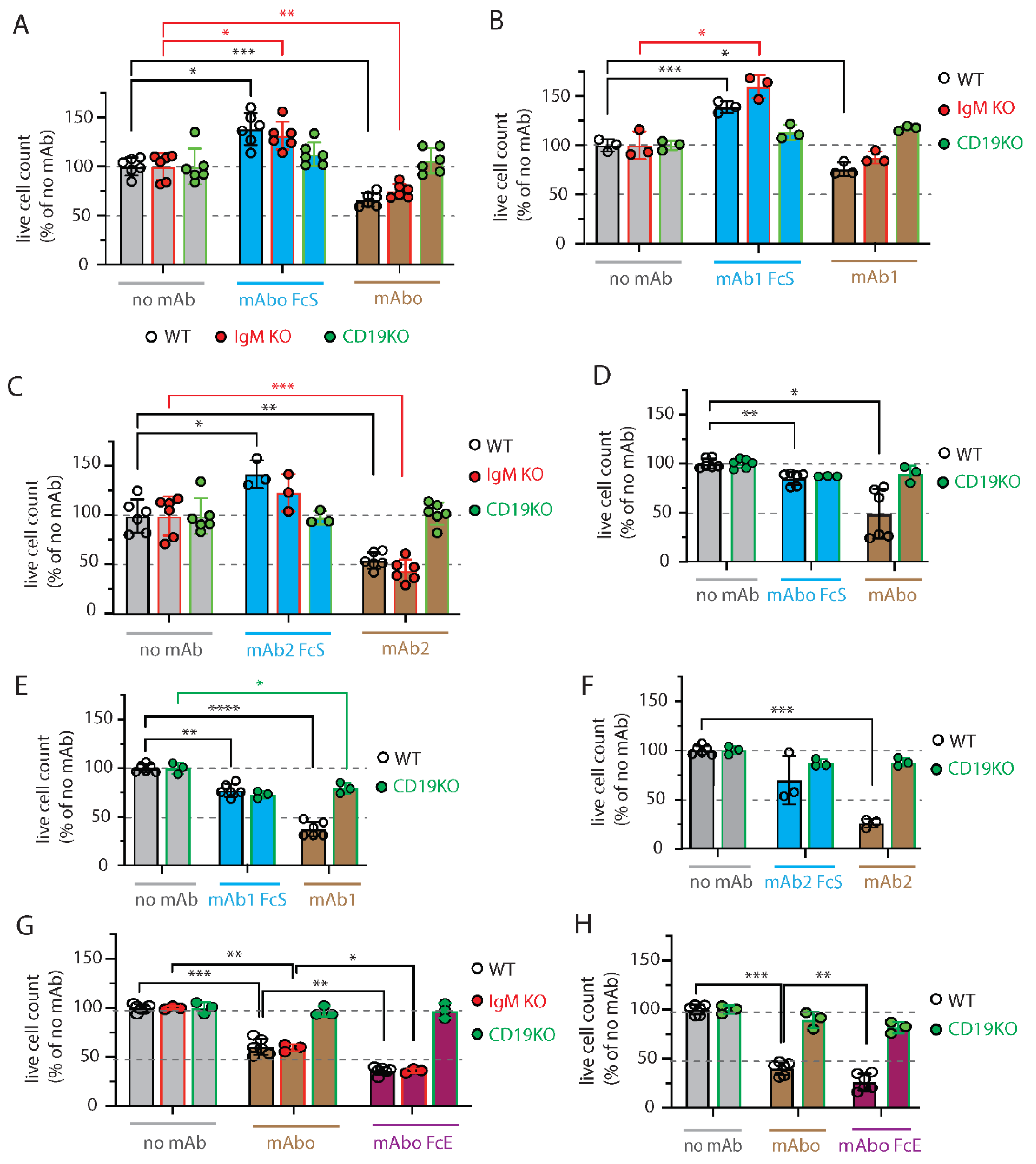
Figure S3C). Expectedly, we found that ADCC in presence mAbo reduced the WT BCWM.1 cells to 68.9±7.2% in comparison to no mAb control 100.4±9.4% (
Figure 3A and
Figure S3C,E)). Similarly, live IgM KO BCWM cells were reduced from 108.7±14.6% in no mAb control to 81.6±7.8% due to mAbo induced ADCC. In contrast CD19 KO cells remained unaffected by mAbo induced ADCC in presence of activated NK cells. Similarly, the mAboFcS treatment failed to kill any of the WT and KO BCWM.1 cells due to lack of NK cell engagement through FcR (
Figure 3A and
Figure S3C,D). Interestingly, normalized percentage of living WT and IgM KO BCWM.1 cells treated with mAboFcS were increased to 143.6±17% and 142.8±15.4%, respectively. This result supports our hypothesis that the failure to ligate a cytotoxic NK could potentially increase survival of lymphoma cells in presence of CD19 mAbs. Notably, the absolute lymphoma cell counts decreases for all BCWM.1 and DHL6 cell types in presence of activated NK cells in 4 hours control experimental ADCC coculture without any mAb (
Figure S3B,C,F). This systemic loss of cell survival under control experimental ADCC coculture is prevented by the addition mAboFcS treatments (
Figure 3A and
Figure S3F).
Similar ADCC responses against different BCWM.1 cell types were seen for other two clonotypes of CD19 antibodies mAb1 and mAb2, and their corresponding FcS forms mAb1FcS and mAb2FcS (
Figure 3B-C). While mAb1 clone reduced the WT BCWM.1 cells to 75.8±7% only, mAb2 treatment reduced to 54.8±8.3% survival, suggesting a variable response to different mAb clonotypes. Similarly, mAb1 and mAb2 induced ADCC reduced the IgM KO cells to 87.5±6.2% and 44.3±11.4% (
Figure 3B-C), respectively. As control, there were no effect of these clonotypes on CD19 KO cells. Interestingly, both mAb1FcS and mAb2FcS caused significant increase in survival of WT BCWM.1 cells up to 138.9±5.8% and 142.5±14.2% (
Figure 3B-C), respectively. Unlike WT and IgM KO cells, there were no increase in survival of CD19 KO BCWM.1 cells upon mAb1FcS or mAb2FcS treatment, demonstrating the role of activated CD19 signaling in survival advantage.
For DHL6 cells, all three different CD19 mAb clones showed relatively higher ADCC efficacy compared to BCWM.1 cells (
Figure 3D-F). The parentages of WT DHL6 survived in ADCC upon mAbo, mAb1 and mAb2 treatments were 49±25.4, 37.4±7.4 and 25.8±4.5, respectively. In contrast to the effects on BCWM.1 cell types, all three clonotypes in their FcS forms failed to cause any survival advantage of DHL6 cells compared to no mAb treatment (
Figure 3D-F and
Figure S3F). In other words, reverting the systemic loss of cell survival under control experimental ADCC coculture were ineffective for DHL6 cells.
To improve on the efficacy of our mAbo clonotype in ADCC, we aimed generating mutant with enhanced FcR binding, as described before [
28,
29]. To this end, we introduced two point-mutations at IgG1 CH2 domain, namely S239D and I332E resulting in mAbo Fc enhanced (mAboFcE) version. We repeated the ADCC assay on BCWM.1 and DHL6 cells with this mAboFcE and compared to original unmutated mAbo treatment (
Figure 3G-H). Expectedly, the mAboFcE treatment enhanced ADCC and drastically reduced survival of WT BCWM.1 cells to 36±3.8% compared to 60.4±8.2% survival upon non-modified mAbo treatment (
Figure 3G). Similarly, survival of WT DHL6 cells were reduced to 26.3±8.4% by mAboFcE treatment compared to 40.2±6.7% upon non-modified mAbo treatment (
Figure 3H). As control, there were no effect of mAboFcE on any CD19 KO cell types. Altogether, these data show differential effects of CD19 mAb clonotypes on ADCC response against lymphoma cells, with highest efficacy caused by mAb2 clonotype against both WM and DLBCL. The clonotype specific variations were more pronounced for BCWM.1 as compared to DHL6 cells, which is indicative to their relative CXCR4 signaling dependency (
Figure 2A). Furthermore, by creating enhanced FcR binding mutant mAboFcE, we could improve on our in-house generated ant-CD19 antibody and attain the similar efficacy as mAb2 clonotype (
Figure 3C and G).
2.5. CXCR4 Antagonizing Peptide Enhances CD19 mAbs Induced ADCC
To down-modulate the CD19 mAb-induced activation of CXCR4 response, we then explored the synergistic potential of CXCR4 inhibition to improve the mAb induced ADCC. The endogenous peptide inhibitor of CXCR4 (EPI-X4) specifically antagonizes CXCR4 and is therefore a promising candidate for drug development for the treatment of CXCR4-dependent diseases [
30]. Optimized EPI-X4 derivatives reduced tumor burden in different cancer models, and specifically the survival of WM cell line in immunocompromised mouse recipient [
26,
31,
32]. We here tested the original EPI-X4, and an optimized version named EPI-X4 JM#21 (hereon referred to as JM#21), which showed 1000-fold increased antagonistic activity compared to the wildtype peptide[
32]. In addition, we included the small molecule CXCR4 antagonist AMD3100 (Plerixafor), which is clinically approved for autologous stem cell transplantation[
33]. Previously, both EPI-X4 and JM#21 treatments were shown to inhibit basal survival signaling pathways by reducing ERK and AKT phosphorylation, and subsequent loss of survival and apoptosis in WM cells [
26]. Therefore, we first tested the dose response of EPI-X4 and JM#21 for 4 hours of treatment as described for ADCC assay and measured the cell survival after 12-18 hours post removal of inhibitors (
Figure S4A). The calculated IC
50 for EPI-X4 and JM#21 treatments were 21.3 and 5.1µM, respectively. Then we tested the inhibitory effect of same concentrations of EPI-X4 and JM#21peptides on CXCL12 induced migration of BCWM.1 cells (
Figure S4B). While >50µM of EPI-X4 peptide was required to significantly reduce the CXCL12 induced migration to 13±1.3%, only 20µM of JM#21 reduced to 10±2.7% compared to PBS treatment resulting 19.3±0.7% migration. Of note, 10µM JM#21 and AMD3100 and 200µM of EPI-X4 was found to be effective for suppressing the CXCL12 induced migration of BCWM.1 cells overexpressing CXCR4 isoform 1, which caused unusual increase in specific migration up to 80% [
26]. Since we were intended to prevent the CXCR4 activation in wildtype BCWM.1 cells expressing endogenous CXCR4, we therefore used 20µM of JM#21 in mAbo induced ADCC and equivalent concentrations of EPI-X4 and AMD3100 (
Figure S4C). While both JM#21 and EPI-X4 treatments improved the mAbo induced ADCC and reduced survival of BCWM.1 significantly compared no inhibitor control, AMD3100 treatment caused no significant difference (
Figure S4C, upper panels). In contrast, all three CXCR4 antagonists caused no significant decrease in survival of DHL6 cells in mAbo induced ADCC assays (
Figure S4C, bottom panels). As control, we also performed mock ADCC coculture experiment with mAboFcS in presence and absence of CXCR4 antagonists. As shown before, mAboFcS treatment did not cause any ADCC alone and survival loss, instead increased the cell survival due to activation of CD19 signaling in absence of NK ligation (
Figure S4C and
Figure 3A-C). However, in presence of all of the CXCR4 antagonists used, survival of mAboFcS treated BCWM.1 cells were significantly decreased (
Figure S4C). And as shown before, DHL6 cells did not have any survival advantage upon mAboFcS treatment in ADCC assay (
Figure S4C and
Figure 3D-F). Despite no added survival advantage, all CXCR4 antagonists except EPI-X4 caused significant decrease in survival in mock ADCC assay in presence of mAboFcS. These results demonstrate the differential effect of CXCR4 antagonists on CD19 mediated activation signal in lymphoma cells and warrants further improvement of combinatorial efficacy in CD19 mAb induced ADCC and enhanced migration. In particular, the peptide antagonist JM#21 effectively enhanced the mAbo induced ADCC in both BCWM.1 and DHL6 cells, as well as prevented the enhanced survival caused by mAboFcS due to lack of NK cell ligation.
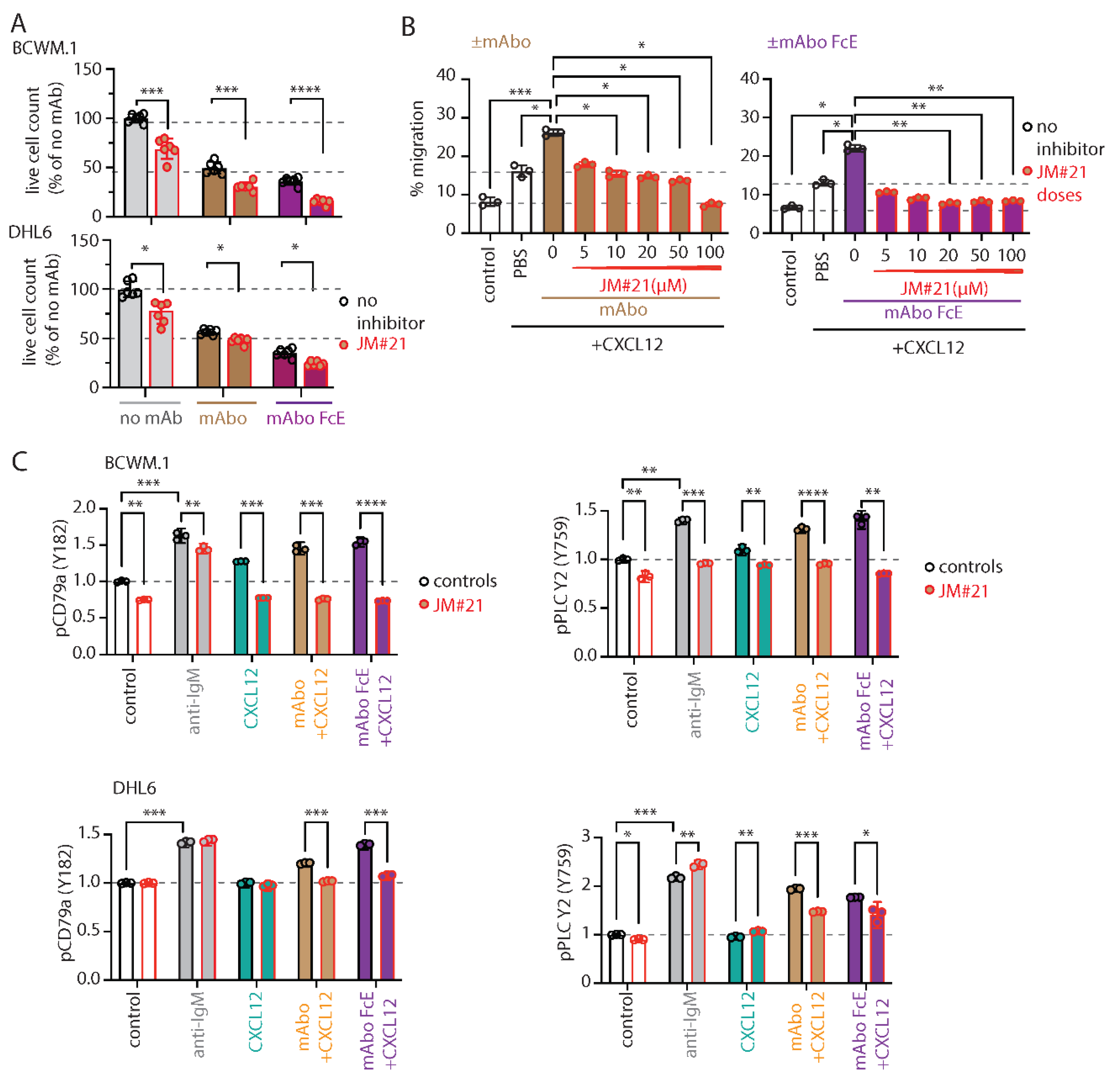
To this end, we tested the effect of JM#21 on the mAboFcE induced ADCC on BCWM.1 and DHL6 cells and compared to mAbo treatment (
Figure 4A). Addition of 20µM JM#21 significantly enhanced the efficacy of mAboFcE causing decreases in survival of BCWM.1 cells to 15.2±2.8% from no inhibitor control 36.1±3.8% (
Figure 4A). In contrast the survival in mAbo induced ADCC was only decreased to 30.9±4.1% from 40.7±5.9%. The survival of DHL6 cells in ADCC assays with mAbo and mAboFcE in presence and absence of 20µM JM#21 were reduced to 47.7±3.3% from control 56.5±2.7% and 24.2±2.3% from 35.2±4.3%, respectively. This result demonstrates the combinatorial efficacy of JM#21 and Fc engineered CD19 mAb that surpasses the individual single treatments. Next, we tested whether JM#21 prevents the CD19 mAb induced enhancement of CXCL12 induced migration (
Figure 4B). As shown before mAbo treatment increased the CXCL12 induced migration of BCWM.1 cells (
Figure 2A). This increased migration was inhibited by JM#21 in a dose dependent manner significantly at doses >10µM (
Figure 4B). Similarly, the enhanced CXCL12 induced migration caused by mAboFcE treatment were significantly blocked by JM#21 doses >20µM. In parallel, we also tested the efficacy of EPI-X4 to prevent the mAbo and mAboFcE induced increase in migration (
Figure S4D). While a minimum of 50µM EPI-X4 was required to significantly prevent mAbo induced increased migration, above 10µM of the same was effectively blocked the mAboFcE induced increase. These results show the efficacy of JM#21 over EPI-X4 against CD19 mAb induced activation of CXCR4 signaling.
Next, we tested the effect of JM#21 on intracellular phosphorylation of BCWM.1 and DHL6 cells (
Figure 4C and
Figure S4E). As shown before, both BCR stimulation with anti-IgM and CXCR4 stimulation with CXCL12 treatment readily induced phosphorylation of pCD79a, pAKT, pERK and pPLC-γ2 in BCWM.1 (
Figure 2E and
Figure S2F). In DHL6 cells, CXCL12 treatment only in presence of mAbo induced pCD79a and pPLC-γ2. Therefore, we first analyzed the effect of mAboFcE in presence of CXCL12 on induction of pCD79a and pPLC-γ2 in both BCWM.1 and DHL6 cells, and then combined with JM#21 treatments (
Figure 4C). Congruent to previous results using mAbo, combination of mAboFcE and CXCL12 (mAboFcE+CXCL12) treatments caused increased pCD79a and pPLC-γ2 in both BCWM.1 and DHL6 cells as compared to CXCL12 only treatments. In contrast, the mAboFcE+CXCL12 treatments induced pAKT and pERK only in BCWM.1 cells similar to that of mAbo +CXCL12 treatments (
Figure S4E). To compare the effect of CXCR4 antagonist, we pretreated cells with 20µM JM#21 similarly as in ADCC assays and then stimulated with anti-IgM, CXCL12, mAbo +CXCL12 and mAboFcE +CXCL12 (
Figure 4C and
Figure S4E). Congruent to previous report by Kaiser et al.[
26], JM#21 treatment caused reduced basal phosphorylation of pAKT and pERK without any addition of stimulation in BCWM.1 cells (
Figure S4E). In addition, we found a decrease in basal pCD79a and pPLC-γ2 level in BCWM.1 cells (
Figure 4C). However, the basal phosphorylation for all four markers remained unchanged in DHL6 cells upon JM#21 treatments (
Figure 4C and
Figure S4E) suggesting a complete CXCR4 signaling independent survival of these cells, which is in resonance to the minimal CXCL12 induced migration (
Figure 1H). Similar to reduced basal phosphorylation, JM#21 pretreatments reduced the anti-IgM and CXCL12 stimulated pCD79a, pAKT, pERK and pPLC-γ2 induction only in BCWM.1 cells but not in DHL6 cells (
Figure 4C and
Figure S4E). In addition, JM#21 pretreatments inhibited the increased pCD79a, pAKT, pERK and pPLC-γ2 in response to mAbo +CXCL12 and mAboFcE +CXCL12 stimulations in both cell types. As such the synergistic increase in phosphorylation in DHL6 cells were less pronounced and therefore the effect of JM#21 pretreatments is minimal as compared to BCWM.1 cells. Altogether these results show that in combination with JM#21 the efficacy of CD19 mAbs including the modified FcR binding enhanced version (mAboFcE) were instantly increased by enhanced ADCC and inhibition of CD19 induced activation signal. Broadly, blocking CXCR4 with optimized EPI-X4 derived peptide antagonist like JM#21 is a promising approach to augment therapeutic effects of CD19 mAbs for the treatment of WM.