Submitted:
27 November 2024
Posted:
28 November 2024
You are already at the latest version
Abstract
Avian necrotic enteritis due to the Gram-positive bacterium Clostridium perfringens has re-emerged following the ban on antibiotic growth promoters in many poultry producing countries. The limited number of previous studies has left important gaps in our understanding of the genetic diversity and virulence traits of the pathogen. To address these knowledge gaps, in this study, we sequenced the genomes of 41 Clostridium perfringens isolates recovered from commercial broiler chicken flocks in Quebec, Canada, including isolates from healthy birds and those affected by necrotic enteritis. We sought to understand the pangenome diversity and interrogated the genomes for key virulence factors involved in necrotic enteritis pathogenesis. On average, the genomes had a GC content of 28% and contained 3,206 coding sequences. A variable presence of toxins, degradative hydrolytic enzymes, and collagen-binding proteins was also found. Through pangenome analysis, we revealed a total of 10,223 genes, 652 (6.4%) of which formed the core genome. Additionally, we identified 17 different plasmids, 12 antibiotic resistance genes, and nine prophage regions. Overall, our results demonstrated a relatively high genetic diversity among chicken Clostridium perfringens isolates collected from the same geographical location, offering new insights into potential virulence mechanisms and adaptation of the pathogen within poultry populations.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection, Isolation of C. perfringens Strains, DNA Extraction, and WGS
2.2. Bioinformatics Workflow for Genome Assembly, Annotation, and Analysis
3. Results
3.1. Genome Assembly and Annotation
3.2. Pangenome Analysis and Genetic Relationships Between Isolates
3.3. In Silico Profiling of Virulence Traits
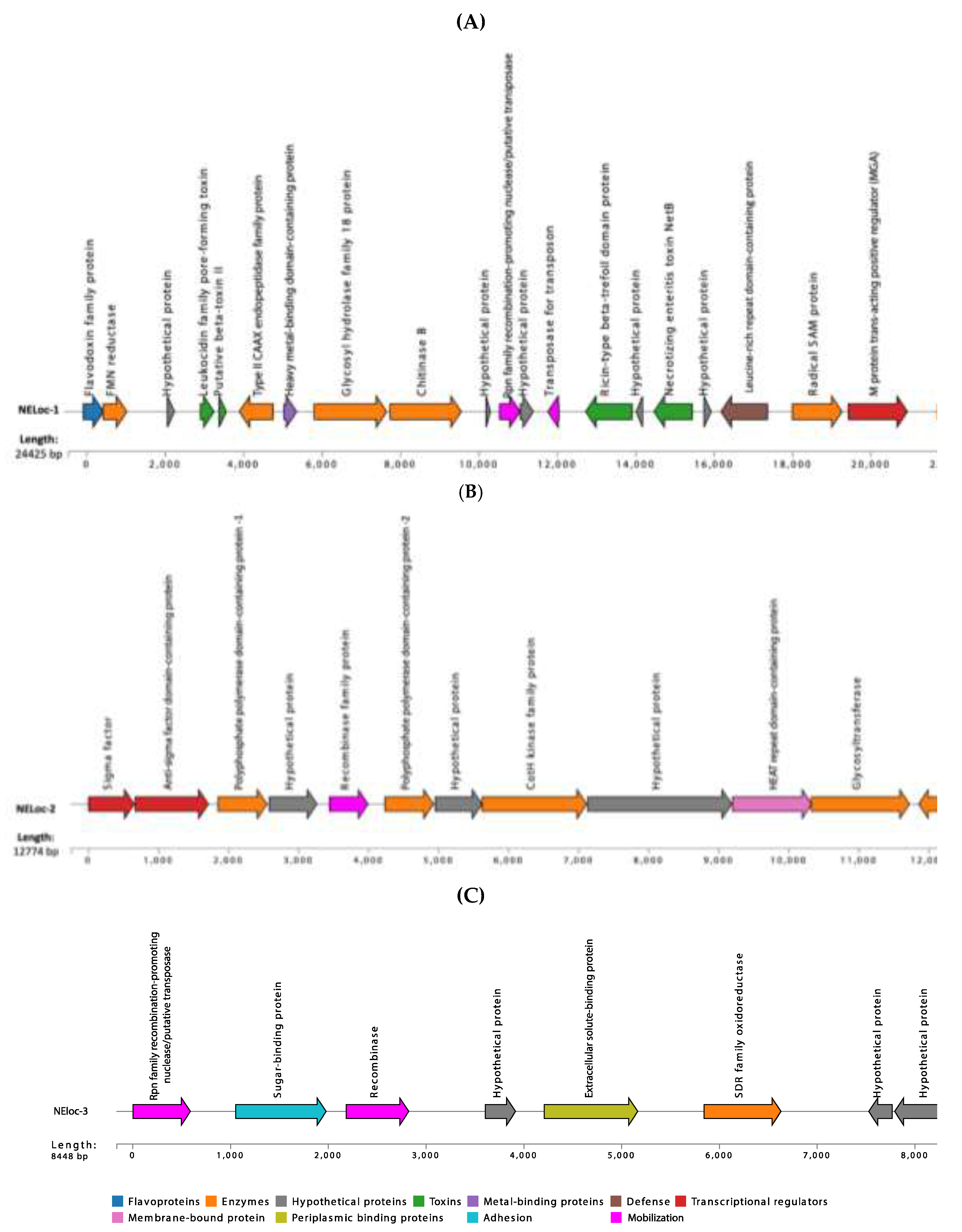
3.4. Characterization of NELoc-1–3
3.5. ARGs Prediction
3.6. Prophage Content of C. perfringens Isolates
3.7. Presence of Plasmids in C. perfringens Isolates
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kiu, R.; Hall, L.J. An update on the human and animal enteric pathogen Clostridium perfringens. Emerg Microbes Infect 2018, 7, 141.
- Mehdizadeh Gohari, I.; A Navarro, M.; Li, J.; Shrestha, A.; Uzal, F.; A McClane, B. Pathogenicity and virulence of Clostridium perfringens. Virulence 2021, 12, 723-753.
- Fathima, S.; Hakeem, W.G.A.; Shanmugasundaram, R.; Selvaraj, R.K. Necrotic Enteritis in Broiler Chickens: A Review on the Pathogen, Pathogenesis, and Prevention. Microorganisms 2022, 10, 1958.
- Prescott, J.F.; Parreira, V.R.; Mehdizadeh Gohari, I.; Lepp, D.; Gong, J. The pathogenesis of necrotic enteritis in chickens: what we know and what we need to know: a review. Avian Pathol 2016, 45, 288-294.
- Chalmers, G.; Bruce, H.L.; Hunter, D.B.; Parreira, V.R.; Kulkarni, R.R.; Jiang, Y.F.; Prescott, J.F.; Boerlin, P. Multilocus sequence typing analysis of Clostridium perfringens isolates from necrotic enteritis outbreaks in broiler chicken populations. J Clin Microbiol 2008, 46, 3957-3964.
- Gholamiandekhordi, A.R.; Ducatelle, R.; Heyndrickx, M.; Haesebrouck, F.; Van Immerseel, F. Molecular and phenotypical characterization of Clostridium perfringens isolates from poultry flocks with different disease status. Vet Microbiol 2006, 113, 143-152.
- Chalmers, G.; Martin, S.W.; Prescott, J.F.; Boerlin, P. Typing of Clostridium perfringens by multiple-locus variable number of tandem repeats analysis. Vet Microbiol 2008, 128, 126-135.
- De Cesare, A.; Borilova, G.; Svobodova, I.; Bondioli, V.; Manfreda, G. Clostridium perfringens occurrence and ribotypes in healthy broilers reared in different European countries. Poult Sci 2009, 88, 1850-1857.
- Lacey, J.A.; Allnutt, T.R.; Vezina, B.; Van, T.T.H.; Stent, T.; Han, X.; Rood, J.I.; Wade, B.; Keyburn, A.L.; Seemann, T.; Chen, H.; Haring, V.; Johanesen, P.A.; Lyras, D.; Moore, R.J. Whole genome analysis reveals the diversity and evolutionary relationships between necrotic enteritis-causing strains of Clostridium perfringens. BMC Genomics 2018, 19, 379.
- Lepp, D.; Gong, J.; Songer, J.G.; Boerlin, P.; Parreira, V.R.; Prescott, J.F. Identification of accessory genome regions in poultry Clostridium perfringens isolates carrying the netB plasmid. J Bacteriol 2013, 195, 1152-1166.
- Gaucher, M.L.; Perron, G.G.; Arsenault, J.; Letellier, A.; Boulianne, M.; Quessy, S. Recurring Necrotic Enteritis Outbreaks in Commercial Broiler Chicken Flocks Strongly Influence Toxin Gene Carriage and Species Richness in the Resident Clostridium perfringens Population. Front Microbiol 2017, 8, 881.
- Park, M.; Deck, J.; Foley, S.L.; Nayak, R.; Songer, J.G.; Seibel, J.R.; Khan, S.A.; Rooney, A.P.; Hecht, D.W.; Rafii, F. Diversity of Clostridium perfringens isolates from various sources and prevalence of conjugative plasmids. Anaerobe 2016, 38, 25-35.
- Parreira, V.R.; Costa, M.; Eikmeyer, F.; Blom, J.; Prescott, J.F. Sequence of two plasmids from Clostridium perfringens chicken necrotic enteritis isolates and comparison with C. perfringens conjugative plasmids. PLoS One 2012, 7, e49753.
- Lepp, D.; Roxas, B.; Parreira, V.R.; Marri, P.R.; Rosey, E.L.; Gong, J.; Songer, J.G.; Vedantam, G.; Prescott, J.F. Identification of novel pathogenicity loci in Clostridium perfringens strains that cause avian necrotic enteritis. PLoS One 2010, 5, e10795.
- Zhou, H.; Lepp, D.; Pei, Y.; Liu, M.; Yin, X.; Ma, R.; Prescott, J.F.; Gong, J. Influence of pCP1NetB ancillary genes on the virulence of Clostridium perfringens poultry necrotic enteritis strain CP1. Gut Pathog 2017, 9, 6.
- Barbara, A.J.; Trinh, H.T.; Glock, R.D.; Glenn Songer, J. Necrotic enteritis-producing strains of Clostridium perfringens displace non-necrotic enteritis strains from the gut of chicks. Vet Microbiol 2008, 126, 377-382.
- Myers, G.S.; Rasko, D.A.; Cheung, J.K.; Ravel, J.; Seshadri, R.; DeBoy, R.T.; Ren, Q.; Varga, J.; Awad, M.M.; Brinkac, L.M.; Daugherty, S.C.; Haft, D.H.; Dodson, R.J.; Madupu, R.; Nelson, W.C.; Rosovitz, M.J.; Sullivan, S.A.; Khouri, H.; Dimitrov, G.I.; Watkins, K.L.; Mulligan, S.; Benton, J.; Radune, D.; Fisher, D.J.; Atkins, H.S.; Hiscox, T.; Jost, B.H.; Billington, S.J.; Songer, J.G.; McClane, B.A.; Titball, R.W.; Rood, J.I.; Melville, S.B.; Paulsen, I.T. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res 2006, 16, 1031-1040.
- Kiu, R.; Caim, S.; Alexander, S.; Pachori, P.; Hall, L.J. Probing Genomic Aspects of the Multi-Host Pathogen Clostridium perfringens Reveals Significant Pangenome Diversity, and a Diverse Array of Virulence Factors. Front Microbiol 2017, 8, 2485.
- Keyburn, A.L.; Sheedy, S.A.; Ford, M.E.; Williamson, M.M.; Awad, M.M.; Rood, J.I.; Moore, R.J. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Infect Immun 2006, 74, 6496-6500.
- Meniaï, I.; Thibodeau, A.; Quessy, S.; Parreira, V.R.; Fravalo, P.; Beauchamp, G.; Gaucher, M.L. Putative antigenic proteins identified by comparative and subtractive reverse vaccinology in necrotic enteritis-causing Clostridium perfringens isolated from broiler chickens. BMC Genomics 2021, 22, 890.
- Coil, D.; Jospin, G.; Darling, A.E. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 2015, 31, 587-589.
- Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068-2069.
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J Mol Biol 1990, 215, 403-410.
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691-3693.
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 2010, 5, e9490.
- Xu, S.; Li, L.; Luo, X.; Chen, M.; Tang, W.; Zhan, L.; Dai, Z.; Lam, T.T.; Guan, Y.; Yu, G. Ggtree: A serialized data object for visualization of a phylogenetic tree and annotation data. Imeta 2022, 1, e56.
- Team, R.C. R: A Language and Environment for Statistical Computing. (R Foundation for Statistical Computing, 2013). Available online: https:// www.r- project. org/ (accessed on 30/09/2024).
- Wishart, D.S.; Han, S.; Saha, S.; Oler, E.; Peters, H.; Grant, J.R.; Stothard, P.; Gautam, V. PHASTEST: faster than PHASTER, better than PHAST. Nucleic Acids Res 2023, 51, W443-W450.
- Roosaare, M.; Puustusmaa, M.; Möls, M.; Vaher, M.; Remm, M. PlasmidSeeker: identification of known plasmids from bacterial whole genome sequencing reads. PeerJ 2018, 6, e4588.
- Inouye, M.; Dashnow, H.; Raven, L.A.; Schultz, M.B.; Pope, B.J.; Tomita, T.; Zobel, J.; Holt, K.E. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 2014, 6, 90.
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; Doshi, S.; Courtot, M.; Lo, R.; Williams, L.E.; Frye, J.G.; Elsayegh, T.; Sardar, D.; Westman, E.L.; Pawlowski, A.C.; Johnson, T.A.; Brinkman, F.S.; Wright, G.D.; McArthur, A.G. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 2017, 45, D566-D573.
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847-2849.
- Camargo, A.; Guerrero-Araya, E.; Castañeda, S.; Vega, L.; Cardenas-Alvarez, M.X.; Rodríguez, C.; Paredes-Sabja, D.; Ramírez, J.D.; Muñoz, M. Intra-species diversity of Clostridium perfringens: A diverse genetic repertoire reveals its pathogenic potential. Front Microbiol 2022, 13, 952081.
- Adams, V.; Han, X.; Lyras, D.; Rood, J.I. Antibiotic resistance plasmids and mobile genetic elements of Clostridium perfringens. Plasmid 2018, 99, 32-39.
- Lacey, J.A.; Johanesen, P.A.; Lyras, D.; Moore, R.J. In silico Identification of Novel Toxin Homologs and Associated Mobile Genetic Elements in Clostridium perfringens. Pathogens 2019, 8, 16.
- Wisniewski, J.A.; Rood, J.I. The Tcp conjugation system of Clostridium perfringens. Plasmid 2017, 91, 28-36.
- Yang, W.Y.; Chou, C.H.; Wang, C. Characterization of toxin genes and quantitative analysis of netB in necrotic enteritis (NE)-producing and non-NE-producing Clostridium perfringens isolated from chickens. Anaerobe 2018, 54, 115-120.
- Ernst, C.M.; Staubitz, P.; Mishra, N.N.; Yang, S.J.; Hornig, G.; Kalbacher, H.; Bayer, A.S.; Kraus, D.; Peschel, A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog 2009, 5, e1000660.
- Ernst, C.M.; Peschel, A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol Microbiol 2011, 80, 290-299.
- Chen, J.; Yu, Z.; Michel, F.C.; Wittum, T.; Morrison, M. Development and application of real-time PCR assays for quantification of erm genes conferring resistance to macrolides-lincosamides-streptogramin B in livestock manure and manure management systems. Appl Environ Microbiol 2007, 73, 4407-4416.
- Matsuoka, M.; Inoue, M.; Nakajima, Y.; Endo, Y. New erm Gene in Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother 2002, 46, 211-215.
- Xu, C.W.; Zhang, A.Y.; Yang, C.M.; Pan, Y.; Guan, Z.B.; Lei, C.W.; Peng, L.Y.; Li, Q.Z.; Wang, H.N. First report of macrolide resistance gene erm(T) harbored by a novel small plasmid from Erysipelothrix rhusiopathiae. Antimicrob Agents Chemother 2015, 59, 2462-2465.
- Varaldo, P.E.; Montanari, M.P.; Giovanetti, E. Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob Agents Chemother 2009, 53, 343-353.
- Woodbury, R.L.; Klammer, K.A.; Xiong, Y.; Bailiff, T.; Glennen, A.; Bartkus, J.M.; Lynfield, R.; Van Beneden, C.; Beall, B.W. Plasmid-Borne erm(T) from invasive, macrolide-resistant Streptococcus pyogenes strains. Antimicrob Agents Chemother 2008, 52, 1140-1143.
- Abril, C.; Brodard, I.; Perreten, V. Two novel antibiotic resistance genes, tet(44) and ant(6)-Ib, are located within a transferable pathogenicity island in Campylobacter fetus subsp. fetus. Antimicrob Agents Chemother 2010, 54, 3052-3055.
- Feng, Y.; Fan, X.; Zhu, L.; Yang, X.; Liu, Y.; Gao, S.; Jin, X.; Liu, D.; Ding, J.; Guo, Y.; Hu, Y. Phylogenetic and genomic analysis reveals high genomic openness and genetic diversity of Clostridium perfringens. Microb Genom 2020, 6, mgen000441.
- Gervasi, T.; Curto, R.L.; Narbad, A.; Mayer, M.J. Complete genome sequence of ΦCP51, a temperate bacteriophage of Clostridium perfringens. Arch Virol 2013, 158, 2015-2017.
- Nariya, H.; Miyata, S.; Tamai, E.; Sekiya, H.; Maki, J.; Okabe, A. Identification and characterization of a putative endolysin encoded by episomal phage phiSM101 of Clostridium perfringens. Appl Microbiol Biotechnol 2011, 90, 1973-1979.
- Gaucher, M.L.; Thibodeau, A.; Fravalo, P.; Archambault, M.; Arsenault, J.; Fournaise, S.; Letellier, A.; Quessy, S. Broiler chicken carcasses and their associated abattoirs as a source of enterotoxigenic Clostridium perfringens: Prevalence and critical steps for contamination. AIMS Microbiol 2018, 4, 439-454.
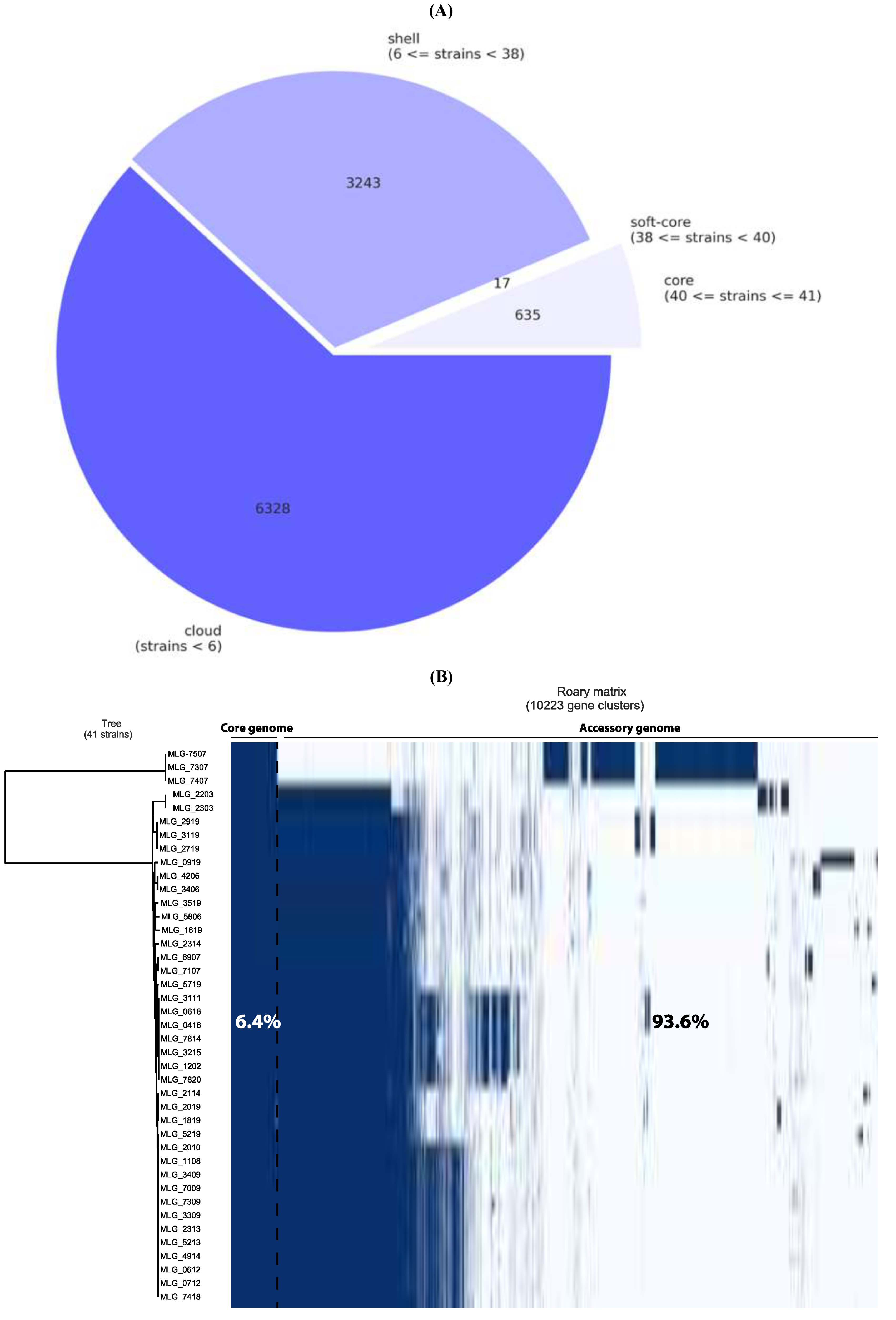
| Description | Description | Nb of strains | Nb of genes | Pangenome (%) | |
|---|---|---|---|---|---|
| Core genome | Core genes | (99% <= strains <= 100%) | 635 | (n = 652), 6.4% | |
| Soft-core genes | (95% <= strains < 99%) | 17 | |||
| Accessory genome | Shell genes | (15% <= strains < 95%) | 3,243 | (n = 9,571), 93.6% | |
| Cloud genes | (0% <= strains < 15%) | 6,328 | |||
| Pangenome | Pangenome | (0% <= strains <= 100%) | 10,223 | 100% |
| Strain | Year of isolation | Type IV pilus profile | PFGE profile | Genome | No of CDSs | BioProject_ID | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | No of Contigs | Coverage (x) | GC (%) | N50 | ||||||
| MLG_7309 | 2010-2011 | 1 | V1 | 3603633 | 204 | 85 | 28 | 51385 | 3403 | PRJNA1188553 |
| MLG_7009 | 2010-2011 | 1 | V2 | 3577078 | 100 | 93 | 28 | 197277 | 3347 | PRJNA1188553 |
| MLG_1108 | 2010-2011 | 1 | V3 | 3546273 | 101 | 88 | 28 | 190425 | 3300 | PRJNA1188553 |
| MLG_3309 | 2010-2011 | 2 | V3 | 3538464 | 144 | 77 | 28 | 58373 | 3298 | PRJNA1188553 |
| MLG_3409 | 2010-2011 | 1 | V3 | 3551654 | 100 | 87 | 28 | 189683 | 3297 | PRJNA1188553 |
| MLG_2010 | 2010-2011 | 2 | V3 | 3551275 | 104 | 92 | 28 | 152768 | 3306 | PRJNA1188553 |
| MLG_2313 | 2010-2011 | 9 | V3 | 3545125 | 88 | 84 | 28 | 177971 | 3290 | PRJNA734442 |
| MLG_2114 | 2010-2011 | 4 | V3 | 3307721 | 49 | 72 | 28.1 | 219784 | 2940 | PRJNA1188553 |
| MLG_7418 | 2010-2011 | 2 | V3 | 3544119 | 82 | 87 | 28 | 219477 | 3286 | PRJNA1188553 |
| MLG_1202 | 2010-2011 | 8 | V5 | 3747051 | 129 | 67 | 27.9 | 114628 | 3572 | PRJNA734442 |
| MLG_3215 | 2010-2011 | 6 | V5 | 3680156 | 96 | 83 | 28 | 170870 | 3498 | PRJNA1188553 |
| MLG_7814 | 2010-2011 | 5 | V6 | 3687085 | 124 | 60 | 28 | 80413 | 3511 | PRJNA734442 |
| MLG_7820 | 2010-2011 | 5 | V7 | 3730847 | 209 | 91 | 27.9 | 48863 | 3570 | PRJNA734442 |
| MLG_2314 | 2010-2011 | 12 | V10 | 3291552 | 43 | 75 | 28.1 | 291371 | 2920 | PRJNA1188553 |
| MLG_4914 | 2010-2011 | 6 | V12 | 3579960 | 152 | 77 | 28 | 82578 | 3345 | PRJNA1188553 |
| MLG_5806 | 2010-2011 | 6 | C1 | 3324484 | 32 | 87 | 28.1 | 474093 | 2963 | PRJNA734442 |
| MLG_4206 | 2010-2011 | 2 | C2 | 3400900 | 48 | 64 | 28 | 141651 | 3040 | PRJNA734442 |
| MLG_3406 | 2010-2011 | 7 | C2 | 3400562 | 55 | 68 | 28 | 126010 | 3039 | PRJNA734442 |
| MLG_5213 | 2010-2011 | 6 | C3 | 3527345 | 92 | 64 | 28 | 158351 | 3269 | PRJNA734442 |
| MLG_0612 | 2010-2011 | 6 | C3 | 3537182 | 94 | 104 | 28 | 217561 | 3283 | PRJNA734442 |
| MLG_0712 | 2010-2011 | 6 | C3 | 3527154 | 171 | 72 | 28 | 49443 | 3287 | PRJNA1188553 |
| MLG_7307 | 2010-2011 | 13 | V1 | 3660280 | 90 | 74 | 28.5 | 97597 | 3296 | PRJNA1188553 |
| MLG_7407 | 2010-2011 | 14 | V1 | 3661751 | 58 | 76 | 28.5 | 172371 | 3293 | PRJNA1188553 |
| MLG_7507 | 2010-2011 | 15 | V1 | 3665112 | 49 | 90 | 28.5 | 233259 | 3292 | PRJNA1188553 |
| MLG_6907 | 2010-2011 | 1 | V2 | 3281089 | 30 | 97 | 28.1 | 699417 | 2921 | PRJNA1188553 |
| MLG_7107 | 2010-2011 | 1 | V2 | 3281533 | 36 | 89 | 28.1 | 222145 | 2920 | PRJNA1188553 |
| MLG_2203 | 2010-2011 | 16 | V3 | 3281091 | 36 | 83 | 28.1 | 219382 | 2880 | PRJNA1188553 |
| MLG_2303 | 2010-2011 | 17 | V3 | 3283967 | 29 | 66 | 28.1 | 721013 | 2881 | PRJNA1188553 |
| MLG_3519 | 2010-2011 | 4 | V5 | 3227573 | 33 | 64 | 28.2 | 255420 | 2867 | PRJNA1188553 |
| MLG_0919 | 2010-2011 | 18 | V6 | 3892868 | 148 | 77 | 28.5 | 62235 | 3661 | PRJNA1188553 |
| MLG_5719 | 2010-2011 | 1 | V7 | 3379598 | 58 | 70 | 28.2 | 133605 | 3023 | PRJNA1188553 |
| MLG_5219 | 2010-2011 | 19 | V8 | 3291656 | 99 | 81 | 28.1 | 62796 | 2953 | PRJNA1188553 |
| MLG_3119 | 2010-2011 | 7 | C1 | 3435369 | 42 | 86 | 28 | 265064 | 3053 | PRJNA734442 |
| MLG_2919 | 2010-2011 | 20 | C1 | 3434769 | 42 | 113 | 28 | 397745 | 3051 | PRJNA734442 |
| MLG_2719 | 2010-2011 | 21 | C1 | 3434532 | 39 | 108 | 28 | 413284 | 3049 | PRJNA734442 |
| MLG_2019 | 2010-2011 | 3 | C2 | 3233362 | 60 | 82 | 28.2 | 127551 | 2880 | PRJNA734442 |
| MLG_1819 | 2010-2011 | 3 | C3 | 3234658 | 48 | 65 | 28.2 | 198347 | 2884 | PRJNA1188553 |
| MLG_1619 | 2010-2011 | 22 | C3 | 3269123 | 39 | 94 | 28.1 | 232490 | 2913 | PRJNA734442 |
| MLG_0418 | 2010-2011 | 5 | C4 | 3788295 | 115 | 94 | 28 | 114406 | 3622 | PRJNA1188553 |
| MLG_0618 | 2010-2011 | 8 | C4 | 3782263 | 143 | 126 | 28 | 71277 | 3624 | PRJNA1188553 |
| MLG_3111 | 2010-2011 | 5 | C5 | 3790123 | 124 | 70 | 28 | 102785 | 3633 | PRJNA1188553 |
| AVERAGE | 3500210 | 86 | 83 | 28 | 200369 | 3206 | ||||
| Strain | Year of isolation | Type IV pilus profile | PFGE profile | Genome | No of CDSs | BioProject_ID | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Length (bp) | No of Contigs | Coverage (x) | GC (%) | N50 | ||||||
| MLG_7309 | 2010-2011 | 1 | V1 | 3603633 | 204 | 85 | 28 | 51385 | 3403 | PRJNA1188553 |
| MLG_7009 | 2010-2011 | 1 | V2 | 3577078 | 100 | 93 | 28 | 197277 | 3347 | PRJNA1188553 |
| MLG_1108 | 2010-2011 | 1 | V3 | 3546273 | 101 | 88 | 28 | 190425 | 3300 | PRJNA1188553 |
| MLG_3309 | 2010-2011 | 2 | V3 | 3538464 | 144 | 77 | 28 | 58373 | 3298 | PRJNA1188553 |
| MLG_3409 | 2010-2011 | 1 | V3 | 3551654 | 100 | 87 | 28 | 189683 | 3297 | PRJNA1188553 |
| MLG_2010 | 2010-2011 | 2 | V3 | 3551275 | 104 | 92 | 28 | 152768 | 3306 | PRJNA1188553 |
| MLG_2313 | 2010-2011 | 9 | V3 | 3545125 | 88 | 84 | 28 | 177971 | 3290 | PRJNA734442 |
| MLG_2114 | 2010-2011 | 4 | V3 | 3307721 | 49 | 72 | 28.1 | 219784 | 2940 | PRJNA1188553 |
| MLG_7418 | 2010-2011 | 2 | V3 | 3544119 | 82 | 87 | 28 | 219477 | 3286 | PRJNA1188553 |
| MLG_1202 | 2010-2011 | 8 | V5 | 3747051 | 129 | 67 | 27.9 | 114628 | 3572 | PRJNA734442 |
| MLG_3215 | 2010-2011 | 6 | V5 | 3680156 | 96 | 83 | 28 | 170870 | 3498 | PRJNA1188553 |
| MLG_7814 | 2010-2011 | 5 | V6 | 3687085 | 124 | 60 | 28 | 80413 | 3511 | PRJNA734442 |
| MLG_7820 | 2010-2011 | 5 | V7 | 3730847 | 209 | 91 | 27.9 | 48863 | 3570 | PRJNA734442 |
| MLG_2314 | 2010-2011 | 12 | V10 | 3291552 | 43 | 75 | 28.1 | 291371 | 2920 | PRJNA1188553 |
| MLG_4914 | 2010-2011 | 6 | V12 | 3579960 | 152 | 77 | 28 | 82578 | 3345 | PRJNA1188553 |
| MLG_5806 | 2010-2011 | 6 | C1 | 3324484 | 32 | 87 | 28.1 | 474093 | 2963 | PRJNA734442 |
| MLG_4206 | 2010-2011 | 2 | C2 | 3400900 | 48 | 64 | 28 | 141651 | 3040 | PRJNA734442 |
| MLG_3406 | 2010-2011 | 7 | C2 | 3400562 | 55 | 68 | 28 | 126010 | 3039 | PRJNA734442 |
| MLG_5213 | 2010-2011 | 6 | C3 | 3527345 | 92 | 64 | 28 | 158351 | 3269 | PRJNA734442 |
| MLG_0612 | 2010-2011 | 6 | C3 | 3537182 | 94 | 104 | 28 | 217561 | 3283 | PRJNA734442 |
| MLG_0712 | 2010-2011 | 6 | C3 | 3527154 | 171 | 72 | 28 | 49443 | 3287 | PRJNA1188553 |
| MLG_7307 | 2010-2011 | 13 | V1 | 3660280 | 90 | 74 | 28.5 | 97597 | 3296 | PRJNA1188553 |
| MLG_7407 | 2010-2011 | 14 | V1 | 3661751 | 58 | 76 | 28.5 | 172371 | 3293 | PRJNA1188553 |
| MLG_7507 | 2010-2011 | 15 | V1 | 3665112 | 49 | 90 | 28.5 | 233259 | 3292 | PRJNA1188553 |
| MLG_6907 | 2010-2011 | 1 | V2 | 3281089 | 30 | 97 | 28.1 | 699417 | 2921 | PRJNA1188553 |
| MLG_7107 | 2010-2011 | 1 | V2 | 3281533 | 36 | 89 | 28.1 | 222145 | 2920 | PRJNA1188553 |
| MLG_2203 | 2010-2011 | 16 | V3 | 3281091 | 36 | 83 | 28.1 | 219382 | 2880 | PRJNA1188553 |
| MLG_2303 | 2010-2011 | 17 | V3 | 3283967 | 29 | 66 | 28.1 | 721013 | 2881 | PRJNA1188553 |
| MLG_3519 | 2010-2011 | 4 | V5 | 3227573 | 33 | 64 | 28.2 | 255420 | 2867 | PRJNA1188553 |
| MLG_0919 | 2010-2011 | 18 | V6 | 3892868 | 148 | 77 | 28.5 | 62235 | 3661 | PRJNA1188553 |
| MLG_5719 | 2010-2011 | 1 | V7 | 3379598 | 58 | 70 | 28.2 | 133605 | 3023 | PRJNA1188553 |
| MLG_5219 | 2010-2011 | 19 | V8 | 3291656 | 99 | 81 | 28.1 | 62796 | 2953 | PRJNA1188553 |
| MLG_3119 | 2010-2011 | 7 | C1 | 3435369 | 42 | 86 | 28 | 265064 | 3053 | PRJNA734442 |
| MLG_2919 | 2010-2011 | 20 | C1 | 3434769 | 42 | 113 | 28 | 397745 | 3051 | PRJNA734442 |
| MLG_2719 | 2010-2011 | 21 | C1 | 3434532 | 39 | 108 | 28 | 413284 | 3049 | PRJNA734442 |
| MLG_2019 | 2010-2011 | 3 | C2 | 3233362 | 60 | 82 | 28.2 | 127551 | 2880 | PRJNA734442 |
| MLG_1819 | 2010-2011 | 3 | C3 | 3234658 | 48 | 65 | 28.2 | 198347 | 2884 | PRJNA1188553 |
| MLG_1619 | 2010-2011 | 22 | C3 | 3269123 | 39 | 94 | 28.1 | 232490 | 2913 | PRJNA734442 |
| MLG_0418 | 2010-2011 | 5 | C4 | 3788295 | 115 | 94 | 28 | 114406 | 3622 | PRJNA1188553 |
| MLG_0618 | 2010-2011 | 8 | C4 | 3782263 | 143 | 126 | 28 | 71277 | 3624 | PRJNA1188553 |
| MLG_3111 | 2010-2011 | 5 | C5 | 3790123 | 124 | 70 | 28 | 102785 | 3633 | PRJNA1188553 |
| AVERAGE | 3500210 | 86 | 83 | 28 | 200369 | 3206 | ||||
| Description | Description | Nb of strains | Nb of genes | Pangenome (%) |
|---|---|---|---|---|
| Core genome | Core genes | (99% <= strains <= 100%) | 635 | (n = 652), 6.4% |
| Soft-core genes | (95% <= strains < 99%) | 17 | ||
| Accessory genome | Shell genes | (15% <= strains < 95%) | 3,243 | (n = 9,571), 93.6% |
| Cloud genes | (0% <= strains < 15%) | 6,328 | ||
| Pangenome | Pangenome | (0% <= strains <= 100%) | 10,223 | 100% |
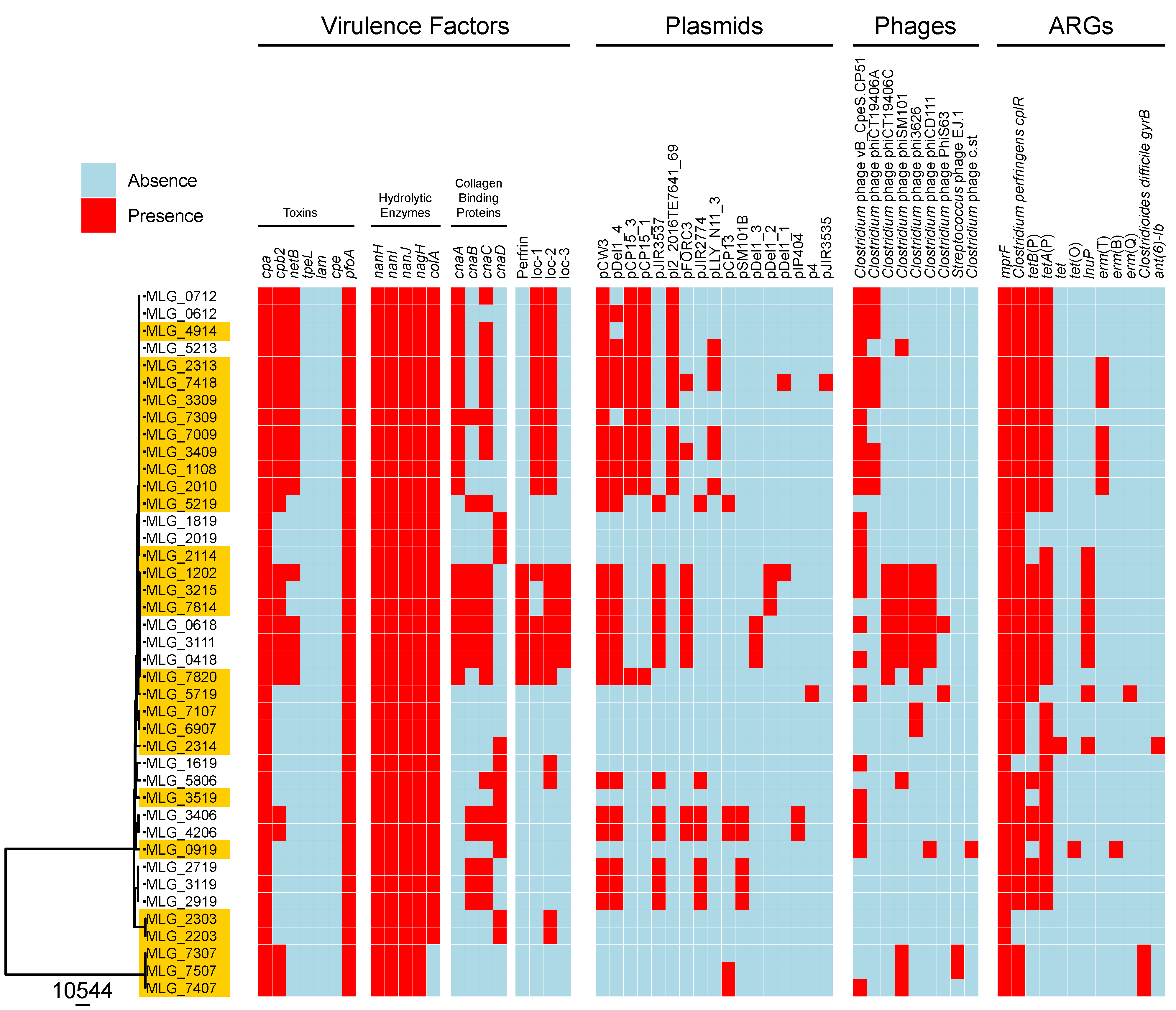
| Toxin | Enzyme | Enzyme | Enzyme | Enzyme | Toxin | Enzyme | Collagen binding protein | Collagen binding protein | Collagen binding protein | Collagen binding protein | Bacteriocin | Toxin | Toxin | NE loci | NE loci | NE loci | Toxin | Toxin | Toxin | |
| Isolate origin | cpa | nanH | nanI | nanJ | nagH | pfoA | colA | cnaA | cnaB | cnaC | cnaD | perfrin | netB | cpb2 |
NE Loc-1 |
NE Loc-2 |
NE Loc-3 |
tpeL | cpe | lam |
| NE-affected (n = 26) | 100% | 100% | 100% | 100% | 100% | 100% | 88% | 50% | 19% | 46% | 23% | 15% | 42% | 65% | 42% | 57% | 11% | 0% | 0% | 0% |
| Healthy (n = 15) | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 40% | 53% | 73% | 40% | 20% | 40% | 53% | 40% | 53% | 20% | 0% | 0% | 0% |
| Total (n = 41) | 100% | 100% | 100% | 100% | 100% | 100% | 92% | 46% | 31% | 56% | 29% | 17% | 41% | 60% | 41% | 56% | 14% | 0% | 0% | 0% |
| ARGs | Description | Occurrence | ||
|---|---|---|---|---|
| NE-affected isolates | Healthy isolates | Total | ||
| mprF | Resistance to cationic peptides | 100% | 100% | 100% |
| C. perfringens cplR | Resistance to pleuromutilin, lincosamide and streptogramin A | 92% | 93% | 92% |
| tetA(P) | Resistance to tetracycline antibiotic | 76% | 86% | 80% |
| tetB(P) | Resistance to tetracycline antibiotic | 57% | 80% | 65% |
| tet | Resistance to tetracycline antibiotic | 3% | 0% | 2% |
| tet(O) | Resistance to tetracycline antibiotic | 3% | 0% | 2% |
| erm(B) | Resistance to macrolide, lincosamide and streptogramin | 3% | 0% | 2% |
| erm(Q) | Resistance to macrolide, lincosamide and streptogramin | 3% | 0% | 2% |
| erm(T) | Resistance to macrolide, lincosamide and streptogramin | 27% | 0% | 17% |
| lnuP | Lincosamide nucleotidyltransferase (LNU) | 23% | 20% | 22% |
| C. difficile gyrB | Resistance to fluoroquinolones | 11% | 0% | 7% |
| ant(6)-Ib | Aminoglycoside nucleotidyltransferase | 3% | 0% | 2% |
| Prophage | Description | Occurrence | ||
|---|---|---|---|---|
| NE-affected isolates | Healthy isolates | Total | ||
| phiCP51 | Holin, amidase, endolysin, stage III sporulation protein D, and putative sporulation sigma factor production | 61% | 66% | 63% |
| phiCT19406A | Endolysin production | 27% | 13% | 22% |
| phiCT19406C | Endolysin production | 15% | 20% | 17% |
| phiSM101 | Endolysin production | 23% | 33% | 26% |
| phi3626 | Endolysin production, stage III sporulation protein D | 23% | 20% | 22% |
| phiCD111 | Endolysin production | 15% | 20% | 17% |
| PhiS63 | Endolysin; glycosyl-hydrolase production | 3% | 6% | 4% |
| Streptococcus phage EJ-1 | Endolysin production | 7% | 0% | 4% |
| Clostridium phage c-st | Endolysin production | 3% | 0% | 2% |
| Name and NCBI Reference Sequence | Size (bp) | Description | Occurrence | ||
|---|---|---|---|---|---|
| NE-affected isolates | Healthy isolates | Total | |||
| pCW3-like plasmid (NC_010937.1) |
47,263 |
tcp conjugation locus Tetracycline-resistance determinant Putative transposase |
53% | 80% | 63% |
| pDel1_4-like plasmid (NZ_CP019580.1) |
49,728 |
tcp conjugation locus Tetracycline-resistance determinant Putative transposase |
46% | 73% | 56% |
| pCP15_3-like plasmid (NZ_CP019570.1) |
3,843 | - | 38% | 20% | 31% |
| pCP15_1-like plasmid (NZ_CP019568.1) |
3,202 | - | 38% | 20% | 31% |
| pJIR3537-like plasmid (NZ_CP025504.1) |
48,779 | Tetracycline-resistance determinant tcp conjugation locus Putative transposase |
15% | 60% | 31% |
| pl2_2016TE7641_69-like plasmid (NZ_MN503253.1) |
82,669 |
tcp conjugation locus Tn3 family transposase Collagen binding domain-containing protein |
30% | 20% | 26% |
| pFORC3-like plasmid (NZ_CP009558.1) |
56,577 |
tcp conjugation locus Tetracycline-resistance determinant Putative transposase |
19% | 33% | 24% |
| pJIR2774-like plasmid (DQ338473.1) |
14,640 | tcp conjugation locus | 3% | 40% | 17% |
| pLLY_N11_3-like plasmid (NZ_CP023413.1) |
72,060 |
tcp conjugation locus Tetracycline-resistance determinant Putative transposase |
19% | 6% | 14% |
| pCP13-like plasmid (AP003515.1) |
54,310 | Probable collagen adhesin Probable transposase Resolvase |
11% | 13% | 12% |
| pSM101B-like plasmid (NC_008264.1) |
12,206 | BhlA/UviB family holin-like peptide | 0% | 33% | 12% |
| pDel1_3-like plasmid (NZ_CP019579.1) |
49,582 | Holin family protein IS630 transposase-related protein Chloramphenicol resistance protein |
0% | 20% | 7% |
| pDel1_2-like plasmid (NZ_CP019578.1) |
69,827 |
tcp conjugation locus Putative transposase |
11% | 0% | 7% |
| pDel1_1-like plasmid (NZ_CP019577.1) |
82,596 |
tcp conjugation locus Putative transposase Collagen binding domain-containing protein |
7% | 0% | 4% |
| pIP404-like plasmid (NC_001388.1) |
10,206 | Bacteriocin UviB | 0% | 13% | 4% |
| p4-like plasmid (NZ_MK275622.1) |
40,125 | IS1595 family transposase | 3% | 0% | 2% |
| pJIR3535-like plasmid (NZ_CP025502.1) |
81,826 |
tcp conjugation locus Tn3 family transposase Collagen binding domain-containing protein |
3% | 0% | 2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





