Submitted:
20 November 2024
Posted:
22 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Cold-Temperature Stress Treatment of Rice Plants
2.3. Assessment of Percent Low-Temperature Seedling Survivability (LTSS) in Rice
2.4. Assessment of Freezing Stress Tolerance in Arabidopsis
2.5. Gene Expression Analysis
2.6. Measurement of Percent Electrolyte Leakage (EL)
2.7. Measurement of Sugar Content in Two-Week-Old Rice and Arabidopsis Seedlings
2.8. Measurement of Chlorophyll Content in Rice Flag Leaves
2.9. Measurement of Leaf Length, Leaf Width, and Yield-Related Phenotypes
2.11. Yeast Two-Hybrid (Y2H) Analyses
2.10. Assessment of Salt Stress Tolerance in Arabidopsis
3. Results
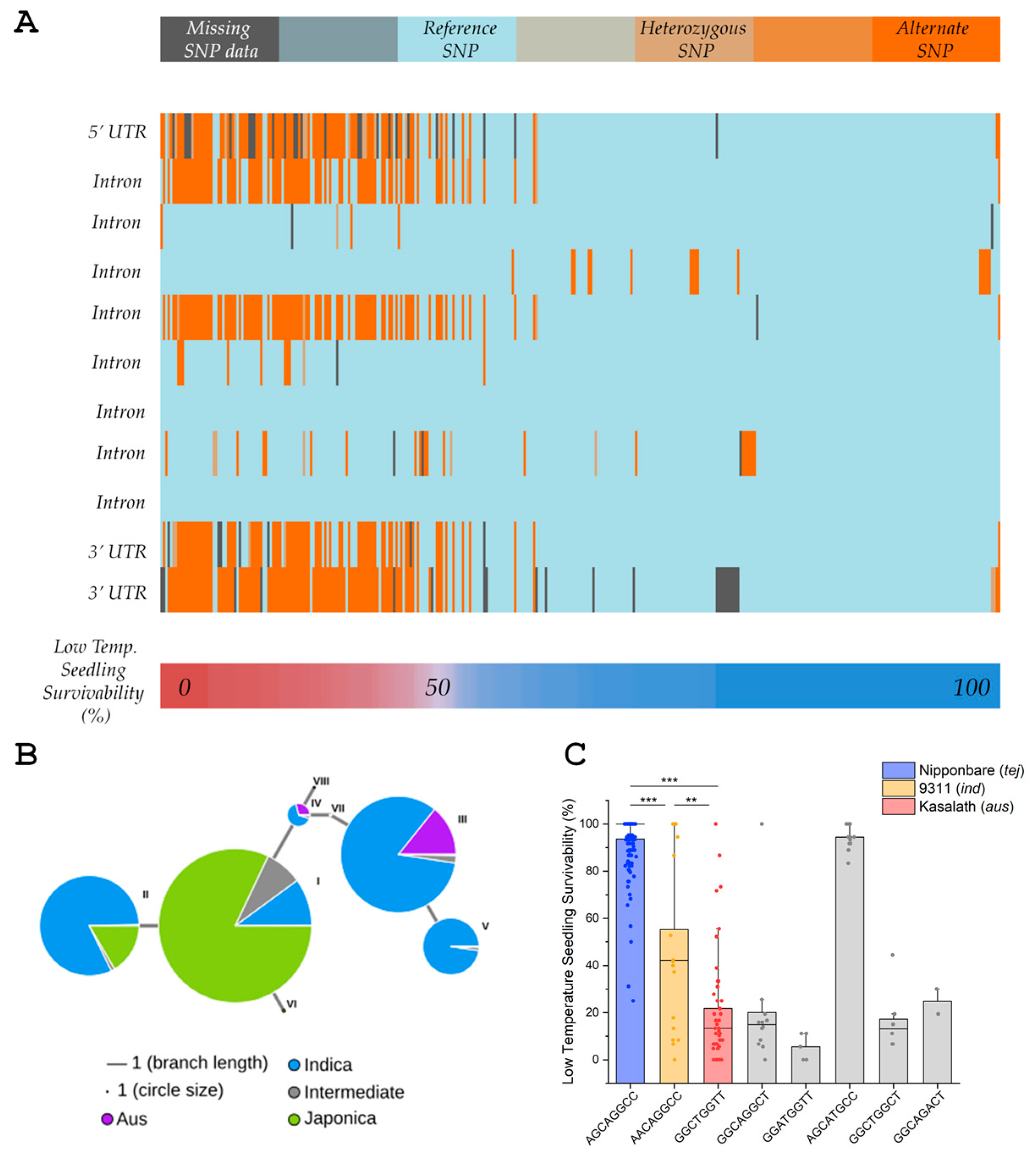
3.1. Identification of LOC_Os07g07240 as a Chilling Tolerance Candidate Gene Associated with qMT7-3
3.2. OsUBC7 Is Upregulated by low Temperature Exposure
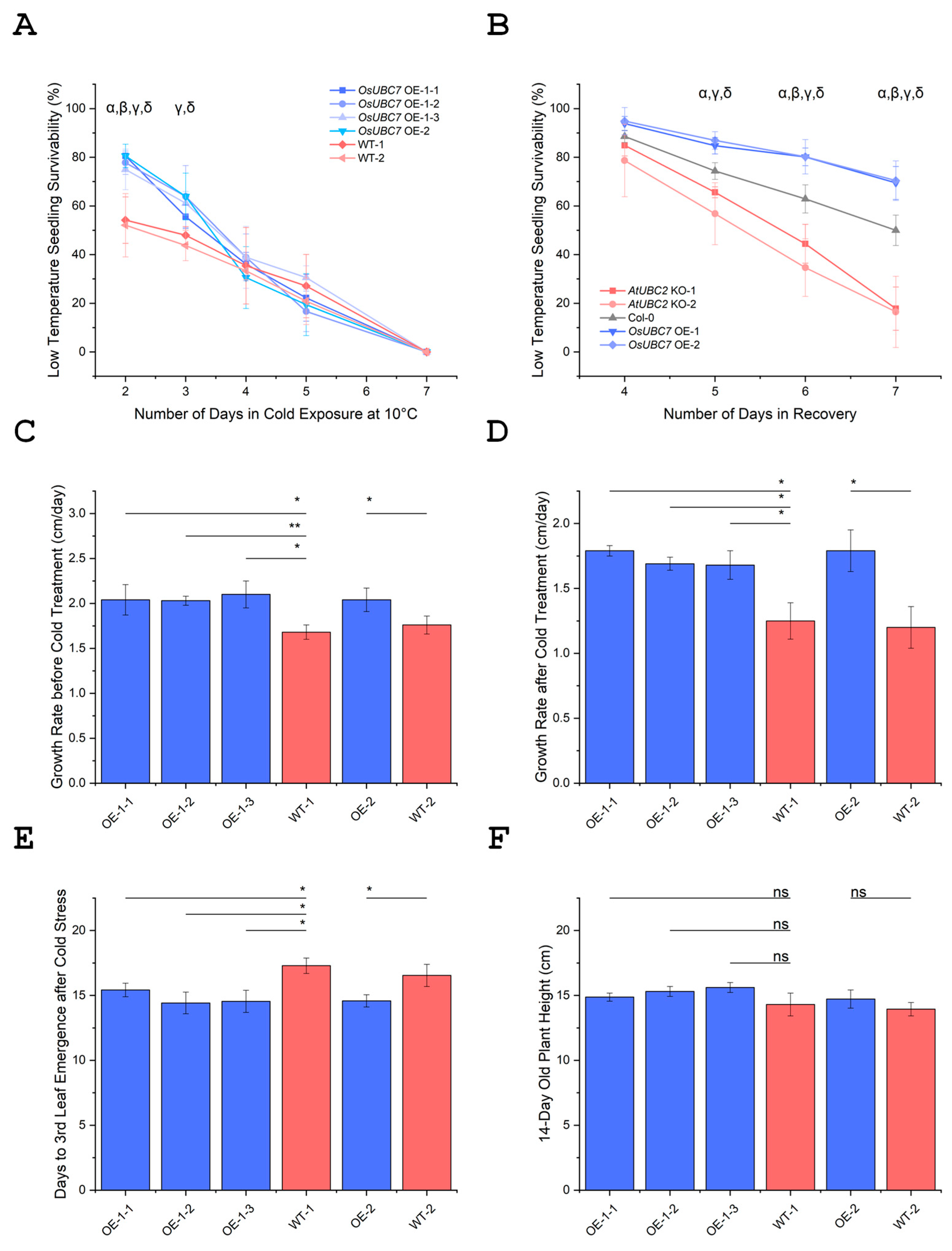
3.3. Overexpression of OsUBC7 Enhances LTSS Scores of Transgenic Rice Lines
3.4. Overexpression of OsUBC7 Increases Freezing Tolerance of Transgenic Arabidopsis Lines
3.5. OsUBC7 OE Promotes Plumule Recovery Growth and Enhances Formation of the Third Leaf After Low Temperature Exposure
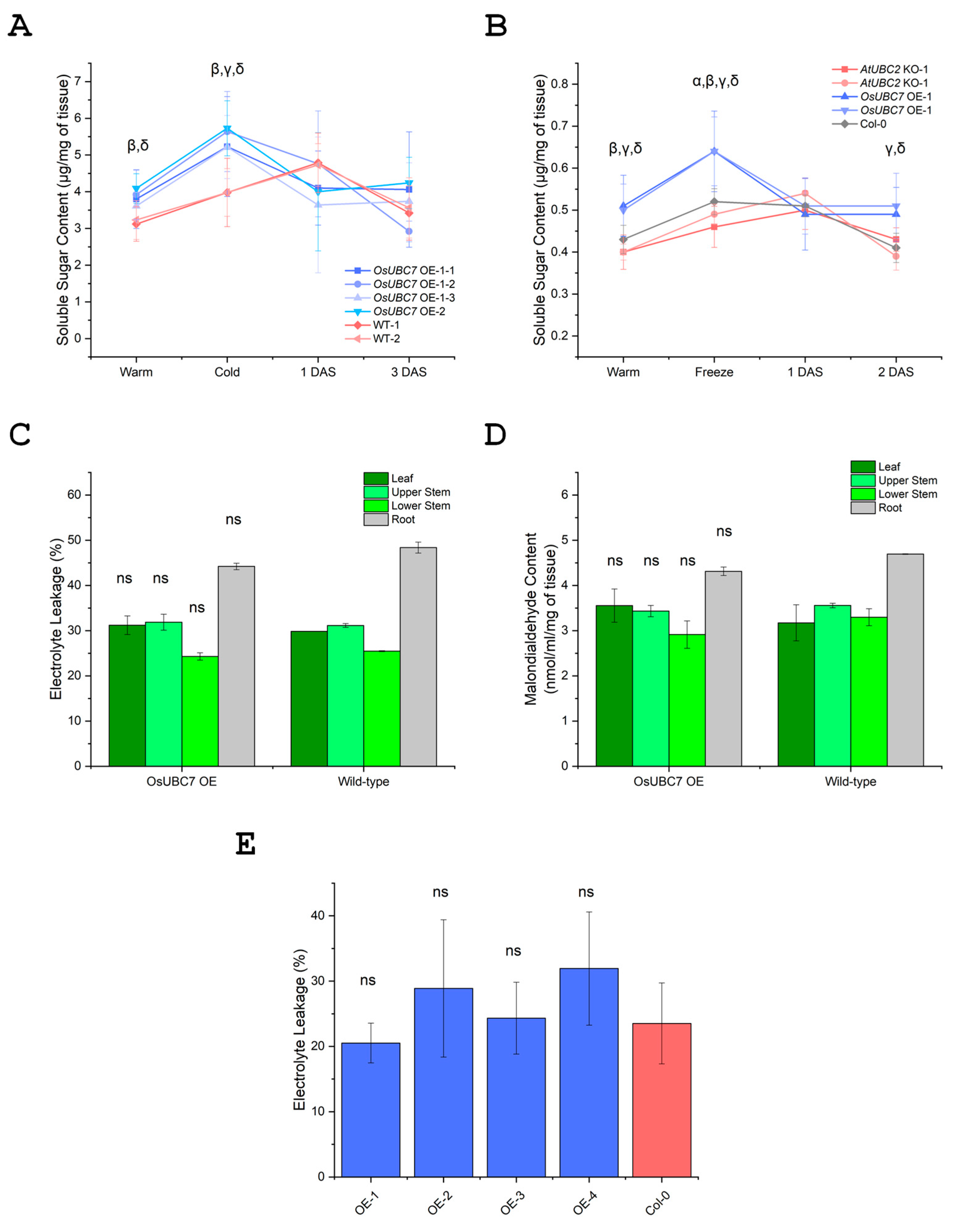
3.6. Overexpression of OsUBC7 in Rice and Arabidopsis Increases Soluble Sugar Content Under Warm, Low-Temperature Exposure, and Recovery Conditions
3.7. Overexpression of OsUBC7 Does Not Improve Membrane Damage After Low Temperature Exposure
3.8. Overexpression of OsUBC7 Does Not Result in Lower Lipid Peroxidation Levels After Low Temperature Exposure
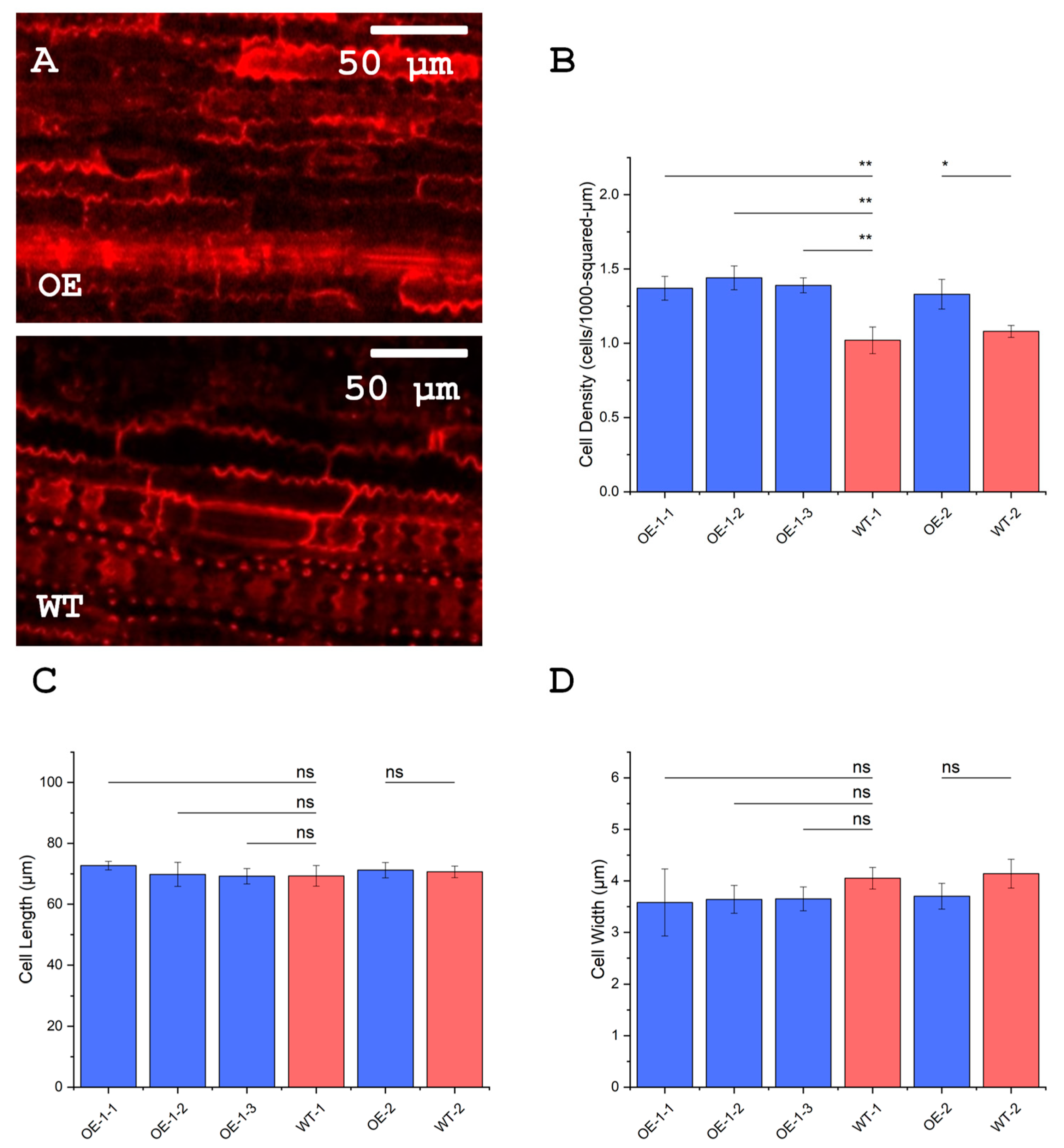
3.9. Leaves of OsUBC7 Overexpression Lines Have a Higher Cell Density Than Wild-Type Plants
3.10. Compared to Other RAD6 Paralogs OsUBC7 Is Highly Expressed at the Flowering Stage
3.11. Overexpression of OsUBC7 Tends to Improve Yield-Related Traits in Rice
3.12. Overexpression of OsUBC7 Does Not Alter Flowering Time
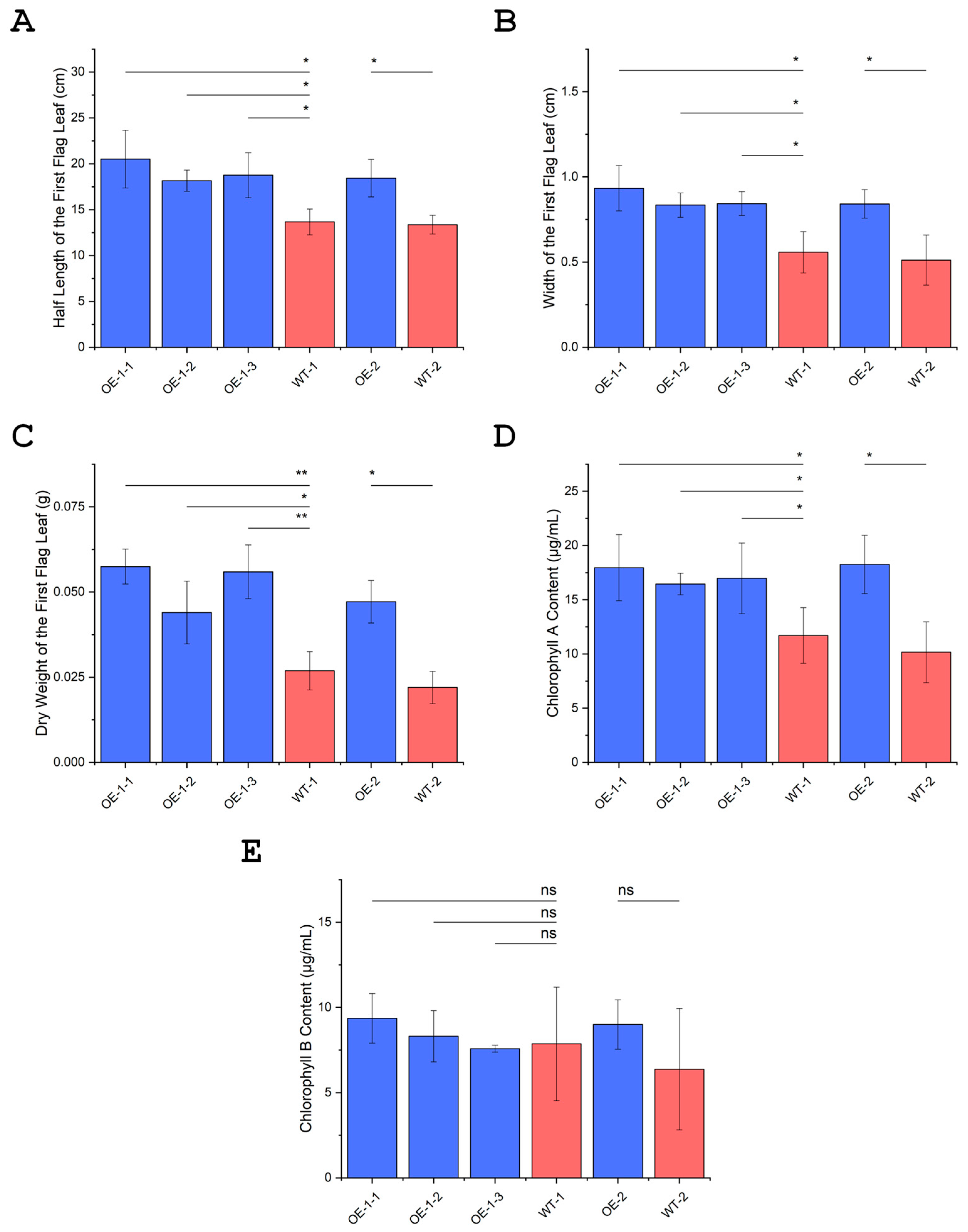
3.13. Overexpression of OsUBC7 Increases Flag Leaf Length, Width, and Dry Weight
3.14. Overexpression of OsUBC7 Leads to an Increase of Chlorophyll Content in Flag Leaves
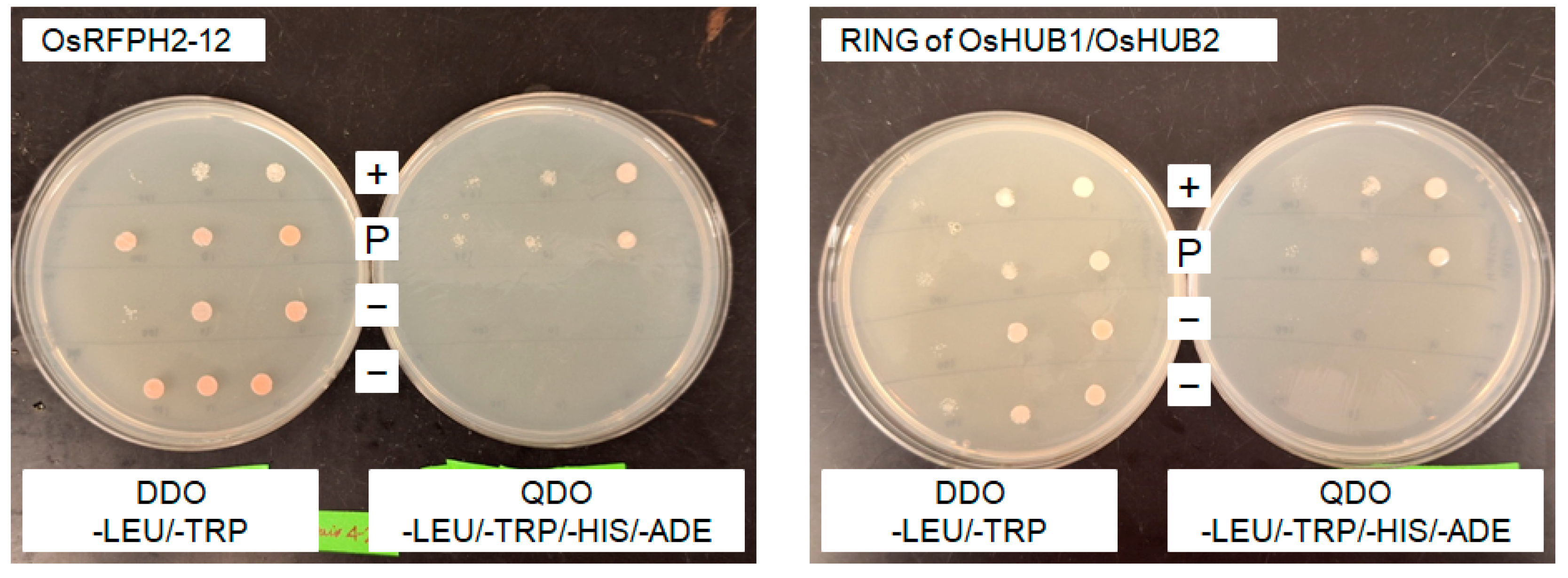
3.15. OsUBC7 Interacts with the E3 Protein OsRFPH2-12 and the RING Finger Domain of the Two E3 Proteins OsHUB1 and OsHUB2
3.16. Overexpression of OsUBC7 Tends to Increase Germination Rates of Arabidopsis Seeds Under High Salinity Stress
4. Discussion
4.1. OsUBC7 May Mediate Cold Stress Tolerance Responses Through Sugar Metabolism and Growth Regulation
4.2. Dynamic Expression of OsUBC7 in Response to Cold Stress and Flowering
4.3. A Role for OsUBC7 in Promoting Rice Cell Proliferation
4.4. A Role for OsUBC7 in Yield Enhancement and Flowering Regulation of Rice
4.5. Interaction of OsUBC7 with E3 Type RING Finger Domain and Potential Target Proteins
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Schläppi, M. R.; Jessel, A. R.; Jackson, A. K.; Phan, H.; Jia, M. H.; Edwards, J. D.; Eizenga, G. C. Navigating Rice Seedling Cold Resilience: QTL Mapping in Two Inbred Line Populations and the Search for Genes. Frontiers in Plant Science 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, N.; Johnson, M.; Beaumont, A.; Schläppi, M. Multiple Cold Tolerance Trait Phenotyping Reveals Shared Quantitative Trait Loci in Oryza Sativa. Rice 2020, 13. [Google Scholar] [CrossRef] [PubMed]
- Dolferus, R. To Grow or Not to Grow: A Stressful Decision for Plants. Plant Science 2014, 229, 247–261. [Google Scholar] [CrossRef] [PubMed]
- Nasim Maghboli Balasjin; Maki, J. S.; Schläppi, M. R. Pseudomonas Mosselii Improves Cold Tolerance of Asian Rice (Oryza Sativa L.) in a Genotype-Dependent Manner by Increasing Proline in Japonica and Reduced Glutathione in Indica Varieties. Canadian Journal of Microbiology 2023, 70, 15–31. [Google Scholar] [CrossRef]
- Li, J.; Zhang, Z.; Chong, K.; Xu, Y. Chilling Tolerance in Rice: Past and Present. Journal of Plant Physiology 2022, 268, 153576. [Google Scholar] [CrossRef]
- Gazquez, A.; Abdelgawad, H.; Baggerman, G.; Van Raemdonck, G.; Asard, H.; Maiale, S. J.; Rodríguez, A. A.; Beemster, G. T. S. Redox Homeostasis in the Growth Zone of the Rice Leaf Plays a Key Role in Cold Tolerance. J. Exp. Bot. 2020, 71(3), 1053–1066. [Google Scholar] [CrossRef]
- Nie, L.; Liu, H.; Zhang, L.; Wang, W. Enhancement in Rice Seed Germination via Improved Respiratory Metabolism under Chilling Stress. Food and Energy Security 2020, 9. [Google Scholar] [CrossRef]
- Arshad, M. S.; Farooq, M.; Asch, F.; Krishna, J. S. V.; Prasad, P. V. V.; Siddique, K. H. M. Thermal Stress Impacts Reproductive Development and Grain Yield in Rice. Plant Physiology and Biochemistry 2017, 115, 57–72. [Google Scholar] [CrossRef]
- Shi, Y.; Phan, H.; Liu, Y.; Cao, S.; Zhang, Z.; Chu, C.; Schläppi, M. R. Glycosyltransferase OsUGT90A1 Helps Protect the Plasma Membrane during Chilling Stress in Rice. J. Exp. Bot. 2020, 71(9), 2723–2739. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, X.; Gao, G.; Ali, I.; Wu, X.; Tang, M.; Chen, L.; Jiang, L.; Liang, T. Effects of Various Seed Priming on Morphological, Physiological, and Biochemical Traits of Rice under Chilling Stress. 2023, 14. [CrossRef]
- Schläppi, M. R.; Jackson, A. K.; Eizenga, G. C.; Wang, A.; Chu, C.; Shi, Y.; Shimoyama, N.; Boykin, D. L. Assessment of Five Chilling Tolerance Traits and GWAS Mapping in Rice Using the USDA Mini-Core Collection. Frontiers in Plant Science 2017, 8. [Google Scholar] [CrossRef]
- Phan, H.; Schläppi, M. Low Temperature Antioxidant Activity QTL Associate with Genomic Regions Involved in Physiological Cold Stress Tolerance Responses in Rice (Oryza Sativa L.). Genes 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Cai, F.; Chen, P.; Chen, L.; Biskup, E.; Liu, Y.; Chen, P.-C.; Chang, J.-F.; Jiang, W.; Jing, Y.; Chen, Y.; Jin, H.; Chen, S. Human RAD6 Promotes G1-S Transition and Cell Proliferation through Upregulation of Cyclin D1 Expression. PLoS ONE 2014, 9. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J. P.; Kanamori, H.; McCombie, W. R.; Ouyang, S.; Schwartz, D. C.; Tanaka, T.; Wu, J.; Zhou, S.; Childs, K. L.; Davidson, R. M.; Lin, H.; Quesada-Ocampo, L.; Vaillancourt, B.; Sakai, H.; Lee, S. S.; Kim, J.; Numa, H.; Itoh, T. Improvement of the Oryza Sativa Nipponbare Reference Genome Using next Generation Sequence and Optical Map Data. Rice 2013, 6. [Google Scholar] [CrossRef]
- Sato, Y. S. ; Hinako Takehisa; Kaori Kamatsuki; Minami, H. ; Nobukazu Namiki; Hiroshi Ikawa; Hajime Ohyanagi; Sugimoto, K.; Antonio, B. A.; Yoshiaki Nagamura. RiceXPro Version 3.0: Expanding the Informatics Resource for Rice Transcriptome. 2012, 41, D1206–D1213. [Google Scholar] [CrossRef]
- Game, J. C.; Williamson, M. S.; Spicakova, T.; Brown, J. M. The RAD6/BRE1 Histone Modification Pathway in Saccharomyces Confers Radiation Resistance through a RAD51-Dependent Process That Is Independent of RAD18. Genetics 2006, 173, 1951–1968. [Google Scholar] [CrossRef]
- Lyakhovich, A.; Shekhar, M. P. V. RAD6B Overexpression Confers Chemoresistance: RAD6 Expression during Cell Cycle and Its Redistribution to Chromatin during DNA Damage-Induced Response. Oncogene 2004, 23, 3097–3106. [Google Scholar] [CrossRef]
- Madura, K.; Prakash, S.; Prakash, L. Expression of theSaccharomyces CerevisiaeDNA Repair GeneRAD6That Encodes a Ubiquitin Conjugating Enzyme, Increases in Response to DNA Damage and in Meiosis but Remains Constant during the Mitotic Cell Cycle. Nucleic Acids Research 1990, 18, 771–778. [Google Scholar] [CrossRef]
- Chao, H.; Zhang, S.; Hu, Y.; Ni, Q.; Xin, S.; Zhao, L.; Ivanisenko, V. A.; Orlov, Y. L.; Chen, M. Integrating Omics Databases for Enhanced Crop Breeding. Journal of integrative bioinformatics 2023, 20, 20230012. [Google Scholar] [CrossRef]
- Kerepesi, I.; Tóth, M.; Boross, L. Water-Soluble Carbohydrates in Dried Plant. Journal of Agricultural and Food Chemistry 1996, 44, 3235–3239. [Google Scholar] [CrossRef]
- Leach, K. A.; Braun, D. M. Soluble Sugar and Starch Extraction and Quantification from Maize (Zea Mays) Leaves. Current Protocols in Plant Biology 2016, 1, 139–161. [Google Scholar] [CrossRef]
- Ritchie, R. J. Consistent Sets of Spectrophotometric Chlorophyll Equations for Acetone, Methanol and Ethanol Solvents. Photosynthesis Research 2006, 89, 27–41. [Google Scholar] [CrossRef]
- Xu, D.; Huang, X.; Xu, Z.-Q.; Schläppi, M. The HyPRP Gene EARLI1 Has an Auxiliary Role for Germinability and Early Seedling Development under Low Temperature and Salt Stress Conditions in Arabidopsis Thaliana. Planta 2011, 234, 565–577. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xin, W.; Wang, Y.; Zhang, D.; Wang, J.; Zheng, H.; Yang, L.; Nie, S.; Zou, D. An Integrated Analysis of the Rice Transcriptome and Lipidome Reveals Lipid Metabolism Plays a Central Role in Rice Cold Tolerance. BMC Plant Biology 2022, 22. [Google Scholar] [CrossRef] [PubMed]
- Hajihashemi, S.; Noedoost, F.; Geuns, J. M. C.; Djalovic, I.; Siddique, K. H. M. Effect of Cold Stress on Photosynthetic Traits, Carbohydrates, Morphology, and Anatomy in Nine Cultivars of Stevia Rebaudiana. Frontiers in Plant Science 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Gammulla, C. G.; Pascovici, D.; Atwell, B. J.; Haynes, P. A. Differential Proteomic Response of Rice (Oryza Sativa ) Leaves Exposed to High- and Low-Temperature Stress. PROTEOMICS 2011, 11, 2839–2850. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, C.; He, Y.; Tian, Z.; Li, J. Rice OVATE Family Protein 6 Regulates Plant Development and Confers Resistance to Drought and Cold Stresses. Journal of Experimental Botany 2017, 68, 4885–4898. [Google Scholar] [CrossRef]
- Yoon, D. H.; Lee, S. S.; Park, H. J.; Lyu, J. I.; Chong, W. S.; Liu, J. R.; Kim, B.-G.; Ahn, J. C.; Cho, H. S. Overexpression ofOsCYP19-4Increases Tolerance to Cold Stress and Enhances Grain Yield in Rice (Oryza Sativa). Journal of Experimental Botany 2015, 67, 69–82. [Google Scholar] [CrossRef]
- Chen, N.; Xu, Y.; Wang, X.; Du, C.; Du, J.; Yuan, M.; Xu, Z.; Chong, K. OsRAN2, Essential for Mitosis, Enhances Cold Tolerance in Rice by Promoting Export of Intranuclear Tubulin and Maintaining Cell Division under Cold Stress. Plant, Cell & Environment 2010, 34, 52–64. [Google Scholar] [CrossRef]
- Ma, Q.; Dai, X.; Xu, Y.; Guo, J.; Liu, Y.; Chen, N.; Xiao, J.; Zhang, D.; Xu, Z.-H.; Zhang, X.; Chong, K. Enhanced Tolerance to Chilling Stress in OsMYB3R-2 Transgenic Rice Is Mediated by Alteration in Cell Cycle and Ectopic Expression of Stress Genes. Plant Physiology 2009, 150, 244–256. [Google Scholar] [CrossRef]
- De Freitas, G. M.; Thomas, J.; Liyanage, R.; Lay, J. O.; Basu, S.; Ramegowda, V.; do Amaral, M. N.; Benitez, L. C.; Bolacel Braga, E. J.; Pereira, A. Cold Tolerance Response Mechanisms Revealed through Comparative Analysis of Gene and Protein Expression in Multiple Rice Genotypes. PLOS ONE 2019, 14, e0218019. [Google Scholar] [CrossRef]
- Zhang, Q.; Chen, Q.; Wang, S.; Hong, Y.; Wang, Z. Rice and Cold Stress: Methods for Its Evaluation and Summary of Cold Tolerance-Related Quantitative Trait Loci. Rice 2014, 7. [Google Scholar] [CrossRef] [PubMed]
- Singh, A. K.; Dhanapal, S.; Yadav, B. S. The Dynamic Responses of Plant Physiology and Metabolism during Environmental Stress Progression. Molecular Biology Reports 2019, 47, 1459–1470. [Google Scholar] [CrossRef] [PubMed]
- Furtauer, L.; Weiszmann, J.; Weckwerth, W.; Nägele, T. Dynamics of Plant Metabolism during Cold Acclimation. International Journal of Molecular Sciences 2019, 20, 5411. [Google Scholar] [CrossRef]
- Yang, M.; Yang, J.; Su, L.; Sun, K.; Li, D.; Liu, Y.; Wang, H.; Chen, Z.; Guo, T. Metabolic Profile Analysis and Identification of Key Metabolites during Rice Seed Germination under Low-Temperature Stress. Plant Science 2019, 289, 110282. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Meguro-maoka, A.; Yoshida, M. Analysis of Sugar Content and Expression of Sucrose Transporter Genes (OsSUTs) in Rice Tissues in Response to a Chilling Temperature. Japan Agricultural Research Quarterly: JARQ 2017, 51, 137–146. [Google Scholar] [CrossRef]
- Tian, Y.; Zhang, H.; Pan, X.; Chen, X.; Zhang, Z.; Lu, X.; Huang, R. Overexpression of Ethylene Response Factor TERF2 Confers Cold Tolerance in Rice Seedlings. Transgenic Research 2010, 20, 857–866. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, Y.; Xiong, L. Characterization of Stress-Responsive CIPK Genes in Rice for Stress Tolerance Improvement. Plant Physiol. 2007, 144(3), 1416–1428. [Google Scholar] [CrossRef]
- Liu, C.; Schläppi, M.; Mao, B.; Wang, W.; Wang, A.; Chu, C. The BZIP73 Transcription Factor Controls Rice Cold Tolerance at the Reproductive Stage. Plant Biotechnology Journal 2019, 17, 1834–1849. [Google Scholar] [CrossRef]
- Xu, L.; Rozenn Ménard; Berr, A.; Fuchs, J.; Valérie Cognat; Meyer, D.; Shen, W.-H. The E2 Ubiquitin-Conjugating Enzymes, AtUBC1 and AtUBC2, Play Redundant Roles and Are Involved in Activation ofFLCExpression and Repression of Flowering inArabidopsis Thaliana. Plant Journal 2009, 57, 279–288. [Google Scholar] [CrossRef]
- Sun, Y.; Zhao, J.; Li, X.; Li, Y. E2 Conjugases UBC1 and UBC2 Regulate MYB42-Mediated SOS Pathway in Response to Salt Stress in Arabidopsis. New Phytologist 2020, 227, 455–472. [Google Scholar] [CrossRef]
- Chen, K.; Tang, W.-S.; Zhou, Y.-B.; Xu, Z.-S.; Chen, J.; Ma, Y.-Z.; Chen, M.; Li, H.-Y. Overexpression of GmUBC9 Gene Enhances Plant Drought Resistance and Affects Flowering Time via Histone H2B Monoubiquitination. Frontiers in Plant Science 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- E, Z.; Zhang, Y.; Li, T.; Wang, L.; Zhao, H. Characterization of the Ubiquitin-Conjugating Enzyme Gene Family in Rice and Evaluation of Expression Profiles under Abiotic Stresses and Hormone Treatments. PLOS ONE 2015, 10, e0122621. [Google Scholar] [CrossRef]
- Bae, H.; Kim, W. T. Classification and Interaction Modes of 40 Rice E2 Ubiquitin-Conjugating Enzymes with 17 Rice ARM-U-Box E3 Ubiquitin Ligases. Biochemical and Biophysical Research Communications 2014, 444, 575–580. [Google Scholar] [CrossRef] [PubMed]
- Ying, Y.; Pang, Y.; Bao, J. Comparative Ubiquitome Analysis Reveals Diverse Functions of Ubiquitination in Rice Seed Development under High-Temperature Stress. Seed Biology 2023, 2. [Google Scholar] [CrossRef]
- Zhu, L.; Cheng, H.; Peng, G.; Wang, S.; Zhang, Z.; Ni, E.; Fu, X.; Zhuang, C.; Liu, Z.; Zhou, H. Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles. Genomics Proteomics & Bioinformatics 2020, 18, 305–320. [Google Scholar] [CrossRef]
- Melo, F. V.; Oliveira, M. M.; Saibo, N. J. M.; Lourenço, T. F. Modulation of Abiotic Stress Responses in Rice by E3-Ubiquitin Ligases: A Promising Way to Develop Stress-Tolerant Crops. Frontiers in Plant Science 2021, 12. [Google Scholar] [CrossRef]
- Dametto, A.; Buffon, G.; Dos Reis Blasi, É. A.; Sperotto, R. A. Ubiquitination Pathway as a Target to Develop Abiotic Stress Tolerance in Rice. Plant Signaling & Behavior 2015, 10, e1057369. [Google Scholar] [CrossRef]
- Byun, M. Y.; Cui, L. H.; Oh, T. K.; Jung, Y.-J.; Lee, A.; Park, K. Y.; Kang, B. G.; Kim, W. T. Homologous U-Box E3 Ubiquitin Ligases OsPUB2 and OsPUB3 Are Involved in the Positive Regulation of Low Temperature Stress Response in Rice (Oryza Sativa L.). Frontiers in Plant Science 2017, 8. [Google Scholar] [CrossRef]
- Cui, L. H.; Min, H. J.; Byun, M. Y.; Oh, H. G.; Kim, W. T. OsDIRP1, a Putative RING E3 Ligase, Plays an Opposite Role in Drought and Cold Stress Responses as a Negative and Positive Factor, Respectively, in Rice (Oryza Sativa L.). Frontiers in Plant Science 2018, 9. [Google Scholar] [CrossRef]
- Cui, L. H.; Min, H. J.; Yu, S. G.; Byun, M. Y.; Oh, T. R.; Lee, A.; Yang, H. W.; Kim, W. T. OsATL38 Mediates Mono-Ubiquitination of the 14-3-3 Protein OsGF14d and Negatively Regulates the Cold Stress Response in Rice. Journal of Experimental Botany 2021, 73, 307–323. [Google Scholar] [CrossRef]
- Fang, H.; Meng, Q.; Xu, J.; Tang, H.; Tang, S.; Zhang, H.; Huang, J. Knock-down of Stress Inducible OsSRFP1 Encoding an E3 Ubiquitin Ligase with Transcriptional Activation Activity Confers Abiotic Stress Tolerance through Enhancing Antioxidant Protection in Rice. Plant Molecular Biology 2015, 87, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Sato, Y. S. ; Hinako Takehisa; Kaori Kamatsuki; Minami, H. ; Nobukazu Namiki; Hiroshi Ikawa; Hajime Ohyanagi; Sugimoto, K.; Antonio, B. A.; Yoshiaki Nagamura. RiceXPro Version 3.0: Expanding the Informatics Resource for Rice Transcriptome. 2012, 41, D1206–D1213. [Google Scholar] [CrossRef]
- Verkade, H.; Teli, T.; Laursen, L.; Murray, J.; O’Connell, M. A Homologue of the Rad18 Postreplication Repair Gene Is Required for DNA Damage Responses throughout the Fission Yeast Cell Cycle. Molecular Genetics and Genomics 2001, 265, 993–1003. [Google Scholar] [CrossRef] [PubMed]
- Bertoli, C.; Skotheim, J. M.; de Bruin, R. A. M. Control of Cell Cycle Transcription during G1 and S Phases. Nature Reviews Molecular Cell Biology 2013, 14, 518–528. [Google Scholar] [CrossRef] [PubMed]
- Moeller, S. J.; Sheaff, R. J. G1 Phase: Components, Conundrums, Context. Results and Problems in Cell Differentiation 1–29. [CrossRef]
- Mansueto, L.; Fuentes, R. R.; Borja, F. N.; Detras, J.; Abriol-Santos, J. M.; Chebotarov, D.; Sanciangco, M.; Palis, K.; Copetti, D.; Poliakov, A.; Dubchak, I.; Solovyev, V.; Wing, R. A.; Hamilton, R. S.; Mauleon, R.; McNally, K. L.; Alexandrov, N. Rice SNP-Seek Database Update: New SNPs, Indels, and Queries. Nucleic Acids Research 2016, 45, D1075–D1081. [Google Scholar] [CrossRef]
- Yonemaru, J.; Yamamoto, T.; Fukuoka, S.; Uga, Y.; Hori, K.; Yano, M. Q-TARO: QTL Annotation Rice Online Database. Rice 2010, 3, 194–203. [Google Scholar] [CrossRef]
- Gu, X.; Jiang, D.; Wang, Y.; Bachmair, A.; He, Y. Repression of the Floral Transition via Histone H2B Monoubiquitination. The Plant Journal 2009, 57, 522–533. [Google Scholar] [CrossRef]
- Cao, Y.; Dai, Y.; Cui, S.; Ma, L. Histone H2B Monoubiquitination in the Chromatin of FLOWERING LOCUS C Regulates Flowering Time in Arabidopsis. Plant Cell 2008, 20(10), 2586–2602. [Google Scholar] [CrossRef]
- Alemu, S. T. Photosynthesis Limiting Stresses under Climate Change Scenarios and Role of Chlorophyll Fluorescence: A Review Article. Cogent Food & Agriculture 2020, 6, 1785136. [Google Scholar] [CrossRef]
- Moradi, F.; Ismail, A. M. Responses of Photosynthesis, Chlorophyll Fluorescence and ROS-Scavenging Systems to Salt Stress during Seedling and Reproductive Stages in Rice. Annals of Botany 2007, 99, 1161–1173. [Google Scholar] [CrossRef]
- Tanaka, A.; Tanaka, R. Chlorophyll Metabolism. Current Opinion in Plant Biology 2006, 9, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Qu, M.; Zheng, G.; Hamdani, S.; Jemâa Essemine; Song, Q.; Wang, H.; Chu, C.; Sirault, X.; Zhu, X. Leaf Photosynthetic Parameters Related to Biomass Accumulation in a Global Rice Diversity Survey. Plant Physiology 2017, 175, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Tang, N.; Li, X.; Xie, Y.; Xiang, D.; Fu, J.; Shen, J.; Yang, J.; Tu, H.; Li, X.; Hu, H.; Xiong, L. Reversible Histone H2B Monoubiquitination Fine-Tunes Abscisic Acid Signaling and Drought Response in Rice. 2019, 12 (2), 263–277. [CrossRef]
- Zhou, W.; Zhu, Y.; Dong, A.; Shen, W.-H. Histone H2A/H2B Chaperones: From Molecules to Chromatin-Based Functions in Plant Growth and Development. The Plant Journal 2015, 83, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Li, X.; Wang, Z.; Ding, M.; Sun, Y.; Dong, F.; Chen, F.; Liu, L.; Doughty, J.; Li, Y.; Yong Xiu Liu. Histone H2B Monoubiquitination Mediated by HISTONE MONOUBIQUITINATION1 and HISTONE MONOUBIQUITINATION2 Is Involved in Anther Development by Regulating Tapetum Degradation-Related Genes in Rice. Plant Physiology 2015, 168, 1389–1405. [Google Scholar] [CrossRef]
- Min, H. J.; Jung, Y. J.; Kang, B. G.; Kim, W. T. CaPUB1, a Hot Pepper U-Box E3 Ubiquitin Ligase, Confers Enhanced Cold Stress Tolerance and Decreased Drought Stress Tolerance in Transgenic Rice (Oryza Sativa L.). Molecules and Cells 2015, 39, 250–257. [Google Scholar] [CrossRef]
- Lourenço, T.; Sapeta, H.; Figueiredo, D. D.; Rodrigues, M.; Cordeiro, A.; Abreu, I. A.; Saibo, N. J. M.; Oliveira, M. M. Isolation and Characterization of Rice (Oryza Sativa L.) E3-Ubiquitin Ligase OsHOS1 Gene in the Modulation of Cold Stress Response. Plant molecular biology 2013, 83, 351–363. [Google Scholar] [CrossRef]
- Lin, W.; Wu, S.; Wei, M. Ubiquitylome Analysis Reveals the Involvement of Ubiquitination in the Cold Responses of Banana Seedling Leaves. Journal of Proteomics 2023, 288, 104994. [Google Scholar] [CrossRef]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR Signaling Cascade and Its Regulation in Plants Responding to Cold Stress. International Journal of Molecular Sciences 2022, 23, 1549. [Google Scholar] [CrossRef]
- Guan, Y.; Delight Hwarari; Harriet Mateko Korboe; Ahmad, B.; Cao, Y.; Movahedi, A.; Yang, L. Low Temperature Stress-Induced Perception and Molecular Signaling Pathways in Plants. Environmental and Experimental Botany 2023, 207, 105190–105190. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Network of Plant Cold-Stress Responses. Trends in Plant Science 2022. [Google Scholar] [CrossRef]
- Sung Don Lim; Hwang, J.-G.; Chang Gyo Jung; Hwang, S.-G.; Moon, J.-C.; Cheol Seong Jang. Comprehensive Analysis of the Rice RING E3 Ligase Family Reveals Their Functional Diversity in Response to Abiotic Stress. DNA research 2013, 20, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Pineiro, M.; Jarillo, J. A. Ubiquitination in the Control of Photoperiodic Flowering. Plant Science 2013, 198, 98–109. [Google Scholar] [CrossRef] [PubMed]
- Hellmann, H.; Estelle, M. Plant Development: Regulation by Protein Degradation. Science 2002, 297, 793–797. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. F-Box Proteins in Flowering Plants. Chinese Science Bulletin 2002, 47, 1497. [Google Scholar] [CrossRef]
- Shi, Y.; Guo, E.; Wang, L.; Li, T.; Jiang, S.; Xiang, H.; Zhang, T.; Cheng, X.; Zhu, X.; Zhou, L.; Ma, H.; Yang, X. Effects of Chilling at the Booting and Flowering Stages on Rice Phenology and Yield: A Case Study in Northeast China. Journal of Agronomy and Crop Science 2021, 208, 197–208. [Google Scholar] [CrossRef]
- Xu, L.; Rozenn Ménard; Berr, A.; Fuchs, J.; Valérie Cognat; Meyer, D.; Shen, W.-H. The E2 Ubiquitin-Conjugating Enzymes, AtUBC1 and AtUBC2, Play Redundant Roles and Are Involved in Activation of FLC Expression and Repression of Flowering inArabidopsis Thaliana. Plant Journal 2009, 57, 279–288. [Google Scholar] [CrossRef]
- Zhao, H.; Yao, W.; Ouyang, Y.; Yang, W.; Wang, G.; Lian, X.; et al. RiceVarMap: a comprehensive database of rice genomic variations. Nucl. Acid Res. 2015, 43, D1018–D1022. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.; Tian, X.; Lin, X.; Han, Y.; Han, Z.; Sha, H.; Liu, J.; Liu, J.; et al. A transposon insertion in the promoter of OsUBC12 enhances cold tolerance during japonica rice germination. Nature Comm. 2024, 15, 2211. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, C.; Sha, H.; Wang, X.; Yu, Y.; Liu, J.; Zhao, G.; Wang, J.; Qiu, G.; Xu, X.; Fang, J. Ubiquitin-Conjugating Enzyme OsUBC11 Affects the Development of Roots via Auxin Pathway. Rice 2023, 16, 9. [Google Scholar] [CrossRef]
- Liu, J.; Nie, B.; Yu, B.; Xu, F.; Zhang, Q.; Wang, Y.; Xu, W. Rice ubiquitin-conjugating enzyme OsUbc13 negatively regulates immunity against pathogens by enhancing the activity of OsSnRK1a. Plant Biotechnol. J. 2023, 21, 1590–1610. [Google Scholar] [CrossRef]
- Wang, Y.; Yue, J.; Yang, N.; Zheng, C.; Zheng, Y.; Wu, X.; Yang, J.; Zhang, L.; Liu, L.; Ning, Y.; Bhadauria, V.; et al. An ERAD-related ubiquitin-conjugating enzyme boosts broad-spectrum disease resistance and yield in rice. Nature Food 2023, 4, 774–787. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhang, S.; Luo, Y; Li, F.; Tan, J.; Wang, B.; Zhao, Z.; Lin, H.; Zhang, T.; Liu, J., Liu, X., Guo, J.; et al. Rice OsUBR7 modulates plant height by regulating histone H2B monoubiquitination and cell proliferation. Rice OsUBR7 modulates plant height by regulating histone H2B monoubiquitination and cell proliferation. Plant Commun. 2022, 3(6), 100412. [Google Scholar] [CrossRef]
| House-Keeping Gene | Hrs of Exposure | Normalized mRNA Abundance | Log2 Fold-Change | ||
| Kasalath | Krasnodars. | Kasalath | Krasnodars. | ||
| 18S ribosomal RNA | 0 | 2.09E-03 ± 1.20E-04 | 1.63E-03 ± 2.90E-04 | 0 | 0 |
| 12 | 3.13E-03 ± 4.06E-03 | 2.30E-03 ± 6.20E-04 | 0.585 | 0.496 | |
| 24 | 3.12E-03 ± 2.80E-04 | 3.86E-03 ± 1.57E-03 | 0.575 | 1.245 | |
| 36 | 4.11E-03 ± 1.25E-03 | 1.10E-02 ± 2.55E-03 | 0.978 | 2.751 | |
| 48 | 4.35E-03 ± 1.26E-03 | 7.51E-03 ± 2.56E-03 | 1.057 | 2.205 | |
| OsACT1 | 0 | 1.13E-01 ± 8.10E-02 | 7.70E-02 ± 1.30E-02 | 0 | 0 |
| 12 | 1.54E-01 ± 4.90E-02 | 1.58E-01 ± 3.60E-02 | 0.444 | 1.029 | |
| 24 | 2.16E-01 ± 9.20E-02 | 3.81E-01 ± 1.92E-01 | 0.941 | 2.305 | |
| 36 | 5.46E-01 ± 1.31E-01 | 6.05E-01 ± 4.67E-01 | 2.275 | 2.973 | |
| 48 | 6.26E-01 ± 2.92E-01 | 7.89E-01 ± 1.38E-01 | 2.470 | 3.356 | |
| OsUBC32 | 0 | 5.15E-02 ± 1.10E-02 | 3.00E-02 ± 1.07E-02 | 0 | 0 |
| 12 | 6.87E-02 ± 2.26E-02 | 5.20E-02 ± 1.48E-02 | 0.411 | 0.791 | |
| 24 | 1.90E-01 ± 3.45E-02 | 2.48E-01 ± 8.90E-02 | 1.880 | 3.046 | |
| 36 | 2.41E-01 ± 3.00E-02 | 3.04E-01 ± 1.50E-01 | 2.227 | 3.338 | |
| 48 | 2.18E-01 ± 9.81E-02 | 3.03E-01 ± 1.48E-01 | 2.084 | 3.333 | |
| OsUBQ5 | 0 | 5.22E-02 ± 1.97E-02 | 4.23E-02 ± 2.55E-02 | 0 | 0 |
| 12 | 2.22E-01 ± 8.00E-03 | 1.40E-01 ± 3.95E-02 | 2.091 | 1.731 | |
| 24 | 2.76E-01 ± 1.11E-01 | 3.28E-01 ± 1.09E-01 | 2.403 | 2.954 | |
| 36 | 2.76E-01 ± 6.04E-02 | 4.96E-01 ± 6.11E-02 | 2.403 | 3.551 | |
| 48 | 3.55E-01 ± 1.31E-01 | 1.07E+00 ± 2.18E-01 | 2.766 | 4.659 | |
| House-Keeping Genes | Target Gene and Development Stage | Relative mRNA Abundance | Log2 Fold-Change | ||
| Kasalath | Krasnodars. | Kasalath | Krasnodars. | ||
| 18S ribosomal RNA | OsUBC7 | ||||
| 2-week-old | 2.09E-03 ± 1.20E-04 | 1.63E-03 ± 2.90E-04 | 0 | 0 | |
| Flowering | 1.78E-02 ± 9.07E-03 | 1.71E-02 ± 4.60E-03 | 3.093 | 3.392 | |
| OsACT1 | 2-week-old | 1.13E-01 ± 8.10E-02 | 7.71E-02 ± 1.34E-02 | 0 | 0 |
| Flowering | 1.78E+00 ± 3.88E-01 | 1.74E+00 ± 4.30E-01 | 3.379 | 4.493 | |
| 18S ribosomal RNA | OsUBC8 | ||||
| 2-week-old | 1.22E-02 ± 4.81E-03 | 1.36E-02 ± 5.92E-03 | 0 | 0 | |
| Flowering | 5.55E-03 ± 1.81E-03 | 4.01E-03 ± 8.80E-04 | -1.120 | -1.737 | |
| OsACT1 | 2-week-old | 1.41E+00 ± 3.49E-01 | 1.20E+00 ± 2.59E-01 | 0 | 0 |
| Flowering | 6.58E-01 ± 1.66E-01 | 6.23E-01 ± 1.00E-01 | -1.089 | -0.943 | |
| 18S ribosomal RNA | OsUBC9 | ||||
| 2-week-old | 5.49E-02 ± 2.12E-02 | 5.44E-02 ± 1.21E-02 | 0 | 0 | |
| Flowering | 1.00E-01 ± 1.96E-02 | 9.04E-02 ± 2.59E-02 | 0.872 | 0.731 | |
| OsACT1 | 2-week-old | 1.69E+01 ± 9.10E+00 | 1.13E+01 ± 1.07E+00 | 0 | 0 |
| Flowering | 2.23E+01 ± 8.68E+00 | 1.94E+01 ± 1.89E+00 | 0.401 | 0.774 | |
| Yield-Related Phenotype | Line | Data from Three Generations | p-value(Two-Way ANOVA) |
| Total Seeds per Plant | OE-1 | 80.3 ± 33.7 | 0.124 |
| WT-1 | 30.7 ± 3.3 | ||
| OE-2 | 66.9 ± 16.5 | 0.072 | |
| WT-2 | 36.3 ± 14.1 | ||
| Number of Seeds per Panicle | OE-1 | 34.4 ± 12.7 | 0.278 |
| WT-1 | 23.5 ± 6.9 | ||
| OE-2 | 32.2 ± 17.5 | 0.274 | |
| WT-2 | 17.0 ± 9.2 | ||
| Fertility (%) | OE-1 | 86.4 ± 1.0 | 0.080 |
| WT-1 | 77.0 ± 4.1 | ||
| OE-2 | 83.7 ± 5.1 | 0.163 | |
| WT-2 | 62.1 ± 17.8 | ||
| Flowering Time (No. of Days to heading) | OE-1-1 | 101.2 ± 1.9 | 0.684 |
| OE-1-2 | 100.3 ± 2.8 | 0.933 | |
| OE-1-3 | 102.2 ± 5.4 | 0.648 | |
| OE-2 | 100.3 ± 1.9 | 0.375 | |
| WT-1 | 100.5 ± 2.2 | ||
| WT-2 | 98.5 ± 2.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





