Submitted:
21 November 2024
Posted:
21 November 2024
You are already at the latest version
Abstract
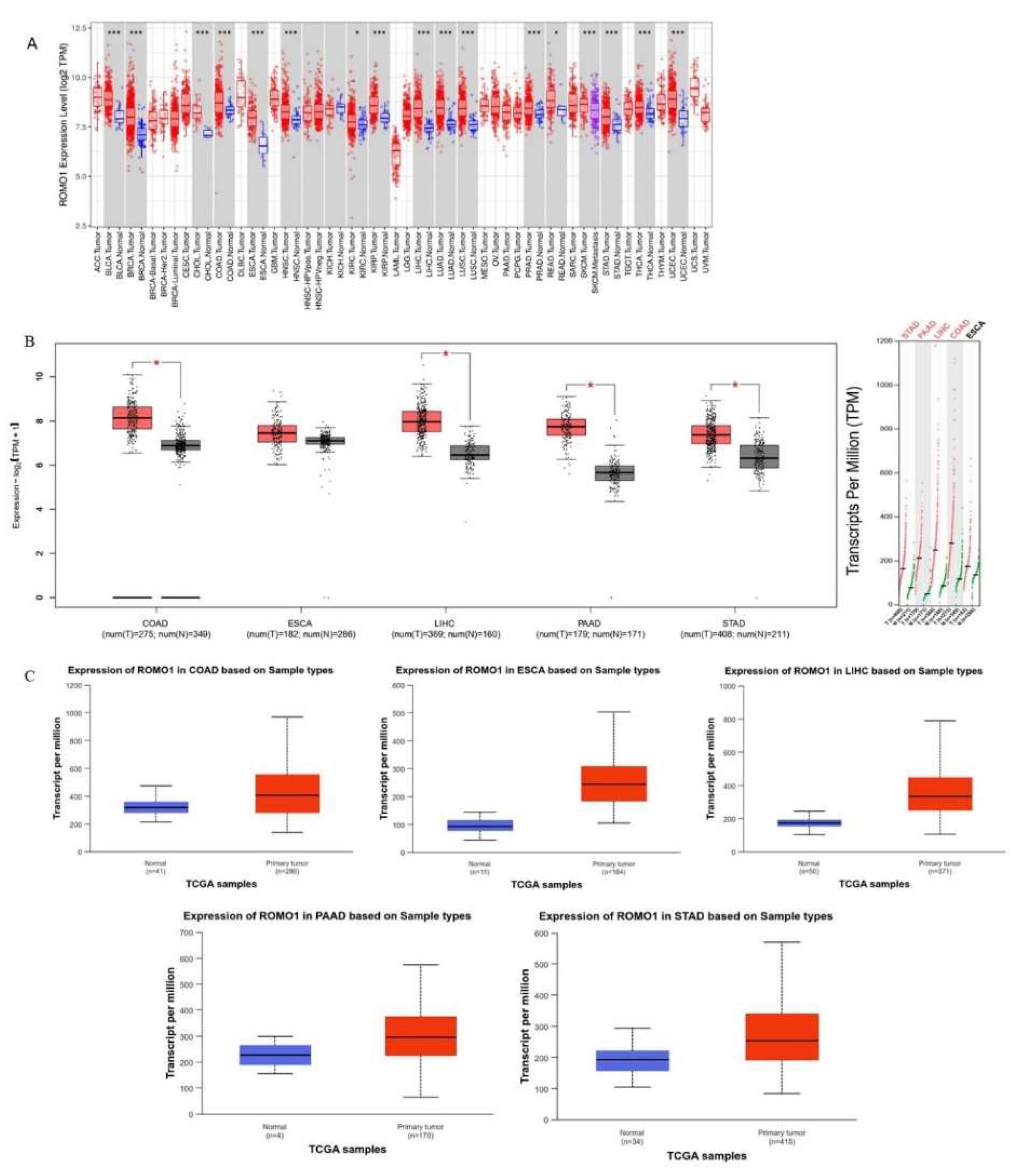
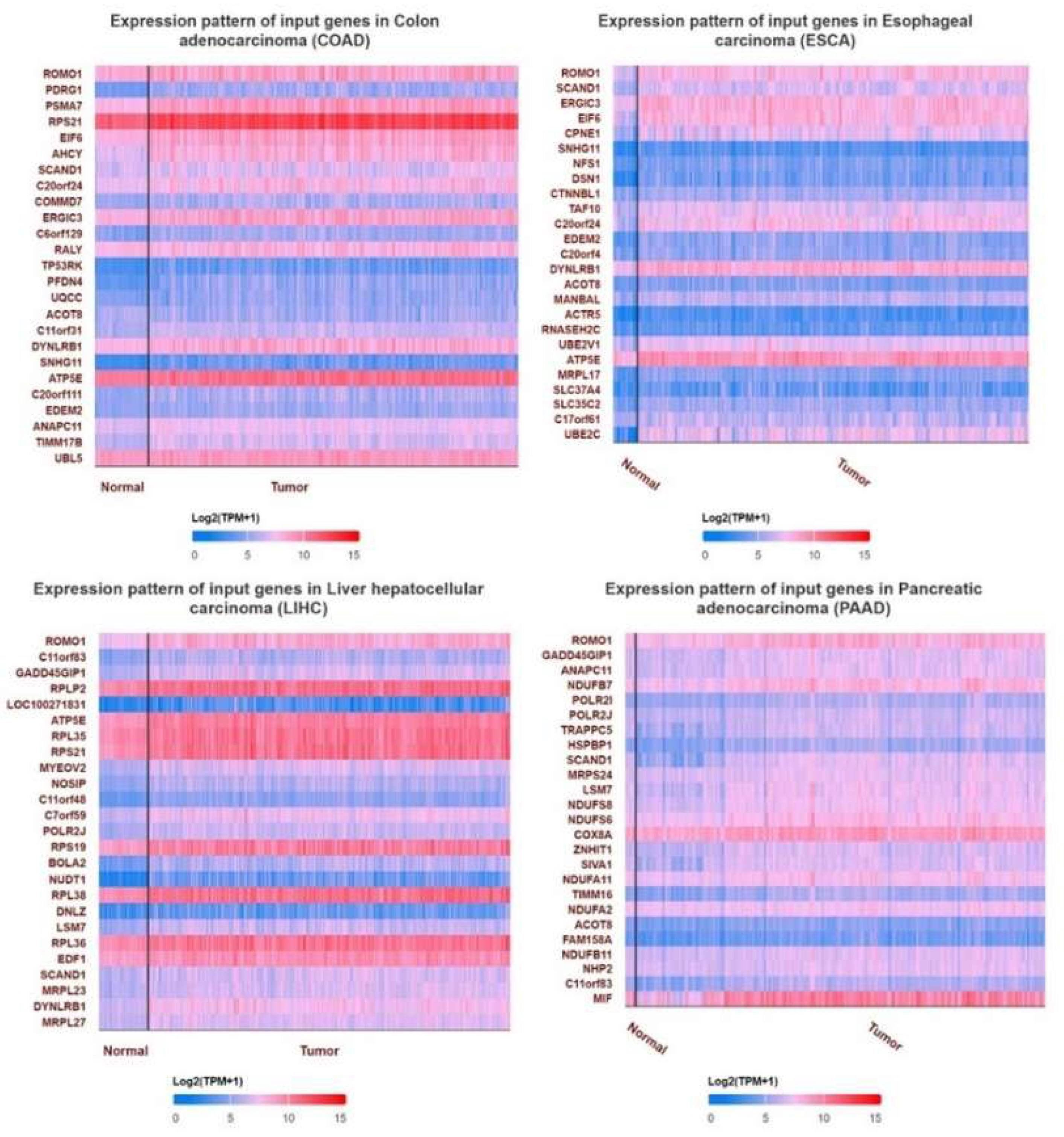
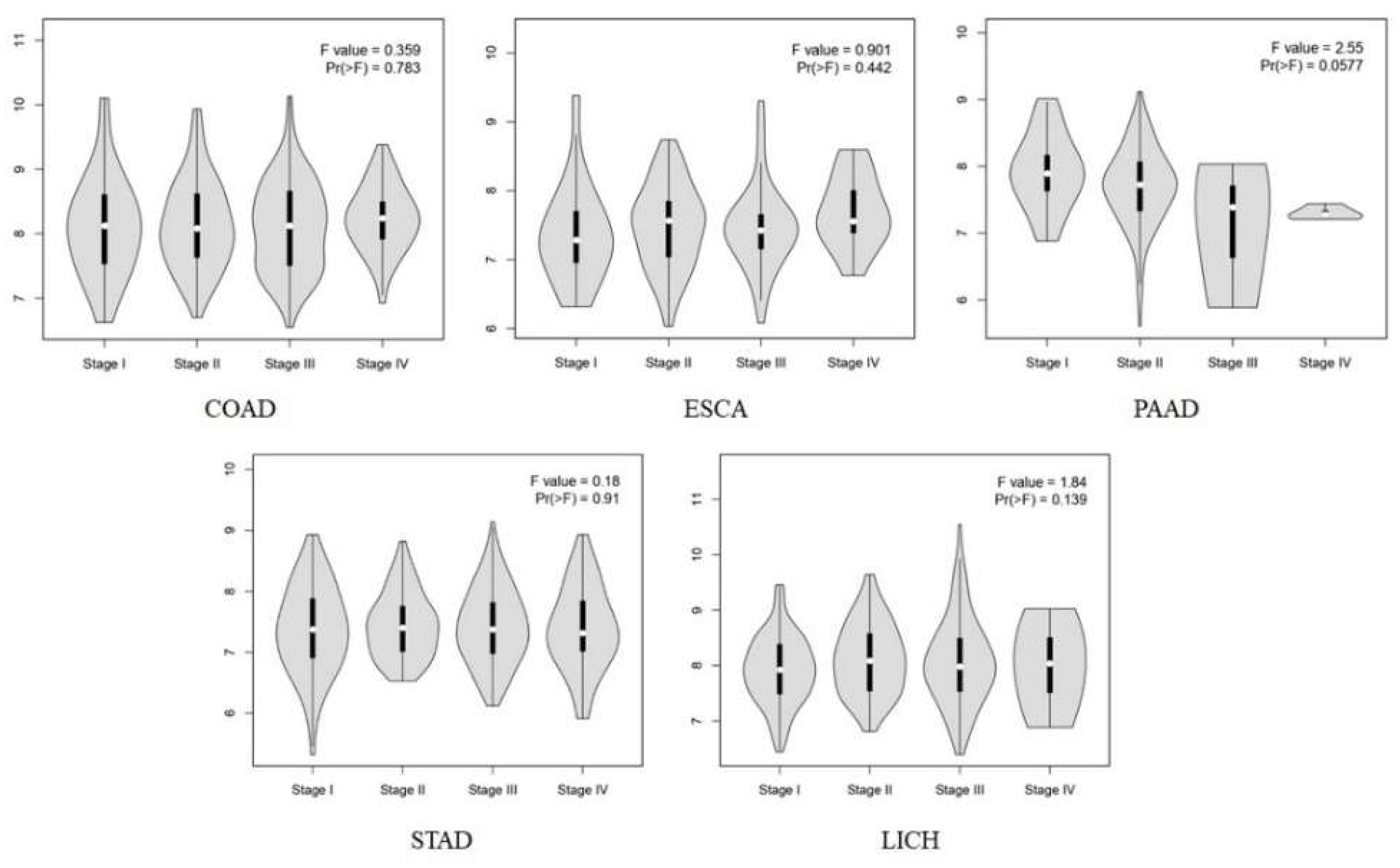
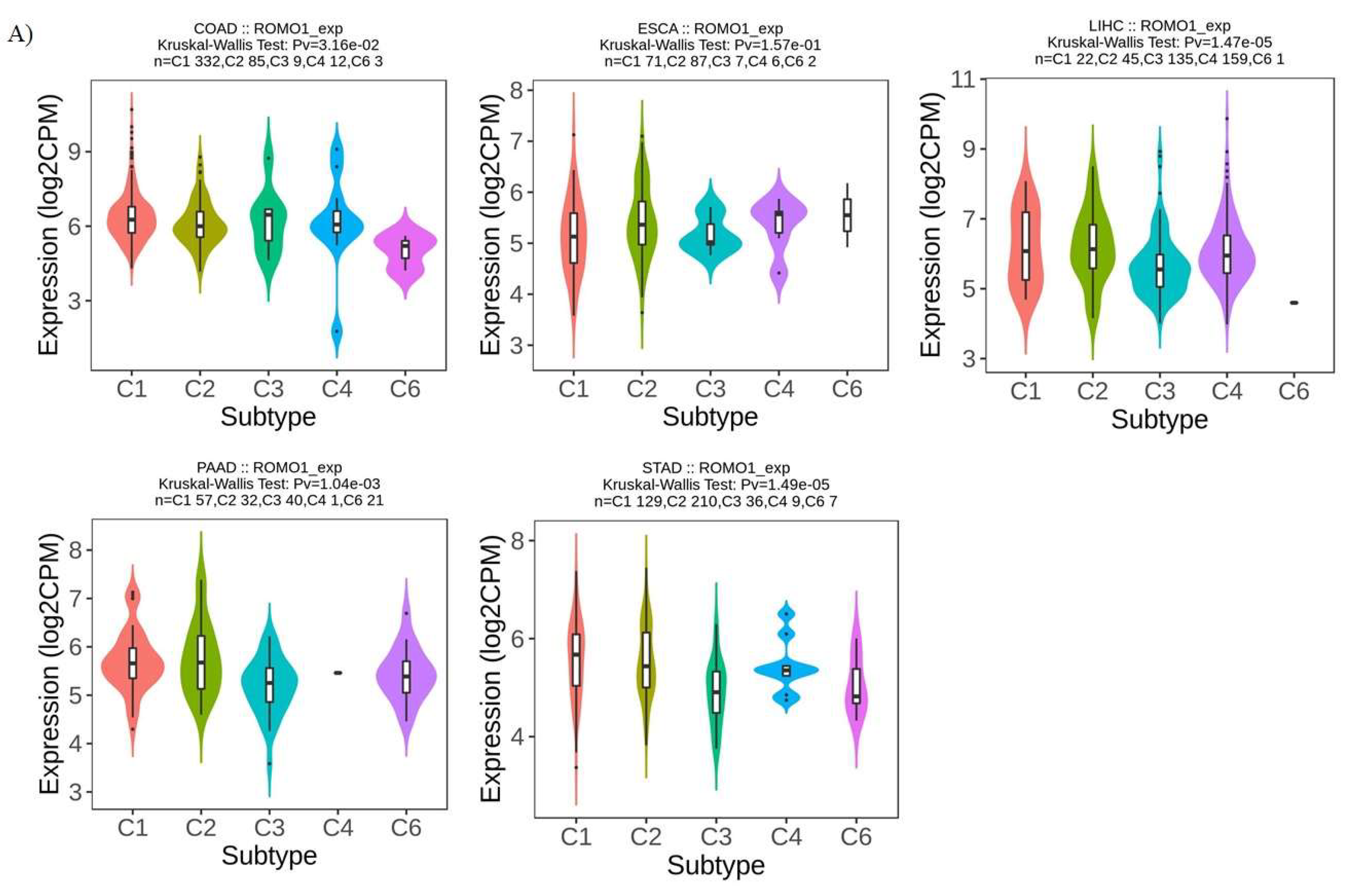
Background:Gastrointestinal tract cancers account for approximately one-third of cancer-related deaths. Early diagnosis and effective treatment are the most important ways to prevent cancer-related morbidity and mortality. ROMO1 has been shown to play an important role in many types of cancer. However, the biological function of ROMO1 is still poorly understood in gastrointestinal system cancers. The aim of this study is to reveal the expression change and oncogenic role of ROMO in gastrointestinal system cancers. Methods:Gene Expression Profiling Interactive Analysis (GEPIA), UALCAN, TIMER, GeneMANIA, TISIDB and STRING were applied to assess the biological function of ROMO1 in gastrointestinal cancers (Colon adenocarcinoma (COAD), Esophageal carcinoma (ESCA), Liver hepatocellular carcinoma (LIHC), Pancreatic adenocarcinoma (PAAD), and Stomach adenocarcinoma (STAD)). Results:ROMO1 is significantly increased in gastrointestinal cancers and overexpression of ROMO1 was associated with clinicopathological features. In addition, ROMO1 has been found to be closely associated with between tumor-infiltrating immune cells in gastrointestinal cancers. ROMO1 is closely related to the inner mitochondrial membrane proteins (TIMM) family. Conclusison: The study revealed that ROMO1 is of significant clinical importance for gastrointestinal cancers and may have potential clinical utility in treatment and prognosis.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Expression Analysis of ROMO1
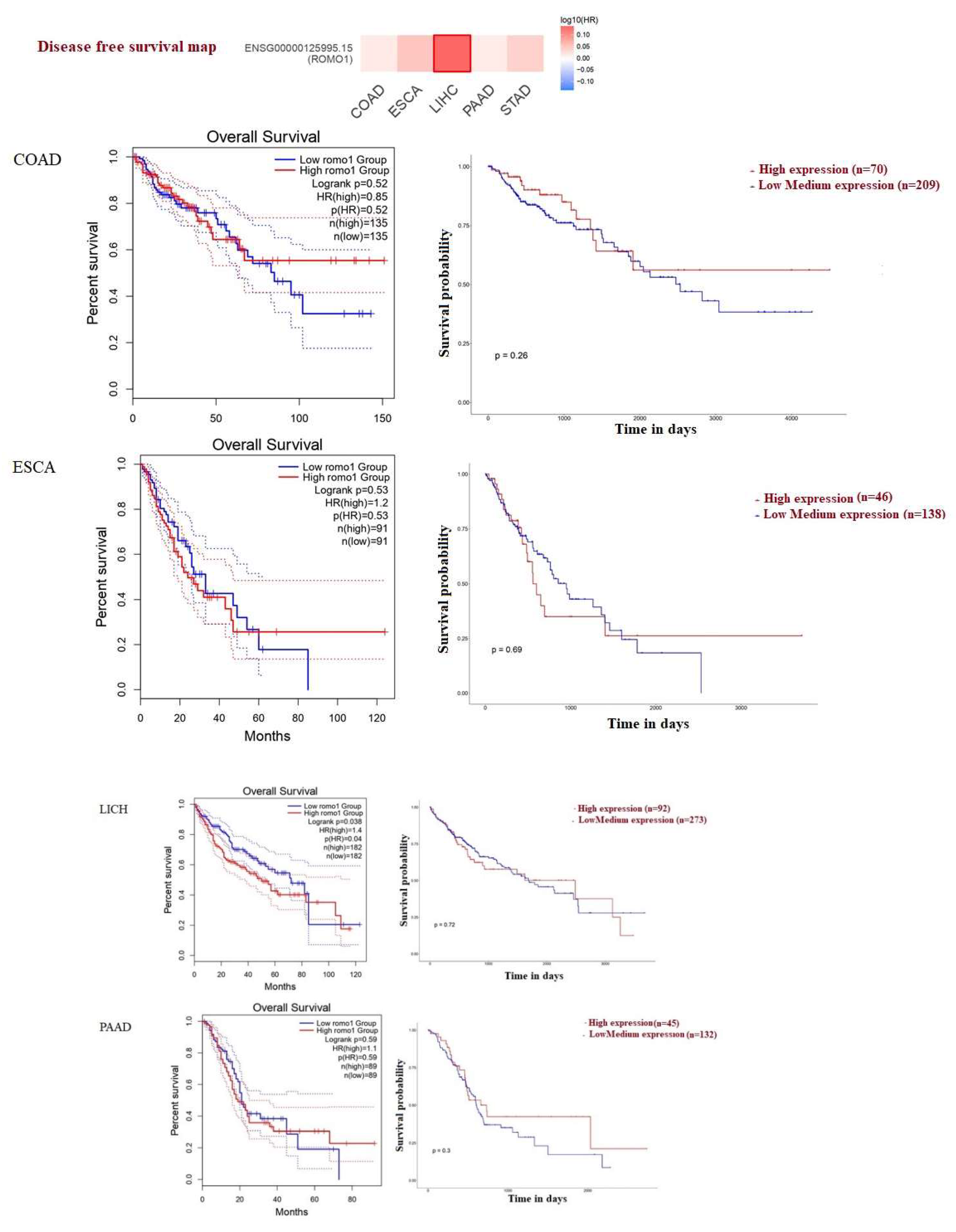
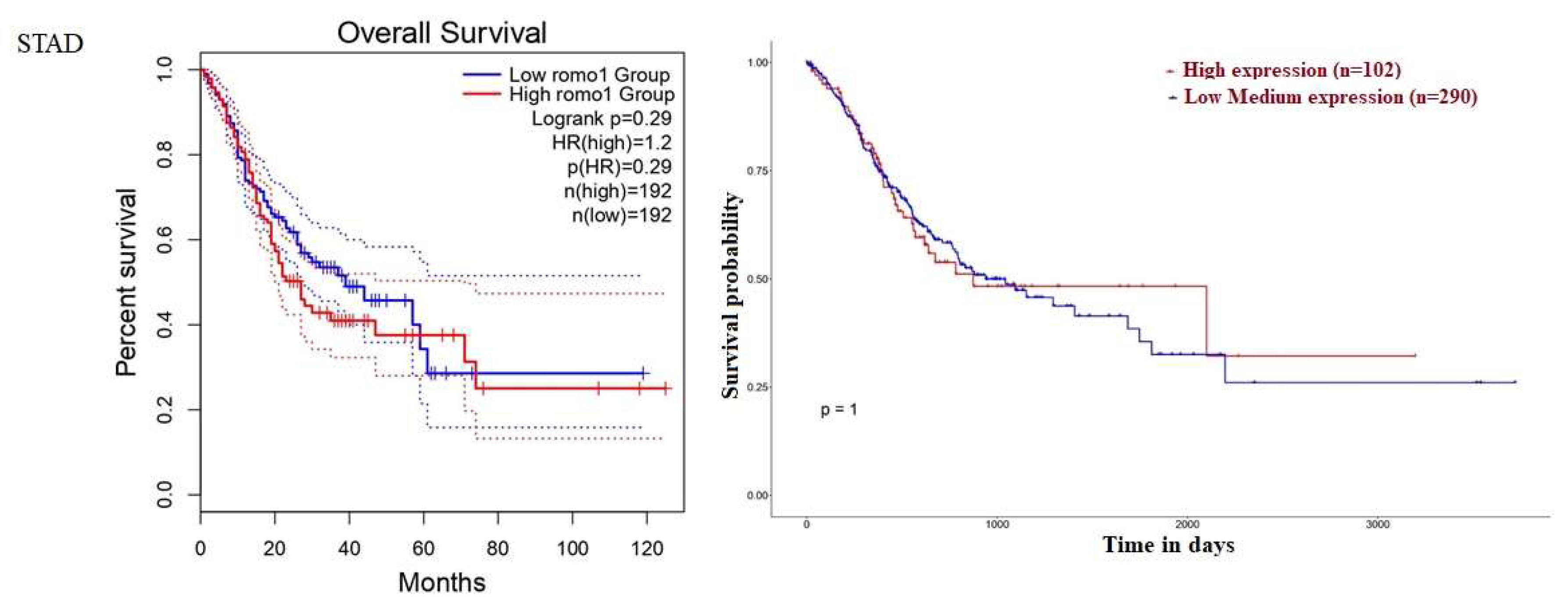
2.2. Survival Analysis of ROMO1
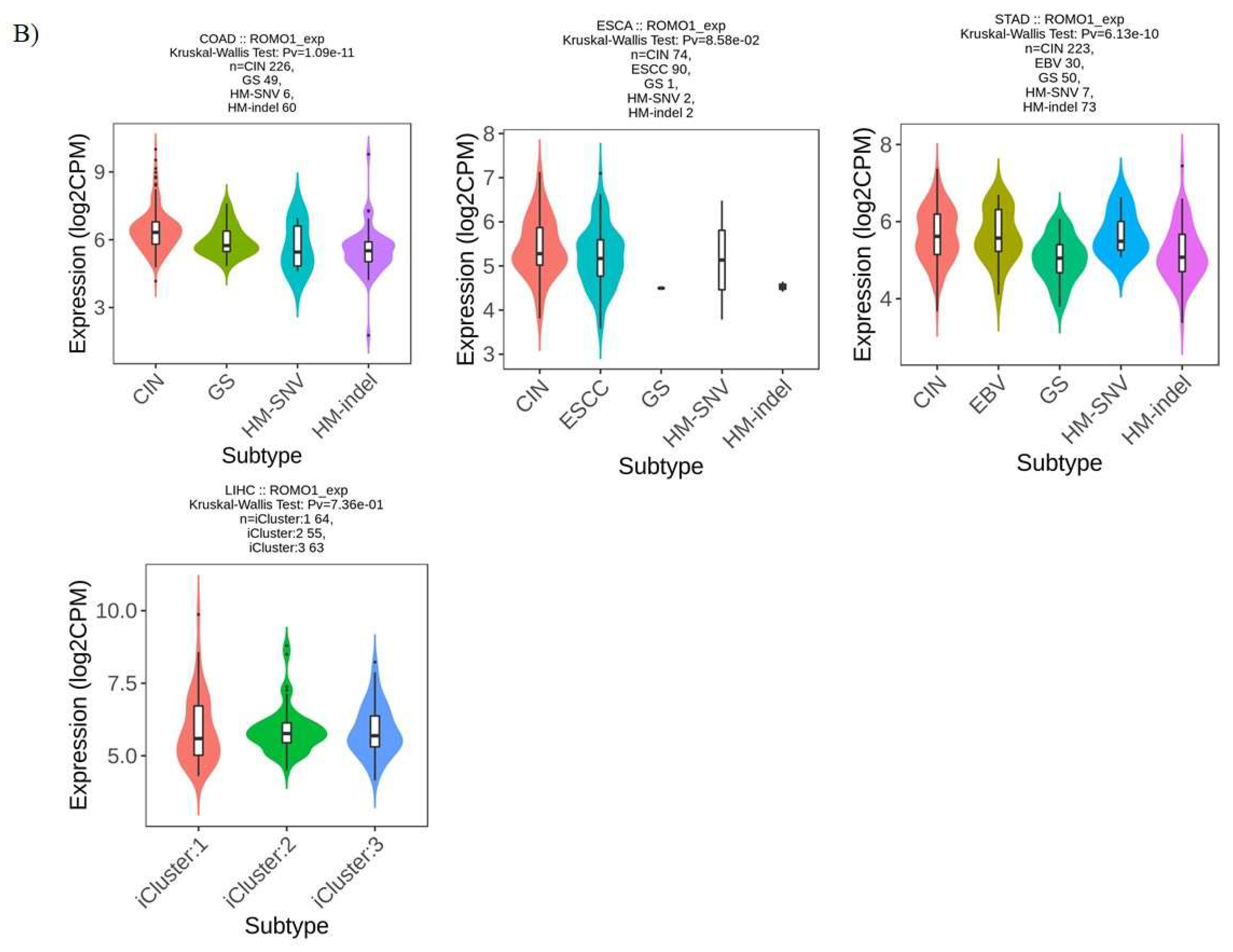
2.3. ROMO1 Expression in Molecular and Immune Subtypes of GI
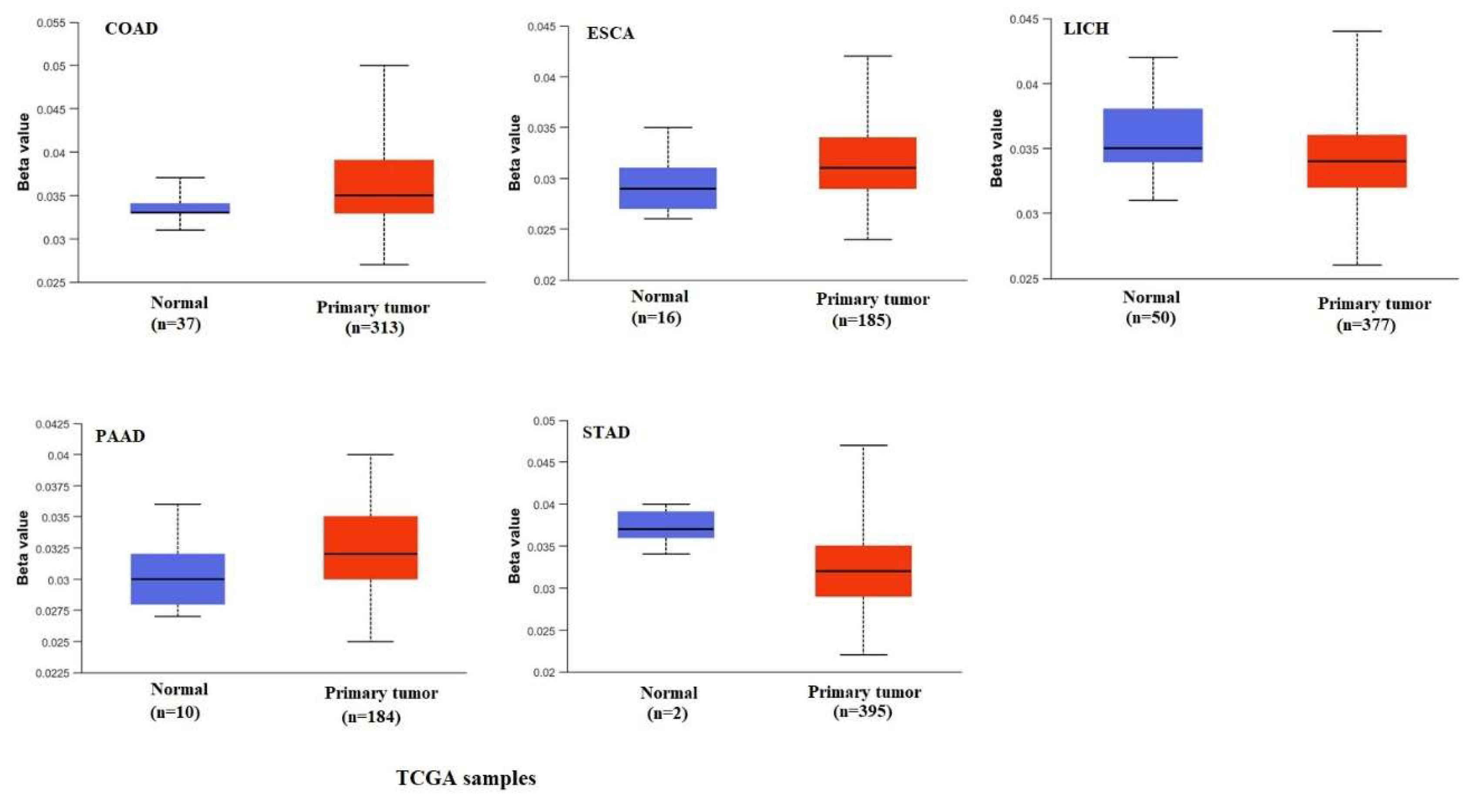
2.4. DNA Methylation Analysis
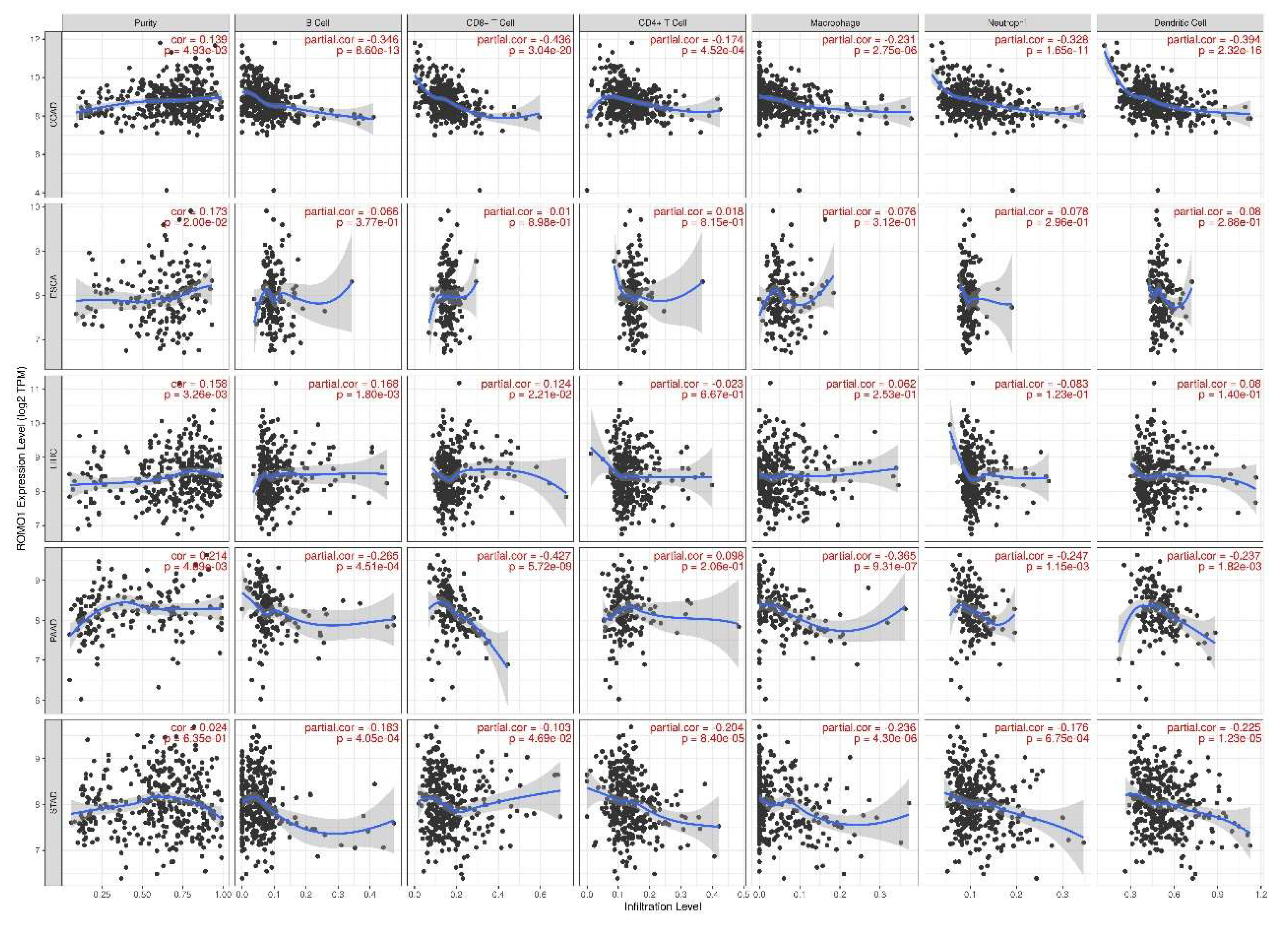
2.5. Correlation Between ROMO1 and Immune Infiltration
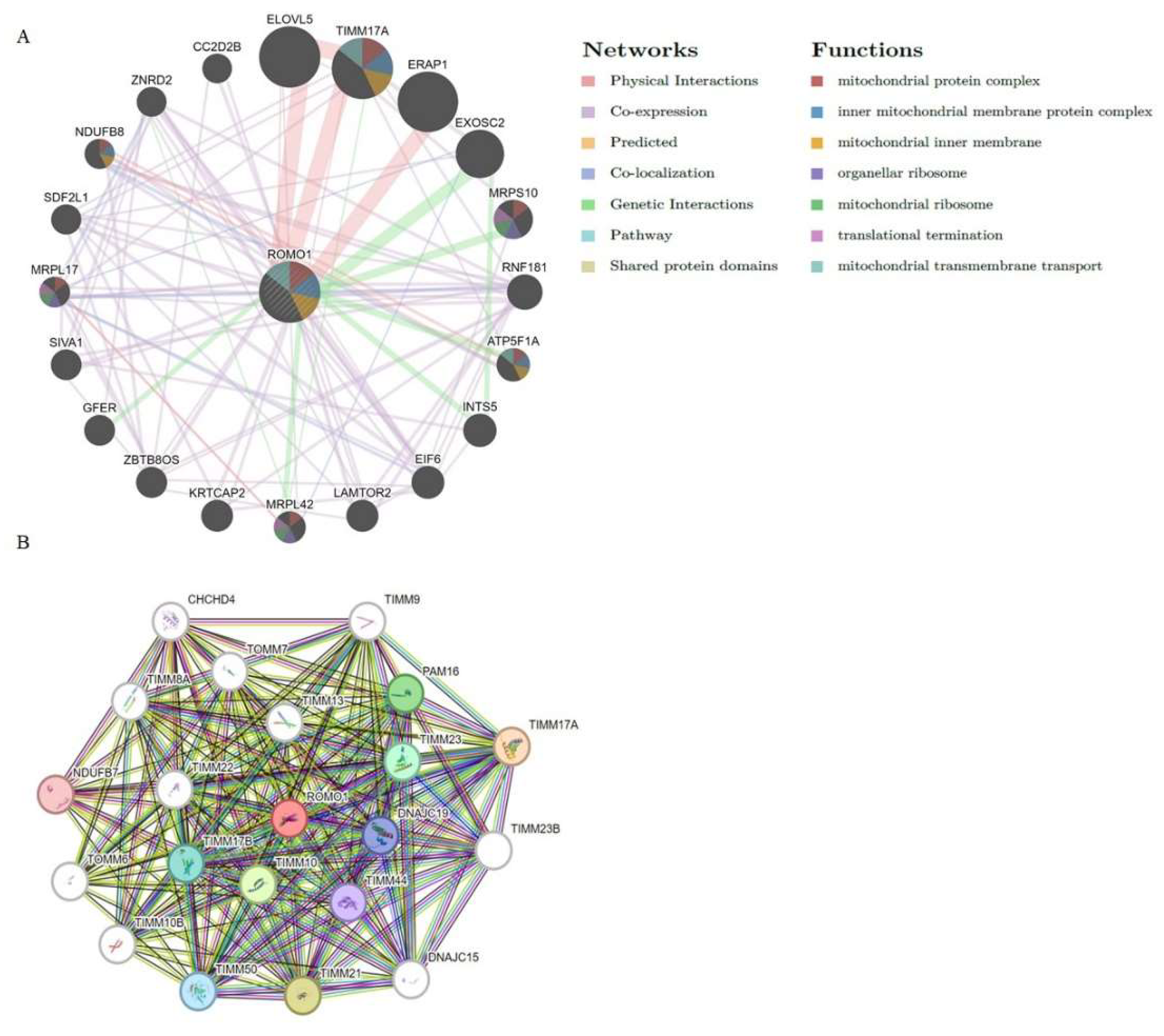
2.6. Analysis of the Gene and Protein That Interact with ROMO1 in Pan-Cancer
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grady WM, Yu M, Markowitz SD. Epigenetic Alterations in the Gastrointestinal Tract: Current and Emerging Use for Biomarkers of Cancer. Gastroenterol. 2021, 160, 690–709. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa V, Patel N, Grover D, et al. Interventional gastroenterology in oncology. CA Cancer J Clin. 2023, 73, 286–319. [Google Scholar] [CrossRef] [PubMed]
- Roshani M, Molavizadeh D, Sadeghi S, et al. Emerging roles of miR-145 in gastrointestinal cancers: A newparadigm. Biomed Pharmacother. 2023, 166, 115264. [Google Scholar] [CrossRef]
- Siegel RL, Miller KD, Fuchs HE, et al. Cancerstatistics, 2022. CA Cancer J Clin. 2022, 72, 7–33. [CrossRef]
- Arnold M, Abnet CC, Neale RE, et al. Global Burden of 5 Major Types of Gastrointestinal Cancer. Gastroenterol. 2020, 159, 335–349.e15. [Google Scholar] [CrossRef]
- Marusawa H, Jenkins BJ. Inflammation and gastrointestinal cancer: an overview. Cancer Lett. 2014, 345, 153–6. [Google Scholar] [CrossRef] [PubMed]
- Roshani M, Jafari A, Loghman A, et al. Applications of resveratrol in the treatment of gastrointestinal cancer. Biomed Pharmacother. 2022, 153, 113274. [Google Scholar] [CrossRef]
- Amini MA, Karimi J, Khodadadi I, et al. Overexpression of ROMO1 and OMA1 are Potentially Biomarkers and Predict Unfavorable Prognosis in Gastric Cancer. J Gastro in test Cancer. 2020, 51, 939–946. [Google Scholar] [CrossRef]
- Ghasemi H, Amini MA, Pegah A, et al. Over expression of reactive oxygen species modulator 1 is associated with advanced grades of bladde rcancer. Mol Biol Rep. 2020, 47, 6497–6505. [Google Scholar] [CrossRef]
- Norton M, Ng AC, Baird S, et al. ROMO1 is an essential redox-dependent regulator of mitochondrial dynamics. Science Signaling. 2014, 7, ra10. [Google Scholar] [CrossRef]
- Amini MA, Talebi SS, Karimi J. Reactive Oxygen Species Modulator 1 (ROMO1), a New Potential Target for Cancer Diagnosis and Treatment. ChonnamMed J. 2019, 55, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Liu D, Liu Y, Xia Z, et al. Reactive oxygen species modulator 1 regulates oxidative stres and induces renal and pulmonary fibrosis in a unilateral ureteral obstructionrat model and in HK-2 cells. Mol Med Rep. 2017, 16, 4855–4862. [Google Scholar] [CrossRef] [PubMed]
- Kim HJ, Jo MJ, Kim BR, et al. Reactive oxygenspecies modulator-1 (Romo1) predicts unfavorable prognosis in colorectal cancer patients. PLoSOne. 2017, 12, e0176834. [Google Scholar] [CrossRef]
- Na, A. R. ,Chung, Y. M., Lee, S. B., Park, S. H., Lee, M. S., & Do Yoo, Y. A critical role for Romo1-derived ROS in cell proliferation. Biochemical and biophysical research communications 2008, 369, 672–678. [Google Scholar]
- You BR, Park WH. The enhancement of propylgallate-induced HeLa cell death by MAPK inhibitors is accompanied by increasing ROS levels. Mol Biol Rep. 2011, 38, 2349–58. [Google Scholar] [CrossRef]
- Lee SH, Min JW, Lee JS, et al. Reactive oxygen species modulator 1 (Romo1) over expression is an independent predictor of poor survival in NSCLC patients who under go surgical resection. Lung Cancer. 2015, 87, 45–52. [Google Scholar] [CrossRef]
- Yu MO, Song NH, Park KJ, et al. Romo1 is associated with ROS production and ellular growth in human gliomas. J Neurooncol. 2015, 121, 73–81. [Google Scholar] [CrossRef]
- Misir S, Ozer Yaman S, Petrović N, et al. Identification of a Novel hsa_circ_0058058/miR-324-5p Axis and Prognostic/Predictive Molecules for Acute Myeloid Leukemia Outcome by Bioinformatics-Based Analysis. Biology 2024, 13, 487. [Google Scholar] [CrossRef]
- Li T, Fan J, Wang B, et al. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. CancerRes. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. NucleicAcidsRes. 2017, 45, W98–W102. [Google Scholar] [CrossRef]
- Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia. 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Ru B, Wong CN, Tong Y,et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk D, Franceschini A, Kuhn M, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Warde-Farley D, Donaldson SL, Comes O, et al. The Gene MANIA prediction server: biological network integration for gene prioritization and predicting gene function. NucleicAcidsRes. 2010, 38, W214–W220. [Google Scholar] [CrossRef]
- Deng L, Jiang A, Zeng H, et al. Comprehensive analyses of PDHA1 that serves as a predictive biomarker for immunotherapy response in cancer. Front Pharmacol. 2022, 13, 947372. [Google Scholar] [CrossRef]
- Li F, Simon MC. CancerCellsDon't Live Alone: Metabolic Communication within Tumor Microenvironments. Dev Cell. 2020, 54, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Wang P, Geng J, Gao J, et al. Macrophage achieves self-protection against oxidative stress-induced ageing through the Mst-Nrf2 axis. NatCommun. 2019, 10, 755. [Google Scholar] [CrossRef]
- Wang L, Liu X, Liu Z, et al. Network models of prostate cancer immune microenvironments identify ROMO1 as heterogeneity and prognostic marker. SciRep. 2022, 12, 192. [Google Scholar] [CrossRef]
- Tsoneva E, Vasileva-Slaveva M, Kostov S, et al. ROMO1 – a potential immuno histochemical prognostic marker for cancer development. Oncologie. 2023, 25, 753–758. [Google Scholar] [CrossRef]
- Kim T, Cha YJ, Park JH, et al. Reactive oxygen species modulator 1 expression predicts lymphnode metastasis and survival in early-stagenon-small cell lung cancer. PLoSOne. 2020, 15, e0239670. [Google Scholar] [CrossRef]
- Chung JS, Park S, Park SH, et al. Over expression of Romo1 promotes production of reactive oxygen species and invasiveness of hepatic tumor cells. Gastroenterology. 2012, 143, 1084–94.e7. [Google Scholar] [CrossRef] [PubMed]
- Yang Y, Zhang F, Huang H, Xie Z, Huang W, Xie H, Wang F. Long noncoding RNA LINC00319 regulates ROMO1 expression and promotes bladder cance rprogression via miR-4492/ROMO1 axis. J Cell Physiol. 2020, 235, 3768–3775. [Google Scholar] [CrossRef] [PubMed]
- Sun G, Cao Y, Qian C, Wan Z, Zhu J, Guo J, Shi L. Romo1 is involved in the immune response of glioblastoma by regulating the function of macrophages. Aging (Albany NY). 2020, 12, 1114–1127. [Google Scholar] [CrossRef]
- Lee S, Park YH, Chung JS, et al. Romo1 andthe NF-κB pathway are involved in oxidative stress-induced tumor cell invasion. Int J Oncol. 2015, 46, 2021–8. [Google Scholar] [CrossRef]
- Muenst S, Läubli H, Soysal SD, et al. The immune system and cancer evasion strategies: therapeutic concepts. J InternMed. 2016, 279, 541–62. [Google Scholar] [CrossRef]
- Kim IG, Kim SY, Kim HA, et al. Disturbance of DKK1 level is partly involved in survival of lung cancer cells via regulation of ROMO1 and γ-radiation sensitivity. Biochem Biophys Res Commun. 2014, 443, 49–55. [Google Scholar] [CrossRef]
- Martinez-Bernabe, T.; Pons, D.G.; Oliver, J.; Sastre-Serra, J. Oxidative Phosphorylation as a Predictive Biomarker of Oxaliplatin Response in Colorectal Cancer. Biomolecules 2024, 14, 1359. [Google Scholar] [CrossRef]
- Wang Z, Li S, Xu F, et al. ncRNAs-mediated high expression of TIMM8A correlates with poor prognosis and act as an oncogene in breast cancer. Cancer Cell Int. 2022, 22, 177. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



