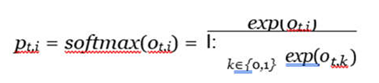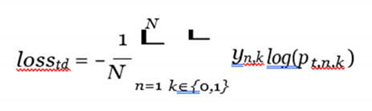Introduction
Breast cancer remains a formidable challenge on the global health landscape, representing one of the most prevalent malignancies and the primary cause of female mortality [
8,
12,
24]. Despite significant advancements in medical science, the battle against breast cancer persists, particularly in regions with limited resources where survival rates plummet below 40% [
16]. Early detection stands as the linchpin in this struggle, offering extended treatment windows and heightened prospects of survival [
15,
21]. Consequently, there arises an imperative for precise and timely diagnosis, particularly in locales grappling with a scarcity of experienced medical professionals [
12].
Breast ultrasound, heralded as a pivotal diagnostic modal- ity, holds promise in discerning between benign, malignant, and normal breast tissue [
22]. Its appeal lies in its cost- effectiveness, non-invasiveness, and absence of ionizing ra- diation, rendering it particularly suitable for regions with constrained healthcare resources [
4,
18,
22]. Yet, the ef- ficacy of ultrasound diagnosis is tethered to the expertise of sonographers, a resource in short supply relative to patient needs [
5,
7]. Therefore, to provide an objective and accurate assessment for the early detection of breast cancer, computer- aided diagnostic systems for breast ultrasound imaging have become necessary.
The development of deep learning in medical image pro- cessing makes the computer-aided breast cancer recognition possible [
3,
10,
14,
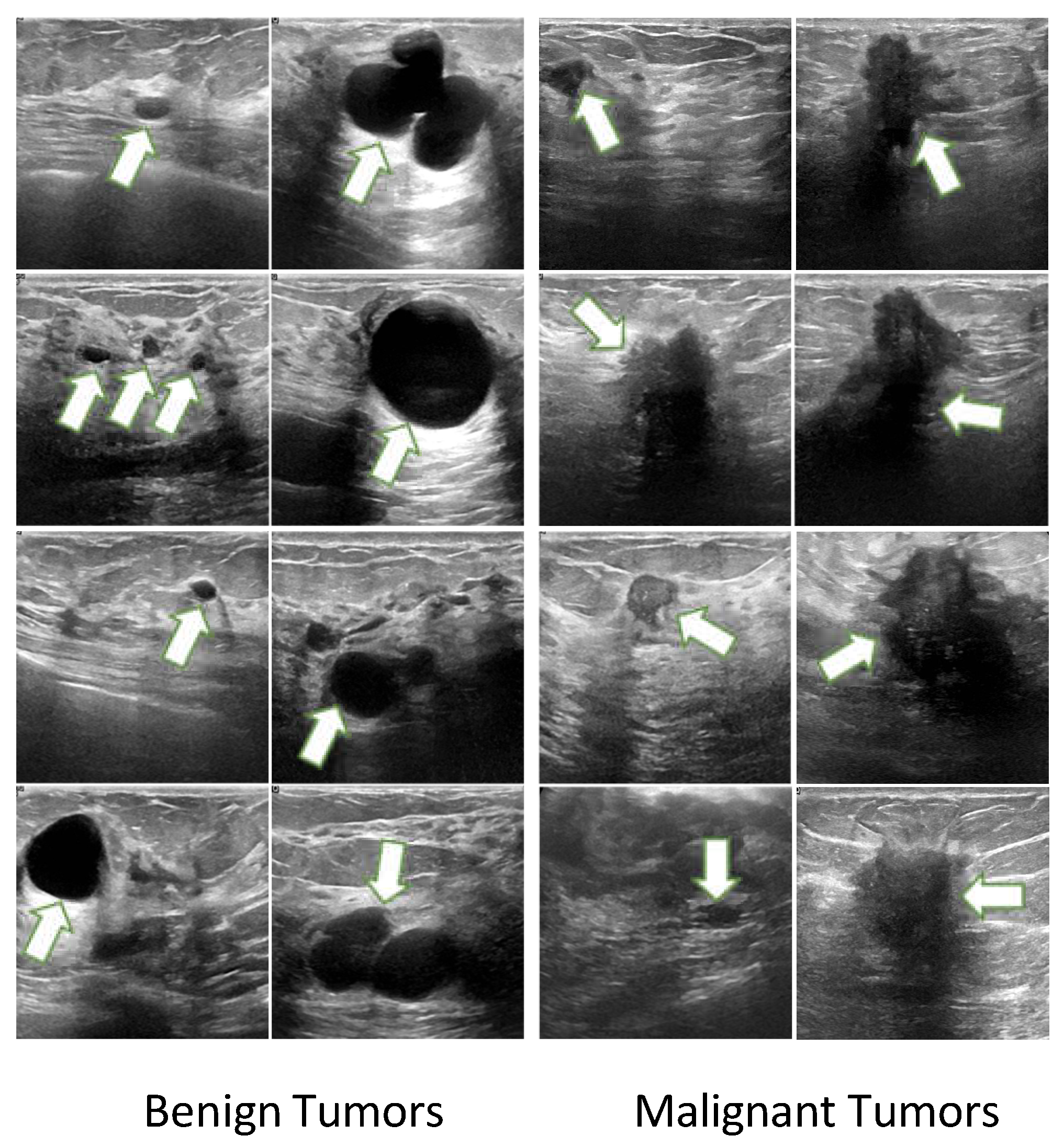
32]. However, a significant challenge faced by most methods lies in the uncertainty regarding tumor locations and proportions within ultrasound images [
11,
34], which can be found in
Figure 1. In order to address this challenge, one line of research incorporates breast cancer char- acteristics by employing labeled Regions of Interest (ROIs) to direct the model’s attention to specific areas. However, these methods cannot diagnose in the absence of ROIs, which is not suitable for real diagnostic scenarios. In contrast, whole-image diagnostic methods can adapt to real diagnostic settings, but they lack the integration of specific breast cancer characteris- tics, leaving room for performance improvement.
Regarding the first approach, predefined ROIs can focus the model’s attention on key areas. For instance, Moon et al. [
20] proposed an image fusion method that combines manually labeled tumor images, segmented tumor images, and tumor shape images into a fused image, then classifies breast cancer tumors from the fused image using an ensemble learning of convolutional neural networks (CNNs). Zhuang et al. [
33] introduced an adaptive multi-model spatial feature fusion method that repeatedly transforms and fuses ultrasound images with manually labeled mask images, then classifies tumor types using a CNN. These methods reduce errors resulting from variations in tumor location and proportions as presented in ultrasound images, however, they are somewhat limited in real diagnostic environments where ROIs of ultrasound images are unavailable.
Whole-image analysis methods are feasible in practical clinical settings. For example, Masud et al. [
17] explored the performance of various pre-trained CNN models combined with different optimization methods in breast cancer diagnosis. Mo et al. [
19] proposed a novel HoVer-Trans method that extracts inter- and intra-layer spatial information of breast tumor tissue transversely and longitudinally. These methods can be directly utilized in real clinical settings without manu- ally marked ROIs and demonstrate acceptable performance. However, due to the uncertain proportion and location of tumors in ultrasound images, there remains room for further improvement.
Besides the aforementioned mainstream research, Zhang et al. [
31] first adopted the proposed REAF method to decouple the diagnosis by fusing extracted confidence features. This method first uses segmentation techniques to extract ROIs, then fuses the segmented images to diagnose breast cancer, which achieves state-of-the-art performance while ensuring usability in actual clinical settings. However, the extracted ROIs cannot be absolutely accurate, and the fusion of erro- neous regional information will affect downstream diagnostic models.
In this paper, we propose a regional prior fusion framework using early and late distillation (RPF-ELD), inspired by the knowledge distillation of the teacher-student framework, for breast cancer diagnosis in ultrasound images. To mitigate the challenges posed by the low proportion and variable positions of tumors, we incorporate the tumor region information as prior knowledge into our framework. To ensure the model’s ap- plicability in real-world scenarios, we propose an early and late feature distillation method, integrating the prior knowledge into the diagnostic model that relies solely on ultrasound image analysis. Specifically, our framework involves the following key steps. First, we fuse the regional prior information of ROIs with ultrasound images to train a prior-fused model as the teacher model. Then, this teacher model is used to distill the prior information into a diagnostic student model based on only the ultrasound image. Finally, through this distillation process, the diagnostic model learns to carry the prior infor- mation and analyze the images without the need for ROIs. By adopting this methodology, our diagnostic model benefits from the inclusion of ROI as prior knowledge while maintaining robustness and applicability in practical clinical settings. We conducted experiments on two publicly available datasets. Experimental results demonstrate that the fusion of regional prior information improves the teacher model significantly, the prior-fused feature distillation enhances the diagnostic model effectively, and the proposed RPF-ELD surpasses current state- of-the-art methods.
The contributions of this paper can be summarized as follows:
We propose a regional prior fusion framework that fuses
unavailable prior information of tumor regions into a diagnostic model by the knowledge distillation paradigm of the teacher-student framework.
We propose an early and late distillation method that distills the prior information from the teacher model to the student model in the levels of contexts and the deep features.
The experimental results, on two publicly released datasets, demonstrate that the proposed framework achieves state-of-the-art performance.
METHOD
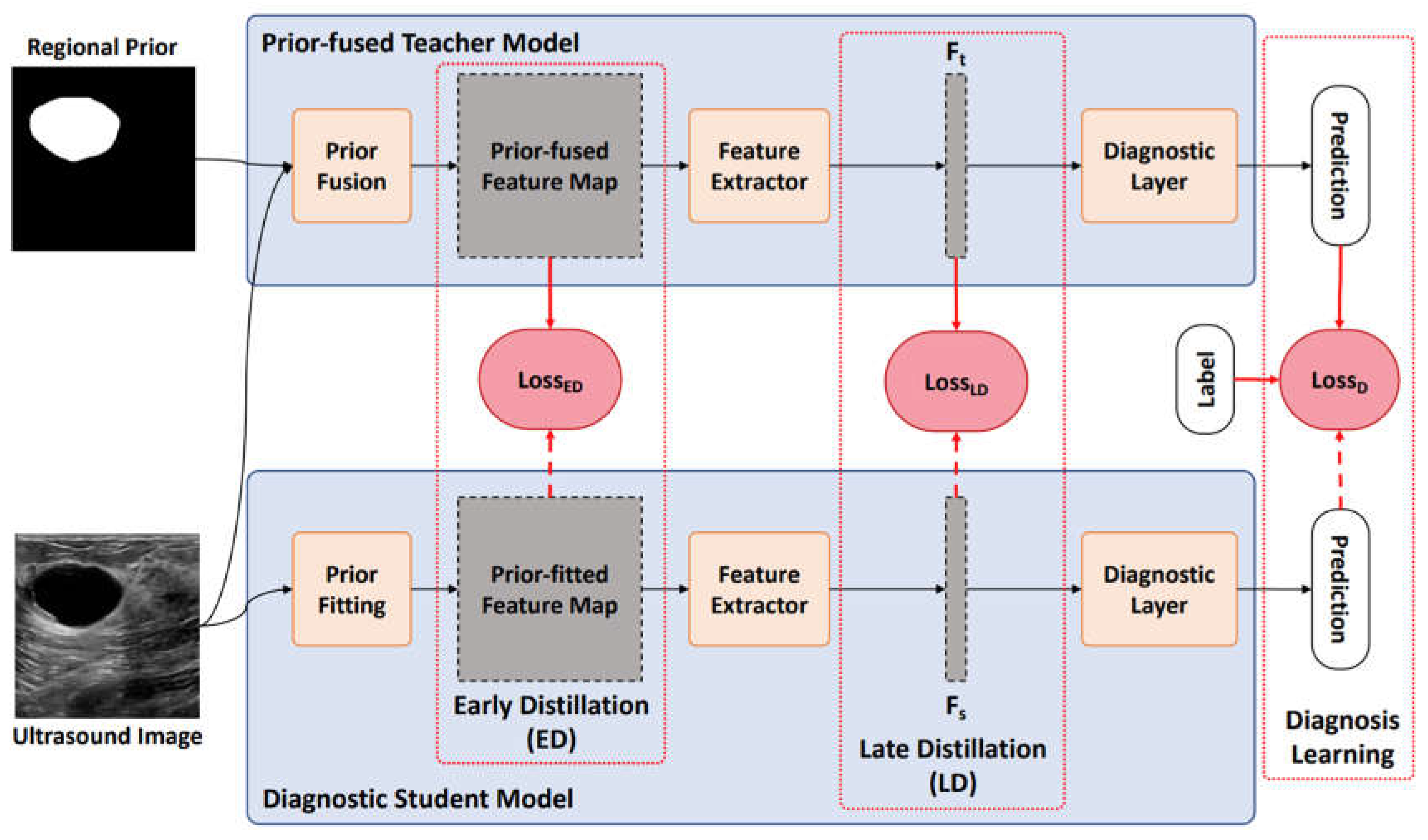
A diagram of the proposed RPF-ELD framework is shown in
Figure 2, which contains two models and three optimization methods. Two models are the prior-fused teacher model and diagnostic student model, and three optimization methods are early distillation, late distillation and diagnosis learning. The prior-fused teacher model learns the tumor pattern from the fusion of ultrasound images and the corresponding regional prior information using diagnosis learning. The diagnostic student model learns the diagnosis from only the ultrasound images using diagnosis learning as well as the early and late distillation from the teacher model.
A. Prior-fused Teacher Model
In ultrasound images, the proportions of tumors are variable, and the locations are also uncertain. These characteristics prevent current methods from high performance using whole images. To solve the interference of tumor uncertainty, we directly fuse the regional prior information of tumor areas into the teacher model. Specifically, during the learning stage, each ultrasound image with the corresponding regional prior information is first sent into the Prior Fusion to obtain a prior-fused feature map. Next, a feature extractor is used to mine deep features from the prior-fused feature map. Finally, a diagnostic layer classifies the type of input tumors.
Prior Fusion: The purpose of Prior fusion is to inte- grate regional prior knowledge of the tumor with ultrasound images, which generates prior-fused feature maps to enhance downstream analysis. To this end, the convolution operation is a suitable approach, which conducts interaction from different channels at the pixel level using trainable kernels. Therefore, the regional prior and the corresponding image can be fused using a convolution:
where
Iu is the ultrasound image,
Ip is the corresponding prior information of the ROI image, [
·] is the concat operation of two images, and
Mfusion is the prior-fused feature map. The
Mfusion, containing meaningful regional prior information of key areas, will be used for early distillation after well-trained.
Feature Extractor: After obtaining the prior-fused fea- ture map, the Feature Extractor aims to effectively represent the features of the input sample for downstream diagnosis. Although various deep models for feature extraction have been proposed, pretrained visual models are still effective methods for visual feature extraction. Therefore, pretrained visual models are selected to be the feature extractor for the teacher model. The feature extraction process can be illustrated as follows:
where
Te is the feature extractor of the teacher model,
Ft is the extracted feature representation for downstream diagnosis as well as the late distillation for the student model. In this study, we employ VGG16 as the feature extractor, which will be further discussed in the experiment.
Diagnostic Layer and Diagnosis Learning: To analyze tumors effectively on the extracted feature representations, we employ a fully connected network as the diagnostic layer to classify the type of tumors as follows:
where
Dt is the diagnostic layer of the teacher model,
ot is the output logit from the teacher model.
ot,k is k-
th dimensional value, where k is 0 or 1 with 0 denoting benign and 1 denoting malignant (in some cases, k is 0, 1 or 2, where 2 denotes the normal).
pt,i is the likelihood of the input sample belonging to type
i from the teacher model.
Finally, the whole prior-fused teacher model is optimized using the cross-entropy loss function as:
where N is the total number of training samples, yn,k is the label for sample n, which is 1 if it belongs to type k or 0 otherwise, and pt,n,k is the likelihood from the teacher model of sample n belonging to type k or not.
B. Diagnostic Student Model
The diagnostic student model learns the tumor pattern from only the ultrasound images using diagnosis learning as well as the proposed distillation method to obtain the tumor regional prior knowledge from the teacher model.
Prior Fitting: The Prior Fitting aims to transform the original input ultrasound image into a more meaningful feature map, which will be guided by the teacher model using early distillation. Similar to the Prior Fusion of the teacher model as shown in Eq. (1), we also use a convolutional operation to obtain a prior-fitted feature map
Mfitted:
The parameters of Prior Fitting will be trained together with the whole student model.
Early Distillation: The Early Distillation aims to guide the Prior Fitting to learn a transformation for more meaningful feature maps from only ultrasound images. The guidance of Early Distillation is a loss function that optimizes the gap between the prior-fitted feature map and the prior-fused feature map as illustrated as follows:
where
// · //1 is the loss of L1 distance, and
lossED is the early distance loss. Using the
lossED of Early Distillation, the prior- fitted feature map can be as near as same as possible to the prior-fused feature map, which provides more information in the context level.
Feature Extractor: Similar to the teacher model, the fea- ture extractor of the diagnostic student model aims to extract effective feature representations from
Mfitted for downstream diagnosis:
where
Se is the feature extractor of the student model, using VGG16 because of its good performance, which will be discussed in the experiment. F
s is the feature representation of the input ultrasound image.
Late Distillation: The Late Distillation aims to guide the student model to extract effective feature representations for input ultrasound images. Specifically, it enforces the extracted feature from the student model as similar as possible to that from the prior-fused teacher model using late distillation loss:
Diagnosis Learning: The diagnosis learning enables the student to learn the tumor patterns, which is the same as that of the teacher model of Eq. (5), using cross-entropy to obtain the loss losssd.
Overall Optimization: The overall optimization of the student model is joint learning from supervision of early distillation, late distillation and diagnosis learning:
The joint learning enforces the student model learns the recognition of cancer tumors as well as the experience of prior information.
Table 1.
Quantitive comparisons of feature extractors among state-of-the-art pretrained visual models on the UDIAT and BUSI. datasets. ’Average’ indicates the mean value of AUC, Accuracy, Specificity, Precision, Recall and F1-score. The values following ± are standard deviations.
Table 1.
Quantitive comparisons of feature extractors among state-of-the-art pretrained visual models on the UDIAT and BUSI. datasets. ’Average’ indicates the mean value of AUC, Accuracy, Specificity, Precision, Recall and F1-score. The values following ± are standard deviations.
| Dataset |
|
|
UDIAT |
|
| Methods |
AUC↑
|
Accuracy↑
|
Specificity↑
|
Precision↑
|
Recall↑
|
F1-score↑
|
Average↑
|
| ViT [30] |
74.95±0.101 |
81.29±0.080 |
79.28±0.094 |
76.24±0.105 |
76.24±0.105 |
77.07±0.101 |
77.51±0.098 |
| Swin-Transformer [13] |
76.47±0.096 |
83.23±0.058 |
81.26±0.063 |
79.24±0.084 |
79.24±0.084 |
79.80±0.076 |
79.87±0.077 |
| MaxViT [27] |
78.00±0.098 |
85.16±0.058 |
83.25±0.063 |
82.24±0.081 |
82.24±0.081 |
82.52±0.076 |
82.24±0.076 |
| Efficientb3 [25] |
80.47±0.042 |
85.16±0.053 |
83.65±0.054 |
81.72±0.082 |
81.72±0.082 |
82.34±0.075 |
82.51±0.065 |
| Res50 [9] |
82.95±0.043 |
85.16±0.053 |
84.05±0.063 |
81.19±0.082 |
81.19±0.082 |
82.16±0.075 |
82.78±0.066 |
| VGG16 [23] |
85.04±0.042 |
85.16±0.053 |
83.97±0.054 |
81.72±0.082 |
81.72±0.082 |
82.29±0.075 |
83.32±0.065 |
| Dataset |
BUSI |
| Methods |
AUC↑
|
Accuracy↑
|
Specificity↑
|
Precision↑
|
Recall↑
|
F1-score↑
|
Average↑
|
| ViT |
88.49±0.038 |
78.58±0.047 |
77.74±0.063 |
87.36±0.028 |
75.51±0.063 |
75.56±0.055 |
80.54±0.049 |
| Swin-Transformer [13] |
89.07±0.033 |
80.13±0.046 |
79.63±0.058 |
88.13±0.024 |
76.61±0.049 |
76.96±0.049 |
81.75±0.043 |
| MaxViT [27] |
89.42±0.036 |
80.39±0.041 |
78.61±0.050 |
88.48±0.027 |
77.56±0.053 |
77.63±0.045 |
82.01±0.042 |
| Efficientb3 [25] |
90.12±0.026 |
80.13±0.047 |
78.02±0.059 |
88.69±0.023 |
78.83±0.042 |
76.96±0.047 |
82.12±0.041 |
| Res50 [9] |
90.12±0.034 |
80.26±0.040 |
78.02±0.048 |
88.69±0.022 |
78.56±0.048 |
77.95±0.043 |
82.27±0.039 |
| VGG16 [23] |
90.42±0.034 |
81.42±0.032 |
79.86±0.055 |
89.04±0.020 |
78.83±0.052 |
78.73±0.039 |
83.05±0.039 |
Table 2.
Comparisons of the Backbone Model with and without using the Regional Prior Fusion on the UDIAT and BUSI datasets. ’Average’ indicates the mean value of AUC, Accuracy, Specificity, Precision, Recall and F1-score. The values following ± are standard deviations. ’RPF’: Regional Prior Fusion.
Table 2.
Comparisons of the Backbone Model with and without using the Regional Prior Fusion on the UDIAT and BUSI datasets. ’Average’ indicates the mean value of AUC, Accuracy, Specificity, Precision, Recall and F1-score. The values following ± are standard deviations. ’RPF’: Regional Prior Fusion.
| Dataset |
|
|
UDIAT |
|
| Methods |
AUC↑
|
Accuracy↑
|
Specificity↑ Precision↑
|
Recall↑
|
F1-score↑
|
Average↑
|
| Backbone |
85.04±0.042 |
85.16±0.053 |
83.97±0.054 81.72±0.082 |
81.72±0.082 |
82.29±0.075 |
83.32±0.065 |
| Backbone+RPF |
99.84±0.003 |
94.62±0.067 |
96.58±0.041 91.67±0.104 |
91.67±0.104 |
93.12±0.088 |
94.58±0.068 |
| Dataset |
|
|
BUSI |
|
|
|
| Methods |
AUC↑
|
Accuracy↑
|
Specificity↑ Precision↑
|
Recall↑
|
F1-score↑
|
Average↑
|
| Backbone |
90.42±0.034 |
81.42±0.032 |
79.86±0.055 89.04±0.020 |
78.83±0.052 |
78.73±0.039 |
83.05±0.039 |
| Backbone+RPF |
100.0±0.000 |
99.35±0.006 |
99.23±0.008 99.71±0.003 |
99.62±0.004 |
99.42±0.006 |
99.56±0.004 |
Table 3.
Ablation study of the proposed RPF-ELD method crossing the combination of early distillation and late distillation on the UDIAT and BUSI datasets. ’Average’ indicates the mean value of AUC, Accuracy, Specificity, Precision, Recall and F1-score. The values following ± are standard deviations. ’B’: Backbone model; ’S’: Student model; ’T’: Teacher model; ’ED’: Early Distillation; ’LD’: Late Distillation.
Table 3.
Ablation study of the proposed RPF-ELD method crossing the combination of early distillation and late distillation on the UDIAT and BUSI datasets. ’Average’ indicates the mean value of AUC, Accuracy, Specificity, Precision, Recall and F1-score. The values following ± are standard deviations. ’B’: Backbone model; ’S’: Student model; ’T’: Teacher model; ’ED’: Early Distillation; ’LD’: Late Distillation.
| Dataset |
|
|
UDIAT |
|
| Methods |
AUC↑
|
Accuracy↑
|
Specificity↑ Precision↑
|
Recall↑
|
F1-score↑
|
Average↑
|
| B |
85.04% |
85.16% |
83.97% 81.72% |
81.72% |
82.29% |
83.32% |
| S Distilled by T(B+ED) |
87.14% |
87.10% |
92.00% 80.00% |
80.00% |
83.15% |
84.90% |
| S Distilled by T(B+LD) |
88.10% |
87.10% |
87.23% 82.62% |
82.62% |
84.34% |
85.34% |
| S Distilled by T(B+ED+LD) |
88.26% |
88.17% |
90.33% 83.41% |
83.41% |
85.41% |
86.50% |
| Dataset |
|
|
BUSI |
|
|
|
| Methods |
AUC↑
|
Accuracy↑
|
Specificity↑
|
Precision↑
|
Recall↑
|
F1-score↑
|
Average↑
|
| B |
90.42% |
81.42% |
79.86% |
89.04% |
78.83% |
78.73% |
83.05% |
| S Distilled by T(B+ED) |
93.13% |
81.29% |
78.56% |
89.97% |
81.75% |
79.13% |
83.97% |
| S Distilled by T(B+LD) |
92.50% |
82.58% |
82.37% |
89.41% |
80.23% |
81.23% |
84.72% |
| S Distilled by T(B+ED+LD) |
93.97% |
86.02% |
83.99% |
92.10% |
84.68% |
84.12% |
87.48% |
Experimental Settings
Datasets
Two publicly released datasets, UDAIT [
29] and BUSI [
1], are adopted to validate the performance of the proposed framework in this paper.
UDAIT is proposed in the research of Yap et al . [
29], which is widely used in many studies [
6,
26,
28]. It contains 163 samples in total, including 109 benign samples and 54 malignant samples. In the experiments, 20% samples of benign and malignant cases are randomly selected to form a test set. The remaining data forms the training set. Finally, the training set contains 88 benign tumors and 44 malignant tumors, 132 samples in total. The test set contains 21 benign tumors and 10 malignant tumors, in a total of 31 samples.
BUSI is released in the study of Al-Dhabyani et al. [
1]. It contains 437 benign tumor samples, 210 malignant tumor samples, and 132 normal samples, in a total of 779 samples. Similarly, 20% of the samples are also randomly selected from benign tumors, malignant tumors, and normal cases into the test set, while the remaining data form the training set. Finally, the training set contains 350 benign tumors, 168 malignant tumors, and 106 normal samples, in a total of 624 samples in total. The test set contains 87 benign tumors, 42 malignant tumors, and 26 normal samples, in a total of 155 samples.
Evaluation Metrics
To evaluate the performance of the proposed framework in quantity, we employ commonly used metrics, including the area under the ROC curve (AUC), accuracy, specificity, precision, recall, and F1-score. To better display the overall performance of different methods, the average value of the six metrics is also included.
Implementation Details
The feature extraction models for both the teacher and student networks are chosen to use VGG16 [
23] due to its good feature representation ability, which will be discussed in the experiments. The teacher model is trained first, then, fix its parameters to train the student model using the proposed distillation method. For the training stage of the teacher model, the total epoch is set as 120, the batch size is set as 16, and the optimizer uses the SGD algorithm, with a learning rate of 1e-3, a weight decay of 5e-4 and a momentum of 0.9. For the training of the student model, the total epoch is set as 200 and the learning rate is 5e-4. Other settings are the same as the training of the teacher model. All the code in this paper is implemented using Python, with the PyTorch library for deep learning. All the experiments are run on a computer with an Ubuntu system, an NVIDIA GeForce RTX 3080 Ti GPU of 12G VRAM.
Results and Analysis
Feature Extractor Comparison
The key idea of this work is fusing the regional prior information indirectly into a diagnostic model using the distil- lation paradigm from a direct prior-fused model. Appropriate feature extraction methods for both the teacher and student models is a crucial adjective. To find high-performance feature extractors for our framework, we quantitatively compare some commonly used state-of-the-art pretrained visual models for breast cancer recognition in ultrasound images on the UDIAT and BUSI datasets, as shown in Table I. These pretrained visual models contain two groups: models based on Vision Transformer (ViT) and models based on Convolutional Neural Networks (CNNs). The ViT-based models include ViT [
30] Swin-Transformer [
13] and MaxViT [
27], while the CNN- based models include Efficientb3 [
25], Res50 [
9] and VGG16 [
23].
On the UDIAT datasets, the AUC of all models surpasses 94%, and the accuracy is greater than 80%. Among the ViT models, the MaxViT model performs the best, followed by the Swin-Transformer and the original ViT model. MaxViT shows excellent performance across various evaluation met- rics, particularly in terms of AUC and accuracy. Although ViT models show strong performance in certain metrics, among CNN models, VGG16 exhibits superior performance across multiple metrics. Comparatively, EfficientB3 and ResNet50 show strong performance but their overall perform slightly below VGG16. On the BUSI datasets, the comparative results show a similar tendency. Among the ViT-based methods, the MaxViT performs best again, with most of the scores higher than that of the other two. The CNN-based methods also show better performance than the ViT-based methods, among which, the VGG16 also outperforms all other methods crossing all the evaluation scores.
A meaningful phenomenon is found that the CNN models overall outperform the ViT models. The reason may lie in the cutting strategy of the ViT series. Due to the low proportion of tumor regions in ultrasound images, the correlation among cut patches finitely contributes to the diagnosis. In contrast, trainable filter kernels of CNNs could better extract local features of tumor regions.
From the quantitatively comparative results, the VGG16 boosts over the other models on both datasets, making it the feature extractor for both the teacher and student models in this work.
Effectiveness of the Regional Prior Information
To validate the idea of fusing the Regional Priors of tumor ROIs, we compare the performance of the selected backbone model with and without using the Regional Prior Fusion on the UDIAT and BUSI datasets as shown in Table. II. The selected Backbone model is VGG16 according to the comparisons of current state-of-the-art pretrained visual models.
From the results on both datasets, after fusing the regional prior information, the performance of the backbone model is significantly enhanced, with nearly over 10% increase in most of the evaluation scores. It indicates that the tumor regions are effective prior information for the breast cancer diagnosis. In the following, we will use the regional prior fused model as the teacher model to guide the diagnostic student model.
Ablation Study
To evaluate the effectiveness of each part of the proposed RPF-ELD method, we conduct the ablation study crossing the combination of early distillation and late distillation on the UDIAT and BUSI datasets as shown in Table III.
On both datasets, after distilling from the teacher model using only early distillation or late distillation, the performance of the backbone model is improved effectively. Furthermore, using both early distillation and late distillation from the teacher model, the diagnostic student model reaches the best performance, with all scores higher than that of the others.
The results demonstrate that both early distillation and late distillation help improve the diagnostic student model. The early distillation enhances the diagnostic student model in the visual context level, while the late distillation improves it in the deep representation level. So the combination of both strategies improves the performance significantly.
Comparison with State-of-the-art Methods
To legitimately validate the performance of our proposed RPF-ELD framework, we compare it with the current state-of- the-art ultrasound-image-based cancer diagnosis methods on the UDIAT and BUSI datasets, as shown in Table IV. The comparative methods contain GBCNet [
2], HoverTrans [
19] and REAF [
31].
On the UDIAT dataset, RPF-ELD achieved the highest scores in AUC, accuracy, and specificity, showing significant improvements in all evaluation metrics. Similarly, on the BUSI dataset, RPF-ELD led in all major performance indicators. The results on both datasets indicate that the RPF-ELD overall outperforms the state-of-the-art cancer diagnosis methods.
The advantage of our proposed framework is the fusion of regional prior information. The GBCNet uses object detection algorithms to select tumor areas first, and then analysis in the selected areas. This method bypasses lots of background information but the selected key regions are not correct abso- lutely, which may lead to mistakes. The HoverTrans considers the tissue characteristics of horizontal and vertical directions in the ultrasound images but lacks regional information. The REAF uses deep learning models to extract ROI information, which is also not correct completely, maybe leading the error accumulation. Compared with these methods, our framework fuses the real regional prior information by knowledge distil- lation methods. So the superior performance is reasonable.
Ultimately, the comparative results demonstrate that the proposed framework surpasses current state-of-the-art cancer diagnosis methods based on ultrasound images.
Conclusion
In this paper, we propose an RPF-ELD framework for breast cancer diagnosis in ultrasound images. This framework first fuses regional prior information of ROIs with ultrasound images to train a prior-fused teacher model. Then, train a diagnostic model based on only the ultrasound image as well as fuse the prior information by the distillation from the prior-fused teacher model. Finally, the diagnostic model analyzes the images without prior information. Two publicly released datasets are used to validate our proposed framework. Experimental results demonstrate that the prior improves the teacher model significantly, the prior-fused feature distillation enhances the diagnostic model effectively, and the proposed RPF-ELD surpasses current state-of-the-art methods.
Due to the variable proportion and uncertain position of tumor region in ultrasound images, the computer-aided diag- nosis of breast cancer struggles to achieve high performance. The main advantage of the proposed framework is combining information on tumor regions. Although previous studies have combined tumor region information, most of them lost capabil- ity in practical clinical settings. Incorporating the tumor region directly into the model will inevitably lead to the model losing its applicability, because the ultrasound images to be diagnosed are usually not manually labeled, that is, lack ROIs. The key to ensuring suitability is the help of knowledge distillation. Fusing regional prior information into the teacher model causes the teacher model to lose its applicability. However, the student model obtained by distillation uncouples the regional information, thus maintaining the applicability. Additionally, early and late distillation integrate regional information into the student model at different levels, further strengthening the performance of the student model. Therefore, the final diagnostic model can achieve good performance.
References
- W. S. Al-Dhabyani, M. M. M. Gomaa, H. Khaled, and A. A. Fahmy. Dataset of breast ultrasound images. Data in Brief, 28, 2019. [CrossRef]
- S. Basu, M. Gupta, P. Rana, P. Gupta, and C. Arora. Surpassing the human accuracy: Detecting gallbladder cancer from usg images with curriculum learning. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pages 20854–20864, 2022.
- A. D. Berenguer, M. Kvasnytsia, M. N. Bossa, T. Mukherjee, N. Deli- giannis, and H. Sahli. Semi-supervised medical image classification via distance correlation minimization and graph attention regularization. Medical image analysis, 94:103107, 2024. [CrossRef]
- T. B. Bevers, M. A. Helvie, E. T. Bonaccio, K. E. Calhoun, M. B. Daly, W. B. Farrar, J. E. Garber, R. Gray, C. C. Greenberg, R. A. Greenup, N. M. Hansen, R. Harris, A. S. Heerdt, T. Helsten, L. Hodgkiss, T. L. Hoyt, J. G. Huff, L. K. Jacobs, C. D. Lehman, B. S. Monsees, B. L. Niell, C. C. Parker, M. D. Pearlman, L. E. Philpotts, L. B. Shepardson, M. L. Smith, M. Stein, L. Tumyan, C. Williams, M. A. Bergman, and R. Kumar. Breast cancer screening and diagnosis, version 3.2018. Journal of the National Comprehensive Cancer Network: JNCCN, 16 11:1362–1389, 2018.
- D. Buonsenso, A. Chiaretti, A. Curatola, R. Morello, M. Giacalone, and N. Parri. Pediatrician performed point-of-care ultrasound for the detection of ingested foreign bodies: case series and review of the literature. Journal of Ultrasound, 24:107–114, 2020. [CrossRef]
- G. Chen, Y. Dai, J. Zhang, and M. H. Yap. Aau-net: An adaptive attention u-net for breast lesions segmentation in ultrasound images. IEEE Transactions on Medical Imaging, 42:1289–1300, 2022. [CrossRef]
- A. Evans, R. M. Trimboli, A. Athanasiou, C. Balleyguier, P. A. Baltzer, U. Bick, J. Camps Herrero, P. Clauser, C. Colin, E. Cornford, et al. Breast ultrasound: recommendations for information to women and referring physicians by the european society of breast imaging. Insights into imaging, 9:449–461, 2018. [CrossRef]
- J. Ferlay, M. Colombet, I. Soerjomataram, C. Mathers, D. M. Parkin, M. Pin˜eros, A. Znaor, and F. Bray. Estimating the global cancer incidence and mortality in 2018: Globocan sources and methods. Inter- national journal of cancer, 144(8):1941–1953, 2019. [CrossRef]
- K. He, X. Zhang, S. Ren, and J. Sun. Deep residual learning for image recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pages 770–778, 2015.
- H. Hu, L. Gong, D. Dong, L. Zhu, M. Wang, J. He, L. Shu, Y. Cai, S. Cai, W. Su, et al. Identifying early gastric cancer under magnifying narrow- band images with deep learning: a multicenter study. Gastrointestinal Endoscopy, 93(6):1333–1341, 2021. [CrossRef]
- V. P. Jackson. The current role of ultrasonography in breast imaging. Radiologic clinics of North America, 33(6):1161–1170, 1995. [CrossRef]
- S. Lei, R. Zheng, S. Zhang, R. Chen, S. Wang, K. Sun, H. Zeng, W. Wei, and J. He. Breast cancer incidence and mortality in women in china: temporal trends and projections to 2030. Cancer biology & medicine, 18(3):900, 2021. [CrossRef]
- Z. Liu, Y. Lin, Y. Cao, H. Hu, Y. Wei, Z. Zhang, S. Lin, and B. Guo. Swin transformer: Hierarchical vision transformer using shifted windows. 2021 IEEE/CVF International Conference on Computer Vision (ICCV), pages 9992–10002, 2021.
- Y. Ma, W. Zhou, R. Ma, E. Wang, S. Yang, Y.-M. Tang, X.-P. Zhang, and X. Guan. Dove: Doodled vessel enhancement for photoacoustic angiography super resolution. Medical image analysis, 94:103106, 2024. [CrossRef]
- N. J. Massat, A. Dibden, D. Parmar, J. M. Cuzick, P. D. Sasieni, and S. W. Duffy. Impact of screening on breast cancer mortality: The uk program 20 years on. Cancer Epidemiology, Biomarkers & Prevention, 25:455 – 462, 2015. [CrossRef]
- M. Masud, A. E. Eldin Rashed, and M. S. Hossain. Convolutional neural network-based models for diagnosis of breast cancer. Neural Computing and Applications, 34(14):11383–11394, 2022. [CrossRef]
- M. Masud, A. E. E. Rashed, and M. S. Hossain. Convolutional neural network-based models for diagnosis of breast cancer. Neural Computing & Applications, 34:11383 – 11394, 2020. [CrossRef]
- P. Mehnati and M. J. Tirtash. Comparative efficacy of four imaging instruments for breast cancer screening. Asian Pacific Journal of Cancer Prevention, 16(15):6177–6186, 2015. [CrossRef]
- Y. Mo, C. Han, Y. Liu, M. Liu, Z. Shi, J. Lin, B. Zhao, C. Huang, B. Qiu, Y. Cui, L. Wu, X. Pan, Z. Xu, X. Huang, Z. Li, Z. Liu, Y. Wang, and Liang. Hover-trans: Anatomy-aware hover-transformer for roi-free breast cancer diagnosis in ultrasound images. IEEE Transactions on Medical Imaging, 42(6):1696–1706, 2023. [CrossRef]
- W. K. Moon, Y.-W. Lee, H.-H. Ke, S. H. Lee, C.-S. Huang, and R.-F. Chang. Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks. Computer methods and programs in biomedicine, 190:105361, 2020. [CrossRef]
- M. Nothacker, V. Duda, M. Hahn, M. Warm, F. Degenhardt, H. Madjar, S. Weinbrenner, and U.-S. Albert. Early detection of breast cancer: benefits and risks of supplemental breast ultrasound in asymptomatic women with mammographically dense breast tissue. a systematic review. BMC cancer, 9:1–9, 2009. [CrossRef]
- C. M. Sehgal, S. P. Weinstein, P. H. Arger, and E. F. Conant. A review of breast ultrasound. Journal of mammary gland biology and neoplasia, 11:113–123, 2006. [CrossRef]
- K. Simonyan and A. Zisserman. Very deep convolutional networks for large-scale image recognition. CoRR, abs/1409.1556, 2014.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, Jemal, and F. Bray. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: a cancer journal for clinicians, 71(3):209–249, 2021. [CrossRef]
- M. Tan and Q. V. Le. Efficientnet: Rethinking model scaling for convolutional neural networks. Proceedings of Machine Learning Research, 97, 2019.
- C. Thomas, M. Byra, R. Mart´ı, M. H. Yap, and R. Zwiggelaar. Bus-set: A benchmark for quantitative evaluation of breast ultrasound segmentation networks with public datasets. Medical physics, 2023.
- Z. Tu, H. Talebi, H. Zhang, F. Yang, P. Milanfar, A. C. Bovik, and Y. Li. Maxvit: Multi-axis vision transformer. In European Conference on Computer Vision, 2022.
- M. H. Yap, M. Goyal, F. Osman, R. Mart´ı, E. R. E. Denton, A. Juette, and R. Zwiggelaar. Breast ultrasound region of interest detection and lesion localisation. Artificial intelligence in medicine, 107:101880, 2020.
- M. H. Yap, G. Pons, J. Mart´ı, S. Ganau, M. Sent´ıs, R. Zwiggelaar, A. K. Davison, and R. Mart´ı. Automated breast ultrasound lesions detection using convolutional neural networks. IEEE Journal of Biomedical and Health Informatics, 22:1218–1226, 2018.
- L. Yuan, Y. Chen, T. Wang, W. Yu, Y. Shi, F. E. H. Tay, J. Feng, and S. Yan. Tokens-to-token vit: Training vision transformers from scratch on imagenet. 2021 IEEE/CVF International Conference on Computer Vision (ICCV), pages 538–547, 2021.
- Z. Zhang, J. W. Lim, Y. Zheng, B. Chen, D. Chen, and Y. Lin. Reaf: Roi extraction and adaptive fusion for breast cancer diagnosis in ultrasound images. In 2023 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 3422–3429, 2023.
- W. Zhou, Y. Yang, C. Yu, J. Liu, X. Duan, Z. Weng, D. Chen, Q. Liang, Q. Fang, J. Zhou, et al. Ensembled deep learning model outperforms human experts in diagnosing biliary atresia from sonographic gallbladder images. Nature communications, 12(1):1259, 2021. [CrossRef]
- Z. Zhuang, Z. Yang, A. N. J. Raj, C. Wei, P. Jin, and S. Zhuang. Breast ultrasound tumor image classification using image decomposition and fusion based on adaptive multi-model spatial feature fusion. Computer Methods and Programs in Biomedicine, 208:106221, 2021. [CrossRef]
- H. M. Zonderland, E. G. Coerkamp, J. Hermans, M. J. van de Vijver, and A. E. van Voorthuisen. Diagnosis of breast cancer: contribution of us as an adjunct to mammography. Radiology, 213(2):413–422, 1999. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).