Submitted:
18 November 2024
Posted:
18 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
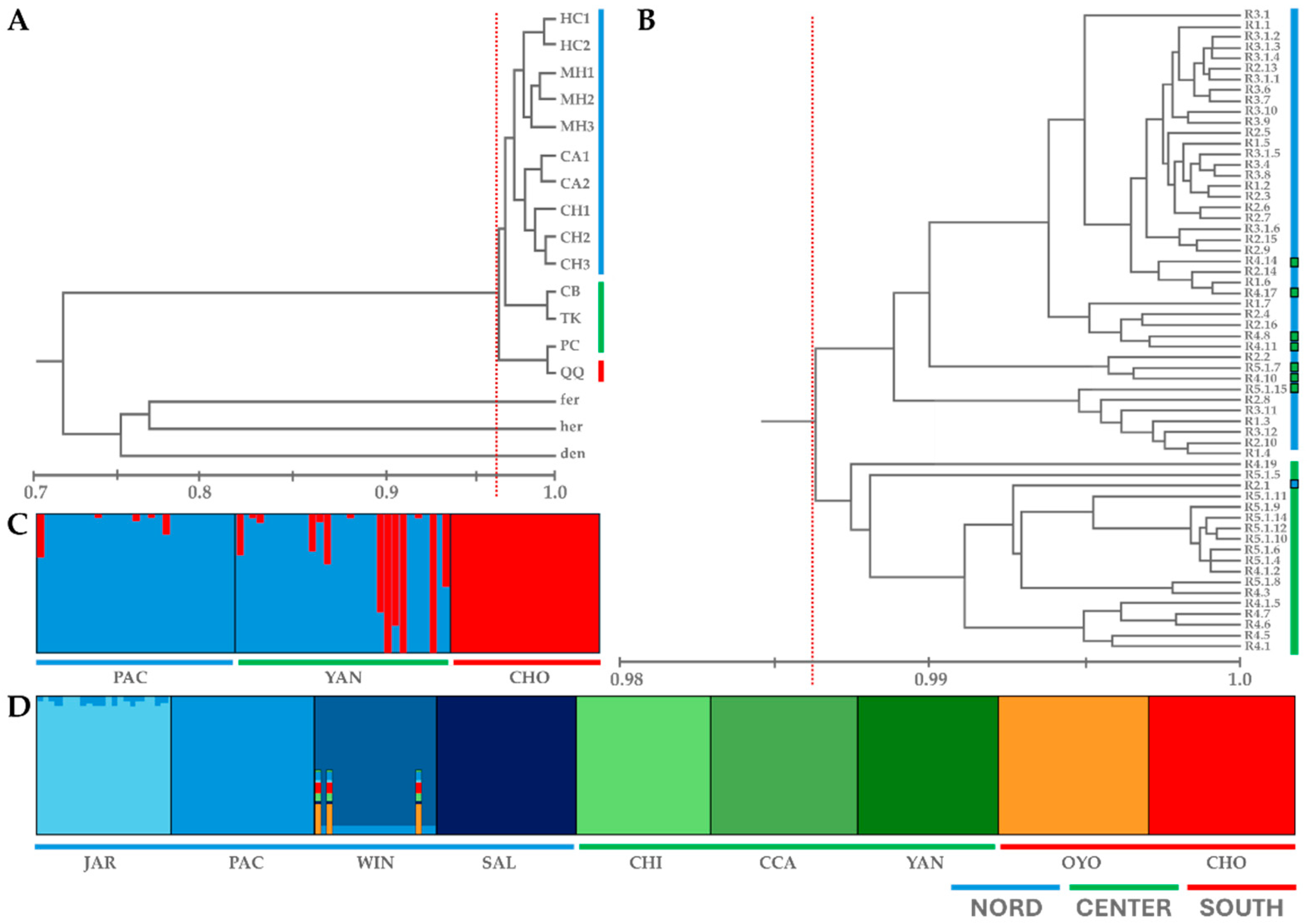
2. Results and Discussion
3. Methodology
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lambe: A. Puya raimondii The IUCN Red List of Threatened Species 2009: E. T168358A6482345. Website https://www.iucnredlist.org/species/168358/6482345 [accessed 15 November 2019] 2009.
- Benzing, D.H. Bromeliaceae: Profile of an Adaptive Radiation; Cambridge University Press, 2000; ISBN 0521430313.
- Hornung-Leoni, C.T.; González-Gómez, P.L.; Troncoso, A.J. Morphology, Nectar Characteristics and Avian Pollinators in Five Andean Puya Species (Bromeliaceae). Acta Oecologica 2013, 51, 54–61. [Google Scholar] [CrossRef]
- Sgorbati, S.; Labra, M.; Grugni, E.; Barcaccia, G.; Galasso, G.; Boni, U.; Mucciarelli, M.; Citterio, S.; Iramátegui, A.B.; Gonzales, L.V. A Survey of Genetic Diversity and Reproductive Biology of Puya raimondii (Bromeliaceae), the Endangered Queen of the Andes. Plant Biol 2004, 6, 222–230. [Google Scholar] [CrossRef] [PubMed]
- Hornung-Leoni, C.T.; Sosa, V.; Simpson, J.; Gil, K. Genetic Variation in the Emblematic Puya raimondii (Bromeliaceae) from Huascarán National Park, Peru. Crop Breeding and Applied Biotechnology 2013, 13, 67–74. [Google Scholar] [CrossRef]
- Tumi, L.; Ge, X.J.; Prado, G.E.; Cosacov, A.; García, V.H.; Arakaki, M.; Suni, M.L. Genetic Diversity and Genetic Structure of Puya raimondii (Bromeliaceae) for Its Conservation in the Peruvian Andes. Rev Peru Biol 2022, 29. [Google Scholar] [CrossRef]
- Liu, L.; James, J.; Zhang, Y.; Wang, Z.; Arakaki, M.; Vadillo, G.; Zhou, Q.; Lascoux, M.; Ge, X. The ‘Queen of the Andes’(Puya raimondii) Is Genetically Fragile and Fragmented: A Consequence of Long Generation Time and Semelparity? New Phytologist 2024. [CrossRef] [PubMed]
- Nei, M.; Li, W.-H. Mathematical Model for Studying Genetic Variation in Terms of Restriction Endonucleases. Proceedings of the National Academy of Sciences 1979, 76, 5269–5273. [Google Scholar] [CrossRef] [PubMed]
- Tumi, L.; Zhang, Y.; Wang, Z.; Suni, M.L.; Burgess, K.S.; Ge, X. Microsatellite Markers for the Endangered Puya raimondii in Peru. Appl Plant Sci 2019, 7, e11308. [Google Scholar] [CrossRef]
- Barcaccia, G. Using Molecular Markers for Characterizing and Preserving Germplasm Resources. In Molecular techniques in crop improvement (2nd edition); Springer Publisher, 2010; pp. 231–255 ISBN 9048129664.
- Ajmone Marsan, P.; Castiglioni, P.; Fusari, F.; Kuiper, M.; Motto, M. Genetic Diversity and Its Relationship to Hybrid Performance in Maize as Revealed by RFLP and AFLP Markers. Theoretical and Applied Genetics 1998, 96, 219–227. [Google Scholar] [CrossRef]
- Blas, R.; Ghislain, M.; del Rosario Herrera, M.; Baudoin, J.-P. Genetic Diversity Analysis of Wild Arracacia Species According to Morphological and Molecular Markers. Genet Resour Crop Evol 2008, 55, 625–642. [Google Scholar] [CrossRef]
- Schulman, A.H. Molecular Markers to Assess Genetic Diversity. Euphytica 2007, 158, 313–321. [Google Scholar] [CrossRef]
- Mba, C.; Tohme, J. Use of AFLP Markers in Surveys of Plant Diversity. In Methods in Enzymology; Elsevier, 2005; Vol. 395, pp. 177–201 ISBN 0076-6879.
- Porcher, E.; Lande, R. Inbreeding Depression under Mixed Outcrossing, Self-Fertilization and Sib-Mating. BMC Evol Biol 2016, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Duthie, A.B.; Reid, J.M. Evolution of Inbreeding Avoidance and Inbreeding Preference through Mate Choice among Interacting Relatives. Am Nat 2016, 188, 651–667. [Google Scholar] [CrossRef]
- Fortier, R.P. Queen of the Andes: The Ecology and Conservation of Puya raimondii. Frontiers in Conservation Science 2024, 5, 1349553. [Google Scholar] [CrossRef]
- Ramirez-Villegas, J.; Cuesta, F.; Devenish, C.; Peralvo, M.; Jarvis, A.; Arnillas, C.A. Using Species Distributions Models for Designing Conservation Strategies of Tropical Andean Biodiversity under Climate Change. J Nat Conserv 2014, 22, 391–404. [Google Scholar] [CrossRef]
- Salazar Castillo, J.; Caceres de Baldarrago, F.; Poma, I.; Raimondo, F.M. Diagnostico Del Estado Actual de Consevación de Puya raimondii En Arequipa–Perù. Quaderni di Botanica ambientale e applicata 2010, 21, 83–91. [Google Scholar]
- Suni, M.L.; Vadillo, G.P.; Arana, C.; Jara-Peña, E.; Salinas, L.; Ponce, M.E.; Ramsay, P.M. Post-Fire Recovery of Puya raimondii, Vegetation and Birds in the Puna of Huascarán National Park, Perú. J Mt Sci 2024, 21, 20–32. [Google Scholar] [CrossRef]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.B.; Kent, J. Biodiversity Hotspots for Conservation Priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.P.; Marengo, J.; Villalba, R.; Halloy, S.; Young, B.; Cordero, D.; Gast, F.; Jaimes, E.; Ruiz, D.; Herzog, S.K. Consequences of Climate Change for Ecosystems and Ecosystem Services in the Tropical Andes. Climate change and biodiversity in the tropical Andes 2011, 1, 1–18. [Google Scholar]
- Barcaccia, G.; Mazzucato, A.; Albertini, E.; Zenoni, S.; Baldoni, L.; Mousavi, S.; Mendes, M.A.; Coimbra, S.; Granell, A.; Pupilli, F. Genetics and Genomics of Plant Reproduction for Crop Breeding, Volume II. Front Plant Sci 2023, 14, 1145208. [Google Scholar] [CrossRef]
- Barcaccia, G. Molecular Markers for Characterizing and Conserving Crop Plant Germplasm. Molecular Techniques in Crop Improvement: 2nd Edition 2009, 231–254.
- Nei, M. The Theory and Estimation of Genetic Distance. 1973.
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution (N Y) 1965, 395–420.
- Palumbo, F.; Galla, G.; Martínez-Bello, L.; Barcaccia, G. Venetian Local Corn (Zea mays L.) Germplasm: Disclosing the Genetic Anatomy of Old Landraces Suited for Typical Cornmeal Mush Production. Diversity (Basel) 2017, 9, 32. [Google Scholar] [CrossRef]
- Palumbo, F.; Galla, G.; Barcaccia, G. Developing a Molecular Identification Assay of Old Landraces for the Genetic Authentication of Typical Agro-Food Products: The Case Study of the Barley ‘Agordino. ’ Food Technol Biotechnol 2017, 55, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Porfiri, O.; Torricelli, R.; Silveri, D.D.; Papa, R.; Barcaccia, G.; Negri, V. The Triticeae Genetic Resources of Central Italy: Collection, Evaluation and Conservation. Hereditas 2001, 135, 187–192. [Google Scholar] [CrossRef]
- Palumbo, F.; Galla, G.; Vitulo, N.; Barcaccia, G. First Draft Genome Sequencing of Fennel (Foeniculum vulgare Mill.): Identification of Simple Sequence Repeats and Their Application in Marker-Assisted Breeding. Molecular Breeding 2018, 38, 1–17. [Google Scholar] [CrossRef]
- Patella, A.; Palumbo, F.; Galla, G.; Barcaccia, G. The Molecular Determination of Hybridity and Homozygosity Estimates in Breeding Populations of Lettuce (Lactuca sativa L.). Genes (Basel) 2019, 10, 916. [Google Scholar] [CrossRef] [PubMed]
- Patella, A.; Palumbo, F.; Ravi, S.; Stevanato, P.; Barcaccia, G. Genotyping by RAD Sequencing Analysis Assessed the Genetic Distinctiveness of Experimental Lines and Narrowed down the Genomic Region Responsible for Leaf Shape in Endive (Cichorium endivia L.). Genes (Basel) 2020, 11, 462. [Google Scholar] [CrossRef] [PubMed]
- Sica, P.; Scariolo, F.; Galvao, A.; Battaggia, D.; Nicoletto, C.; Maucieri, C.; Palumbo, F.; Franklin, D.; Cabrera, M.; Borin, M. Molecular Hallmarks, Agronomic Performances and Seed Nutraceutical Properties to Exploit Neglected Genetic Resources of Common Beans Grown by Organic Farming in Two Contrasting Environments. Front Plant Sci 2021, 12, 674985. [Google Scholar] [CrossRef]
- Palumbo, F.; Galvao, A.C.; Nicoletto, C.; Sambo, P.; Barcaccia, G. Diversity Analysis of Sweet Potato Genetic Resources Using Morphological and Qualitative Traits and Molecular Markers. Genes (Basel) 2019, 10, 840. [Google Scholar] [CrossRef]
- Palumbo, F.; Qi, P.; Pinto, V.B.; Devos, K.M.; Barcaccia, G. Construction of the First SNP-Based Linkage Map Using Genotyping-by-Sequencing and Mapping of the Male-Sterility Gene in Leaf Chicory. Front Plant Sci 2019, 10, 276. [Google Scholar] [CrossRef]
- Hmmam, I.; Mariotti, R.; Ruperti, B.; Cultrera, N.; Baldoni, L.; Barcaccia, G. Venetian Olive (Olea europaea) Germplasm: Disclosing the Genetic Identity of Locally Grown Cultivars Suited for Typical Extra Virgin Oil Productions. Genet Resour Crop Evol 2018, 65, 1733–1750. [Google Scholar] [CrossRef]
- Nicolè, S.; Barcaccia, G.; Erickson, D.L.; Kress, J.W.; Lucchin, M. The Coding Region of the UFGT Gene Is a Source of Diagnostic SNP Markers That Allow Single-Locus DNA Genotyping for the Assessment of Cultivar Identity and Ancestry in Grapevine (Vitis vinifera L.). BMC Res Notes 2013, 6, 1–13. [Google Scholar] [CrossRef]
- Draga, S.; Palumbo, F.; Miracolo Barbagiovanni, I.; Pati, F.; Barcaccia, G. Management of Genetic Erosion: The (Successful) Case Study of the Pear (Pyrus communis L.) Germplasm of the Lazio Region (Italy). Front Plant Sci 2023, 13, 1099420. [Google Scholar] [CrossRef] [PubMed]
- Borin, M.; Palumbo, F.; Vannozzi, A.; Scariolo, F.; Sacilotto, G.B.; Gazzola, M.; Barcaccia, G. Developing and Testing Molecular Markers in Cannabis sativa (Hemp) for Their Use in Variety and Dioecy Assessments. Plants 2021, 10, 2174. [Google Scholar] [CrossRef] [PubMed]
- Barcaccia, G.; Arzenton, F.; Sharbel, T.F.; Varotto, S.; Parrini, P.; Lucchin, M. Genetic Diversity and Reproductive Biology in Ecotypes of the Facultative Apomict Hypericum perforatum L. Heredity (Edinb) 2006, 96, 322–334. [Google Scholar] [CrossRef] [PubMed]

| No. | No. | DNA | Genetic diversity statistics* | ||||||
| Authors | pops | genotypes | analysis | Ht | Hs | Dst | Fis | Fst | Nm |
| Sgorbati et al. [4] | 8 | 160 | AFLP | 0.295 | 0.011 | 0.284 | n.a. | 0.961 | 0.02 |
| Hornung-Leoni et al. [5] | 5 | 60 | AFLP | 0.230 | 0.197 | 0.033 | n.a. | 0.144 | 2.97 |
| Tumi et al. [6] | 3 | 84 | SSR | 0.378 | 0.217 | 0.161 | 0.776 | 0.426 | 0.34 |
| Liu et al. [7] | 9 | 200 | WGS | 0.114 | 0.009 | 0.105 | 0.700 | 0.88‒0.92 | 0.02‒0.03 |
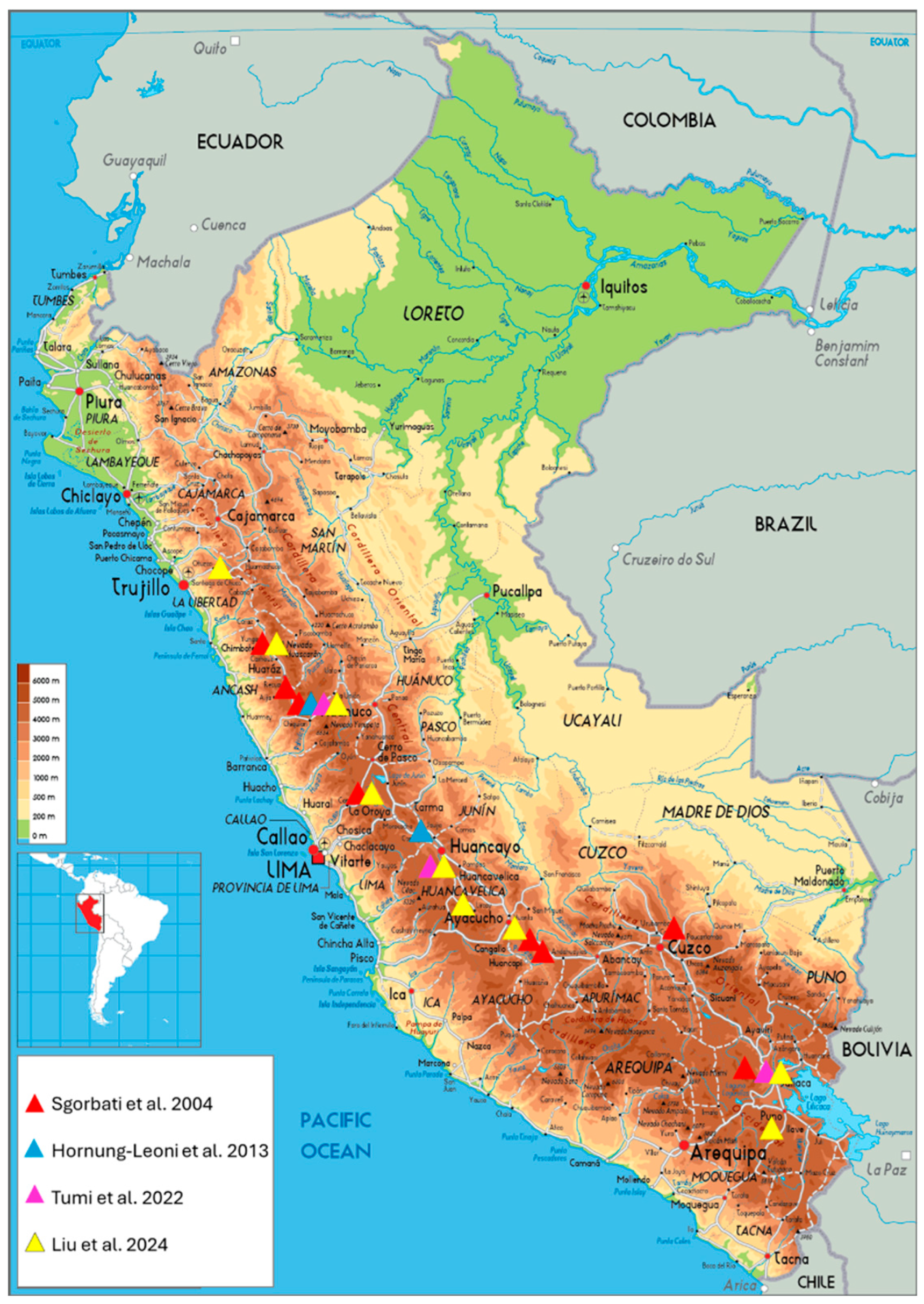
| Authors | Population/Individuals | Location | Geographic coordinates |
| Sgorbati et al. [4] | Huinchos (HC)/20 | Pueblo Libre, Ancash | -9.1333°, -77.8667° |
| Minas Huinac (MH)/20 | Aija, Ancash | -9.7000°, -77.6667° | |
| Carpa (CA)/20 | Recuay, Ancash | -9.8833°, -77.2833° | |
| Cerro Huaypian (CH)/20 | Huaros, Lima | -11.3833°, -76.5333° | |
| Pampa-corral (PC)/20 | Lares, Cusco | -13.1500°, -71.9833° | |
| Carabamba (CB)/20 | Chiara, Ayacucho | -13.4500°, -74.1333° | |
| Titankayocc, (TK)/20 | Vischongo, Ayacucho | -13.5667°, -73.9833° | |
| Quello Quello (QQ)/20 | Lampa, Puno | -15.2500°, -70.3500° | |
| Hornung-Leoni et al. [5] | R1/7 | N.P. Huascarán | -9.87697°, -77.27311° |
| R2/14 | N.P. Huascarán | -9.88333°, -77.25722° | |
| R3/14 | N.P. Huascarán | -9.89056°, -77.28000° | |
| R4/13 | Canchayllo | -11.83225°, -75.71535° | |
| R5/10 | Canchayllo | -11.83000°, -75.69556° | |
| Tumi et al. [6] | Yanacancha (YAN)/15 | Cachi, Chupaca, Yanacancha | -12.247°, -75.475° |
| Yanacancha (YAN)/14 | Huáscar, Chupaca, Yanacancha | -12.236°, -75.440° | |
| Pacahapaqui (PAC)/28 | Bolognesi, Aquia | -9.958°, -77.088° | |
| Lampa (CHO)/27 | Choconchaca, Lampa | -15.258°, -70.088° | |
| Liu et al. [7] | OYO/23 | Ichuna, General Sanchez Cerro, Moquegua | -16.167°, -70.5825° |
| CHO/23 | Lampa, Lampa, Puno | -15.2581°, -70.0883° | |
| CHI/22 | Chiara, Huamanga, Ayacucho | -13.2743°, -74.2054° | |
| CCA/23 | Huancavelica, Huancavelica | -12.8254°, -75.0678° | |
| YAN/23 | Cachi and Huáscar, Junin, Chupaca, Yanacancha | -12.2471°, -75.4755° | |
| JAR/22 | Huaros, Canta, Lima | -11.3913°, -76.5601° | |
| PAC/23 | Bolognesi, Aquia, Ancash | -9.958°, -77.0881° | |
| WIN/22 | Huaylas, Pueblo Libre, Ancash | -9.1059°, -77.8677° | |
| SAL/22 | Otuzco, Salpo, La Libertad | -8.0685°, -78.5744° |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





