1. Introduction
Worldwide, medical imaging technology plays a vital role in the diagnosis and treatment of diseases. X-ray, CT and MRI have become the core tools for clinicians to detect, diagnose and plan treatment, especially in the early screening of diseases such as pneumonia and brain tumors, medical images provide reliable and non-invasive diagnostic means. However, with the increasing demand for medical care, manual analysis of a large number of medical images is not only time-consuming but also easily influenced by subjective factors, which may lead to misdiagnosis or missed diagnosis. Therefore, how to improve the automatic processing level of medical images, and then improve the accuracy and efficiency of diagnosis, has become an important research direction in the medical and computer science fields.
Especially during the epidemic in COVID-19, the shortage of medical resources prompted all walks of life to accelerate the promotion of medical imaging automation technology to reduce the burden on medical workers and improve screening efficiency. In this context, the "Convolutional Neural Network (CNN)" in computer vision technology rises rapidly and becomes the mainstream method of medical image classification and segmentation. With its powerful automatic feature extraction ability, CNN can effectively process complex image data and show excellent performance in the identification of pneumonia, tumors and other diseases.
In recent years, CNN-based medical image processing, particularly with models like UNet and ResNet, has significantly advanced classification and segmentation tasks. UNet’s U-shaped structure enables effective global and local feature extraction, while ResNet's residual connections address gradient disappearance, enhancing classification. Despite these successes, challenges persist: obtaining high-quality annotated data remains difficult due to reliance on medical experts, which limits model generalization. Additionally, segmentation and classification accuracy need improvement for complex, heterogeneous lesions. Interpretability is also critical, as clinicians require not only accurate predictions but insights into the model's decision-making to build trust in its outputs.
This study seeks to optimize CNN models for efficient, multi-task image processing of X-ray and MRI scans for pneumonia and brain tumor diagnoses. By developing a multi-task CNN model, the research aims to enhance classification and segmentation accuracy with limited labeled data, and improve clinical credibility through interpretability techniques. Using a pre-trained ResNet50 model, adaptively adjusted for classification and segmentation, the study will employ real medical datasets and measure performance through metrics like accuracy, recall, and F1 score. This approach promises reliable diagnostic tools to aid clinicians, reducing misdiagnosis and advancing intelligent medical technology.
2. Related Work
Classification and segmentation of medical images play an important role in medical diagnosis, especially in the fields of pneumonia and tumor detection. With the introduction of deep learning, especially convolutional neural network (CNN), the accuracy and efficiency of automated medical image analysis have been significantly improved [
1]. CNN is widely used in disease detection, lesion segmentation and other tasks because of its powerful feature extraction ability, which can effectively process complex image data [
2]. The following is an overview of the main developments in this field:
Olaf Ronneberger, Philipp Fischer, and Thomas Brox introduced the UNet model, designed for medical image segmentation with a U-shaped structure for precise segmentation [
3]. It achieved over 92% accuracy in the ISBI cell tracking challenge, demonstrating its effectiveness, though it may struggle with large, complex tumor images.
UNet++ improves upon UNet by enhancing multi-scale feature fusion through a nested, dense skip-connection structure, leading to more precise segmentation edges [
4]. Proposed by Zongwei Zhou, UNet++ achieves high accuracy, especially for complex lesion shapes, with a Dice coefficient of 83.7% in lung nodule segmentation, outperforming the original UNet [
5]. Additionally, it performs well in segmenting lung and liver lesions.
The ResNet model, developed by He et al., addresses gradient disappearance in deep networks with residual connections, enabling training of very deep layers [
6]. Though initially for classification, ResNet’s robust feature extraction has proven effective in medical image classification and segmentation, achieving over 80% accuracy on the ChestX-ray14 dataset. Its scalability and stability make ResNet a reliable choice across diverse medical datasets for classification and feature extraction tasks.
DenseNet model was put forward by Gao Huang, Zhuang Liu and others [
7]. It enhanced the transmission of information and gradient and alleviated the problem of gradient disappearance by establishing a dense connection between each layer and the output of all previous layers. This innovative structure makes DenseNet perform well in the task of medical image classification and segmentation. Compared with ResNet, DenseNet reduces redundant information by sharing features, with fewer parameters and higher calculation efficiency [
8]. For example, DenseNet achieved an accuracy of 85.5% in the task of skin cancer detection, which was significantly improved compared with other depth models. Its efficient feature sharing mechanism made it the preferred model when data was scarce [
9,
10].
3. Pneumonia Type Recognition Model
3.1. Transfer Learning
In this study, the strategy of transfer learning is used to improve the performance of the model, especially in the task of medical image classification with small data, transfer learning can effectively improve the convergence speed and accuracy of the model. More specifically, we use ResNet50 as the pre-training model, which has been trained on large-scale ImageNet data sets and has strong feature extraction ability. ImageNet data set contains 1000 kinds of natural images, and the model can extract low-level to high-level image features by learning the common features in these categories [
11]. In the process of migration learning, the first layers of ResNet50 remain frozen as feature extractors, and the last fully connected layer is replaced by a new three-category output layer to adapt to the classification task of pneumonia categories (normal, bacterial pneumonia and viral pneumonia) in this study. By freezing the previous convolution layer, we can make use of the effective features learned by the pre-training model on a large number of images, avoid training the deep neural network from scratch, and then reduce the dependence on the amount of training data [
12].
3.2. Mathematical Ideas of Resnet
ResNet(Residual Network) was originally designed to solve the degradation problem caused by the increase of the depth of neural network, that is, when the number of network layers increases, the performance of deep network decreases. The traditional neural network tries to directly fit the complex mapping function, while ResNet makes the network learn the residual function F(x) by introducing the "Residual Block" instead of learning the expected mapping function directly. Mathematically, the output of residual connection can be expressed as:
where F(x) represents the output after passing through the convolution layer and the nonlinear activation function, and is the input. Through this residual learning method, the network can avoid the problem of gradient disappearance and allow the model to obtain stronger expression ability through more layers. ResNet50 uses 50 residual blocks, and each residual block directly transmits the input information through jump connection. In this study, the residual structure of ResNet50 plays a key role in pneumonia image classification, especially when dealing with complex X-ray images, which can extract effective image features. In addition, the combination of Batch Normalization and ReLU activation function further improves the stability and training efficiency of the network [
13].
3.3. Model Training and Parameter Setting
The main structure of the model is based on ResNet50 pre-training model. In order to adapt to the task of pneumonia type identification, we modified the output layer of ResNet50, replacing its last fully connected layer with a three-category output layer to meet the needs of three categories of classification in this study. We also define a self-defined model wrapper class COVID_Detector, which transforms the output of ResNet50 into a class probability distribution through the Softmax layer to ensure that the model output can be used for classification tasks.
The model is trained using the SGD optimizer with a learning rate scheduler and cross-entropy loss function. Key parameters include 20 epochs, an initial learning rate of 0.1 (decaying by 0.1 every 10 epochs), a momentum of 0.9, and a batch size of 32. Throughout training, loss and accuracy are monitored in real-time for both the training and validation sets. At each epoch’s end, validation performance is evaluated, and the model with the highest validation accuracy is saved as the best model. During evaluation, the model is set to evaluation mode to assess loss and accuracy without updating gradients.
In the process of model optimization, stochastic gradient descent (SGD) optimizer is adopted. SGD is a classic optimization algorithm, which is widely used in the training process of deep learning models, especially on large-scale data sets. By calculating the gradient on each training sample or small batch of data and updating the model parameters, the loss function value is gradually reduced, and the model gradually approaches the global optimum in the training process. The updating formula is . In this study, SGD also introduces momentum, which can accelerate the convergence process of gradient descent. The momentum method uses the formula to avoid large oscillation on the complex loss surface, especially when the gradient update oscillates in a certain direction.
4. Result
4.1. Data set
Lung X-ray imaging data is the dataset used in this experiment, i.e., the dataset used to differentiate the diagnostic accuracy of COVID-19 pneumonia, normal pneumonia and viral pneumonia. During the training process, we explicitly divided the dataset into two parts: the training set and the test set. In addition, in order to optimize the model training process and further improve the generalization ability of the model, we randomly partitioned the training data into a training subset and a validation subset based on the ratio of 85:15, and validated the actual performance of the model with an independent test set.
4.2. Model Training Results
As can be seen from the classification report in the figure, the model performs well in the classification task, and the main indicators include Precision, Recall and F1-Score. The accuracy rate measures the proportion of samples predicted by the model as actually belonging to a certain category. The formula is: Precision=TP/(TP+FP), TP(True Positive) indicates the number of samples that are truly positive and correctly classified as positive, and FP(False Positive) indicates the number of samples that are actually negative but wrongly classified as positive. The recall rate measures the proportion of samples that actually belong to a certain category that are correctly classified, and the formula is: Recall=TP/(TP+FN), where FN(False Negative) represents the number of samples that are actually positive but wrongly classified as negative. F1 score is the harmonic average of precision and recall, which can comprehensively measure the accuracy and completeness of the model. The formula is: F1-score = 2 (precision× recall)/(precision+recall).
The model excels in classifying normal lungs, bacterial, and viral pneumonia. For normal lungs, both precision and recall reach 1.00, indicating perfect classification. Bacterial pneumonia has an accuracy of 1.00 and a recall of 0.99, with only one misclassified sample. Viral pneumonia achieves a perfect recall of 1.00, with minimal accuracy deviation, demonstrating overall excellent performance across all tasks.
Table 1.
Model classification result.
Table 1.
Model classification result.
| |
Precision |
Recall |
F1-score |
Support |
| 0 |
1.00 |
1.00 |
1.00 |
111 |
| 1 |
1.00 |
0.99 |
0.99 |
71 |
| 2 |
0.99 |
1.00 |
0.99 |
69 |
| Accuracy |
|
|
1.00 |
251 |
| Macro avg |
1.00 |
1.00 |
1.00 |
251 |
| Weighted avg |
1.00 |
1.00 |
1.00 |
251 |
4.3. Loss Function and Accuracy
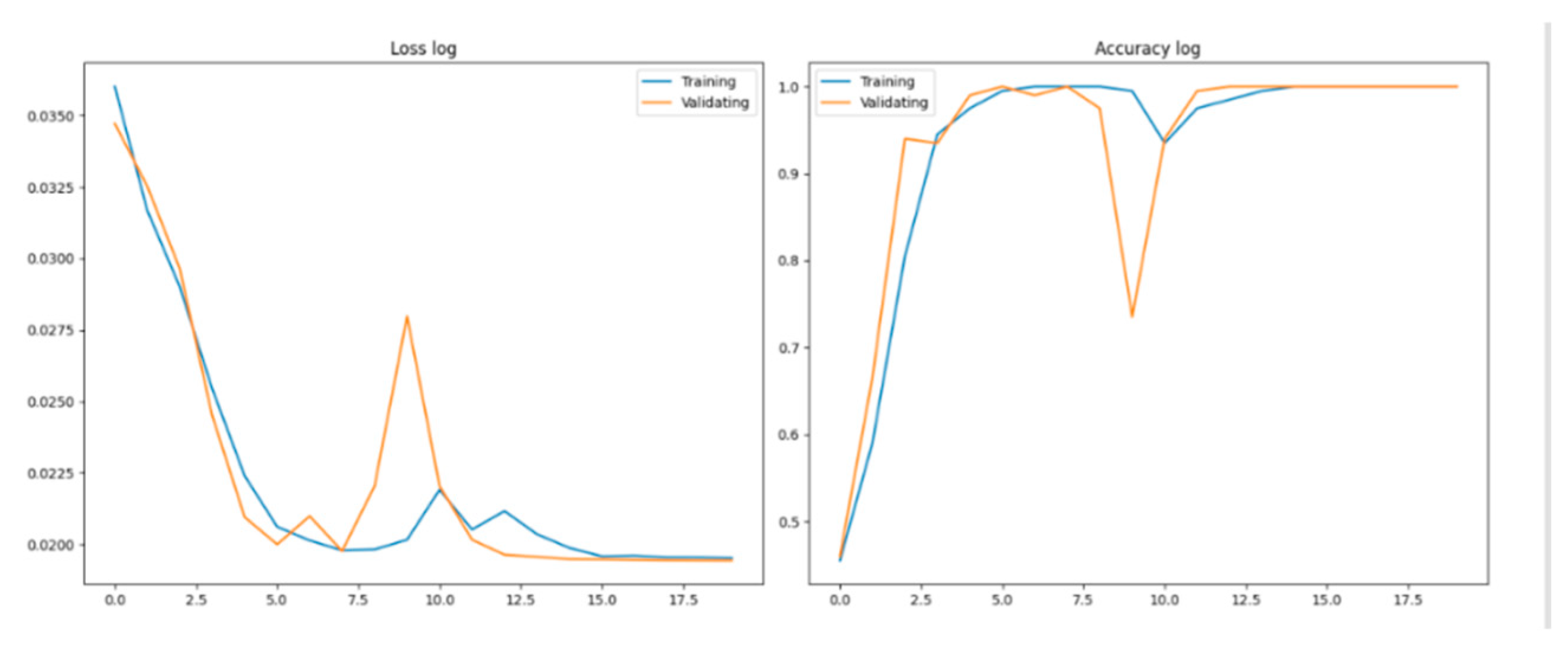
The loss function measures the error between model predictions and true labels, with lower values indicating closer alignment. The Loss log curve shows a rapid initial decrease, stabilizing as the model converges well without overfitting, as validation loss closely follows training loss. Accuracy, a key metric, rises quickly, reflecting the model’s effective learning. By the fifth round, validation accuracy exceeds 90% and eventually nears 100%, indicating strong generalization in classifying the three pneumonia types with consistent performance across both training and validation sets.
Figure 2.
Loss curve and Accuracy cure.
Figure 2.
Loss curve and Accuracy cure.
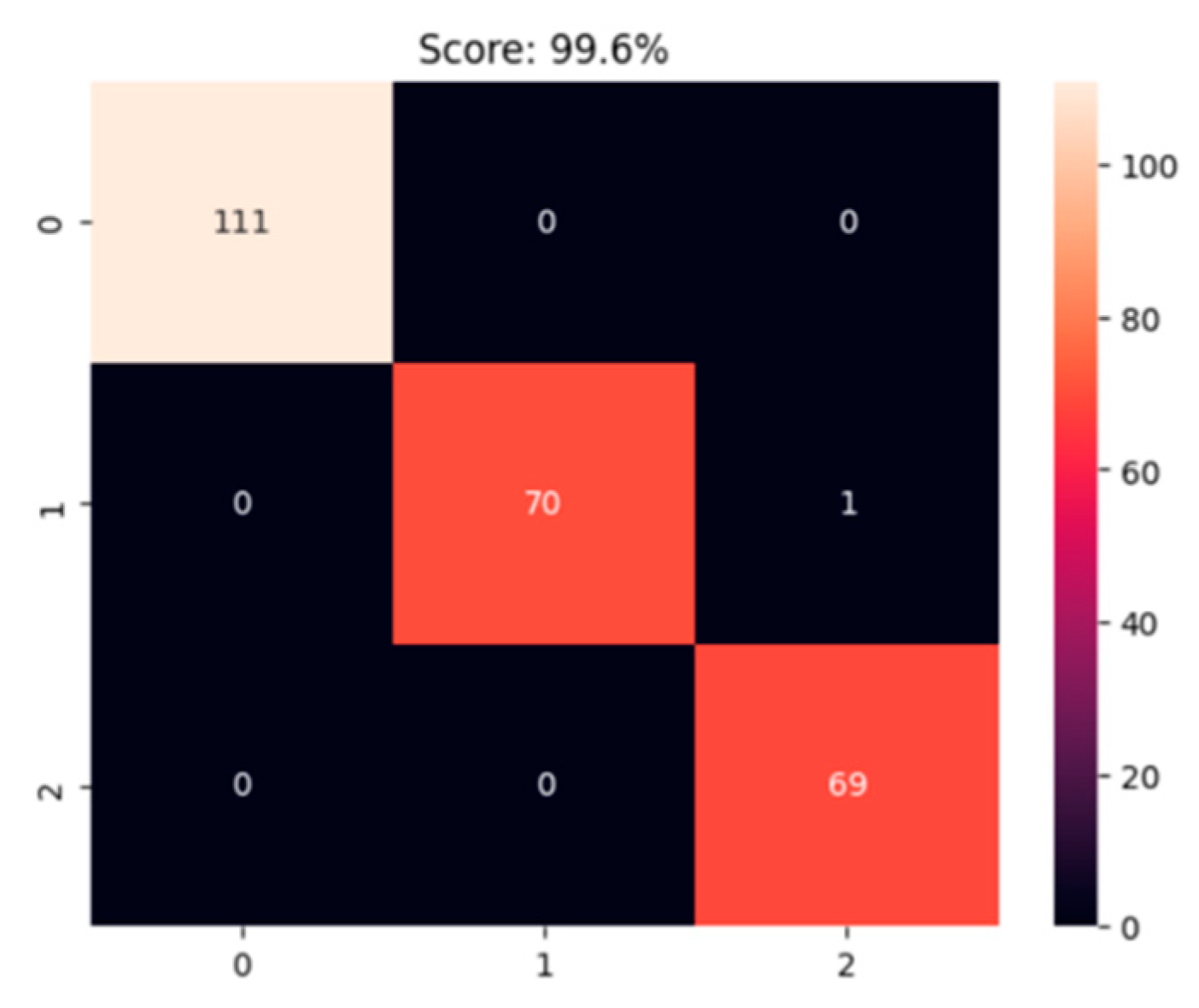
4.4. Confusion Matrix Analysis
The confusion matrix shows the model’s strong classification ability for three pneumonia types. For normal lungs (category 0), all 111 samples were correctly classified, achieving 100% accuracy. Bacterial pneumonia (category 1) had 70 correct classifications with one misclassified sample, resulting in near-perfect precision and recall. Viral pneumonia (category 2) also had 100% accuracy with no misclassifications among 69 samples. Overall, the model’s accuracy is 99.6%, effectively distinguishing between the three categories, with minor misclassifications in bacterial pneumonia not impacting overall performance significantly.
Figure 3.
Confusion Matrix.
Figure 3.
Confusion Matrix.
5. Conclusions
In this study, a deep learning model based on transfer learning is proposed, which adopts the pre-trained ResNet50 architecture and successfully realizes the efficient classification of pneumonia types. By making full use of the feature extraction advantages of transfer learning and fine-tuning at the final classification level, the model has achieved more than 90% accuracy in both verification set and test set. This shows that the model has strong adaptability and robustness in the task of medical image classification, and can effectively assist clinical diagnosis.
The performance of the model benefits from various optimization work. Firstly, by standardizing the image data set and applying various data enhancement techniques (such as random rotation, scaling, translation, etc.), the model effectively avoids the problem of over-fitting, and improves the ability to identify the characteristics of different forms of pneumonia. Secondly, through the learning rate attenuation strategy and momentum gradient optimization, the model converges rapidly in the training process, and has achieved relatively stable training and verification performance in the later stage.
Nevertheless, there are still some limitations in this study. First of all, although the model performs well in the overall classification task, there are still some misclassification cases in distinguishing the subtle differences between bacterial pneumonia and viral pneumonia. Secondly, the scale of the data set is relatively small and the sample source is relatively single, which may limit the generalization ability of the model. Future research should focus on expanding the diversity of data sets, especially by introducing more pneumonia image data from different medical institutions and different populations, so as to enhance the universality of the model.
Future research should refine lesion feature extraction, explore advanced data enhancement and regularization to reduce overfitting, and assess this model’s applicability to other medical imaging tasks. This study underscores the effectiveness of transfer learning in pneumonia classification and offers valuable insights for broader medical image analysis and multi-disciplinary applications.
References
- Ronneberger, O., Fischer, P., & Brox, T. (2015). U-Net: Convolutional networks for biomedical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention.
- Chen, Y. C., Hong, D. J. K., Wu, C. W., & Mupparapu, M. (2019). The use of deep convolutional neural networks in biomedical imaging: A review. Journal of Orofacial Sciences, 11(1), 3-10.
- Weng, W., & Zhu, X. (2021). INet: convolutional networks for biomedical image segmentation. Ieee Access, 9, 16591-16603.
- Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2018). UNet++: A nested U-Net architecture for medical image segmentation. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support.
- Barzekar, H., & Yu, Z. (2022). C-Net: A reliable convolutional neural network for biomedical image classification. Expert Systems with Applications, 187, 116003.
- Huang, G., Liu, Z., Van Der Maaten, L., & Weinberger, K. Q. (2017). Densely connected convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition.
- Ma, T., Dalca, A. V., & Sabuncu, M. R. (2022). Hyper-convolution networks for biomedical image segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (pp. 1933-1942).
- Xu, X., Lu, Q., Yang, L., Hu, S., Chen, D., Hu, Y., & Shi, Y. (2018). Quantization of fully convolutional networks for accurate biomedical image segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 8300-8308).
- Simonyan, K., & Zisserman, A. (2015). Very Deep Convolutional Networks for Large-Scale Image Recognition. International Conference on Learning Representations (ICLR).
- Anwar, S. M., Majid, M., Qayyum, A., Awais, M., Alnowami, M., & Khan, M. K. (2018). Medical image analysis using convolutional neural networks: a review. Journal of medical systems, 42, 1-13.
- Ioffe, S., & Szegedy, C. (2015). Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. Proceedings of the 32nd International Conference on Machine Learning (ICML), 37, 448-456.
- Abraham, G. K., Jayanthi, V. S., & Bhaskaran, P. (2020). Convolutional neural network for biomedical applications. In Computational Intelligence and Its Applications in Healthcare (pp. 145-156). Academic Press.
- Kingma, D. P., & Ba, J. (2014). Adam: A Method for Stochastic Optimization. arXiv preprint arXiv:1412.6980.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).