Submitted:
09 November 2024
Posted:
12 November 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Biological Samples
2.2. RNA Extraction, Quantification, and Quality Control
2.3. Transcriptomic Study
2.4. Statistical Analysis
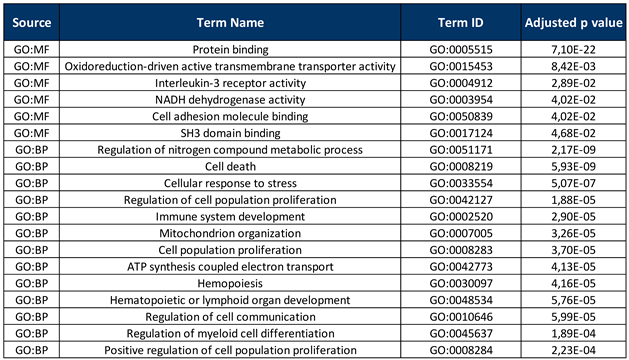
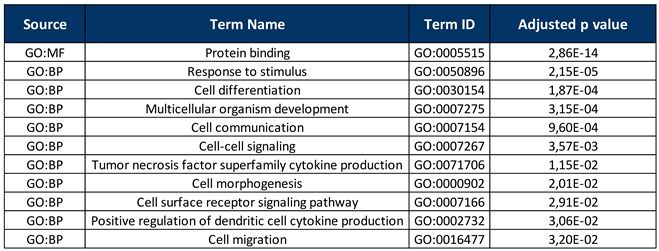
2.5. Gene Enrichment Analysis
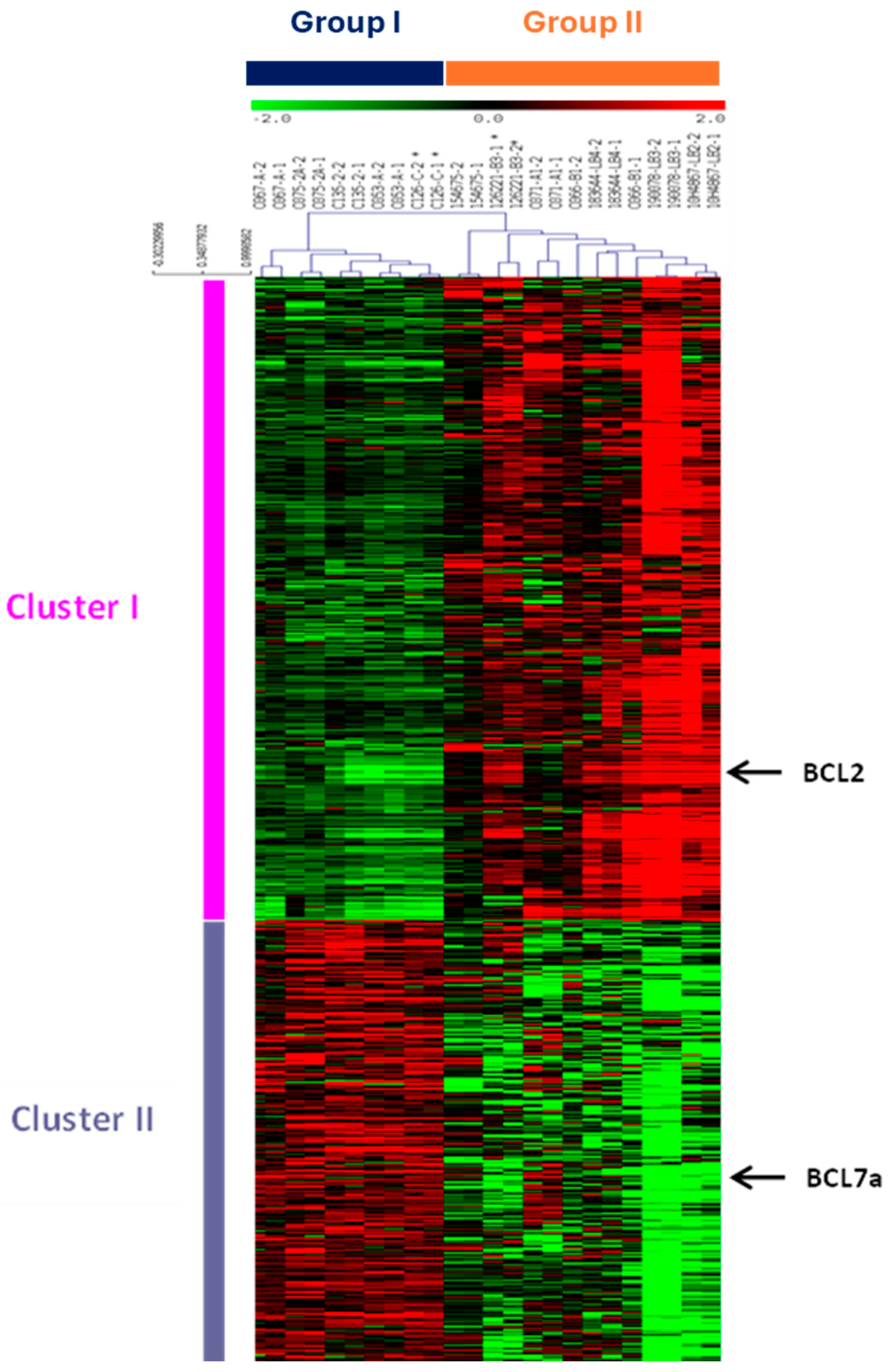
3. Results
4. Discussion
5. Conclusions
Funding
Acknowledgments
References
- International Non-Hodgkin's Lymphoma Prognostic Factors P. A predictive model for aggressive non-Hodgkin's lymphoma. N Engl J Med. Sep 30 1993:329(14):987-94. [CrossRef]
- Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. Feb 3 2000;403(6769):503-11. [CrossRef]
- Karmali R, Gordon LI. Molecular Subtyping in Diffuse Large B Cell Lymphoma: Closer to an Approach of Precision Therapy. Curr Treat Options Oncol. Feb 2017;18(2):11. [CrossRef]
- Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. Jan 1 2004;103(1):275-82. [CrossRef]
- Muris JJ, Meijer CJ, Vos W, et al. Immunohistochemical profiling based on Bcl-2, CD10 and MUM1 expression improves risk stratification in patients with primary nodal diffuse large B cell lymphoma. J Pathol. Apr 2006;208(5):714-23. [CrossRef]
- Younes A, Sehn LH, Johnson P, et al. Randomized Phase III Trial of Ibrutinib and Rituximab Plus Cyclophosphamide, Doxorubicin, Vincristine, and Prednisone in Non-Germinal Center B-Cell Diffuse Large B-Cell Lymphoma. J Clin Oncol. May 20 2019;37(15):1285-1295. [CrossRef]
- Wilson WH, Wright GW, Huang DW, et al. Effect of ibrutinib with R-CHOP chemotherapy in genetic subtypes of DLBCL. Cancer Cell. Dec 13 2021;39(12):1643-1653 e3. [CrossRef]
- Rosenwald A, Staudt LM. Gene expression profiling of diffuse large B-cell lymphoma. Leuk Lymphoma. 2003;44 Suppl 3:S41-7. [CrossRef]
- Thieblemont C, Briere J, Mounier N, et al. The germinal center/activated B-cell subclassification has a prognostic impact for response to salvage therapy in relapsed/refractory diffuse large B-cell lymphoma: a bio-CORAL study. J Clin Oncol. Nov 1 2011;29(31):4079-87. [CrossRef]
- Rutherford SC, Leonard JP. DLBCL Cell of Origin: What Role Should It Play in Care Today? Oncology (Williston Park). Sep 15 2018;32(9):445-9.
- Boltezar L, Prevodnik VK, Perme MP, Gasljevic G, Novakovic BJ. Comparison of the algorithms classifying the ABC and GCB subtypes in diffuse large B-cell lymphoma. Oncol Lett. May 2018;15(5):6903-6912. [CrossRef]
- Alaggio R, Amador C, Anagnostopoulos I, et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia. Jul 2022;36(7):1720-1748. [CrossRef]
- Alaggio R, Amador C, Anagnostopoulos I, et al. Correction: "The 5th edition of The World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms" Leukemia. 2022 Jul;36(7):1720-1748. Leukemia. Sep 2023;37(9):1944-1951. [CrossRef]
- Carbone A, Chadburn A, Gloghini A, Vaccher E, Bower M. Immune deficiency/dysregulation -associated lymphoproliferative disorders. Revised classification and management. Blood Rev. Mar 2024;64:101167. [CrossRef]
- Lurain K, Ramaswami R, Yarchoan R. The role of viruses in HIV-associated lymphomas. Semin Hematol. Oct 2022;59(4):183-191. [CrossRef]
- Dolcetti R, Gloghini A, Caruso A, Carbone A. A lymphomagenic role for HIV beyond immune suppression? Blood. Mar 17 2016;127(11):1403-9. [CrossRef]
- Vaccher E, Gloghini A, Carbone A. HIV-related lymphomas. Curr Opin Oncol. Sep 1 2022;34(5):439-445. [CrossRef]
- Thapa DR, Li X, Jamieson BD, Martinez-Maza O. Overexpression of microRNAs from the miR-17-92 paralog clusters in AIDS-related non-Hodgkin's lymphomas. PLoS One. 2011;6(6):e20781. [CrossRef]
- Torne AS, Robertson ES. Epigenetic Mechanisms in Latent Epstein-Barr Virus Infection and Associated Cancers. Cancers (Basel). Feb 29 2024;16(5). [CrossRef]
- Ramos JC, Sin SH, Staudt MR, et al. Nuclear factor kappa B pathway associated biomarkers in AIDS defining malignancies. Int J Cancer. Jun 1 2012;130(11):2728-33. [CrossRef]
- Capello D, Scandurra M, Poretti G, et al. Genome wide DNA-profiling of HIV-related B-cell lymphomas. Br J Haematol. Jan 2010;148(2):245-55. [CrossRef]
- Rinaldi A, Mian M, Chigrinova E, et al. Genome-wide DNA profiling of marginal zone lymphomas identifies subtype-specific lesions with an impact on the clinical outcome. Blood. Feb 3 2011;117(5):1595-604. [CrossRef]
- Chapman JR, Bouska AC, Zhang W, et al. EBV-positive HIV-associated diffuse large B cell lymphomas are characterized by JAK/STAT (STAT3) pathway mutations and unique clinicopathologic features. Br J Haematol. Sep 2021;194(5):870-878. [CrossRef]
- Scott DW, Wright GW, Williams PM, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. Feb 20 2014;123(8):1214-7. [CrossRef]
- Madan R, Gormley R, Dulau A, et al. AIDS and non-AIDS diffuse large B-cell lymphomas express different antigen profiles. Mod Pathol. Mar 2006;19(3):438-46. [CrossRef]
- Patrone L, Henson SE, Teodorovic J, et al. Gene expression patterns in AIDS versus non-AIDS-related diffuse large B-cell lymphoma. Exp Mol Pathol. Apr 2003;74(2):129-39. [CrossRef]
- Wang W, Lopez McDonald MC, Kim C, et al. The complementary roles of STAT3 and STAT1 in cancer biology: insights into tumor pathogenesis and therapeutic strategies. Front Immunol. 2023;14:1265818. [CrossRef]
- Trujillo-Ochoa JL, Kazemian M, Afzali B. The role of transcription factors in shaping regulatory T cell identity. Nat Rev Immunol. Dec 2023;23(12):842-856. [CrossRef]
- Ning S, Pagano JS, Barber GN. IRF7: activation, regulation, modification and function. Genes Immun. Sep 2011;12(6):399-414. [CrossRef]
- Liu X, Song J, Zhang H, et al. Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell. Feb 13 2023;41(2):272-287 e9. [CrossRef]
- Cai M, Chen N. The Roles of IRF-8 in Regulating IL-9-Mediated Immunologic Mechanisms in the Development of DLBCL: A State-of-the-Art Literature Review. Front Oncol. 2022;12:817069. [CrossRef]
- Natoli A, Lupertz R, Merz C, et al. Targeting the IL-4/IL-13 signaling pathway sensitizes Hodgkin lymphoma cells to chemotherapeutic drugs. Int J Cancer. Oct 15 2013;133(8):1945-54. [CrossRef]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. Jan 2010;2(1):a001008. [CrossRef]
- Duffy MJ, O'Donovan N, McDermott E, Crown J. Validated biomarkers: The key to precision treatment in patients with breast cancer. Breast. Oct 2016;29:192-201. [CrossRef]
- Adams CM, McBride A, Michener P, et al. Identifying Targetable Vulnerabilities to Circumvent or Overcome Venetoclax Resistance in Diffuse Large B-Cell Lymphoma. Cancers (Basel). Jun 3 2024;16(11). [CrossRef]
- Takahara T, Nakamura S, Tsuzuki T, Satou A. The Immunology of DLBCL. Cancers (Basel). Jan 29 2023;15(3). [CrossRef]
- Mu W, Patankar V, Kitchen S, Zhen A. Examining Chronic Inflammation, Immune Metabolism, and T Cell Dysfunction in HIV Infection. Viruses. Jan 31 2024;16(2). [CrossRef]
- Isaguliants M, Bayurova E, Avdoshina D, Kondrashova A, Chiodi F, Palefsky JM. Oncogenic Effects of HIV-1 Proteins, Mechanisms Behind. Cancers (Basel). Jan 15 2021;13(2). [CrossRef]
- Ramos-Medina R, Montes-Moreno S, Maestre L, et al. BCL7A protein expression in normal and malignant lymphoid tissues. Br J Haematol. Jan 2013;160(1):106-9. [CrossRef]
- Scott DW, Gascoyne RD. The tumour microenvironment in B cell lymphomas. Nat Rev Cancer. Aug 2014;14(8):517-34. [CrossRef]
 |
 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





