Submitted:
25 October 2024
Posted:
29 October 2024
You are already at the latest version
Abstract
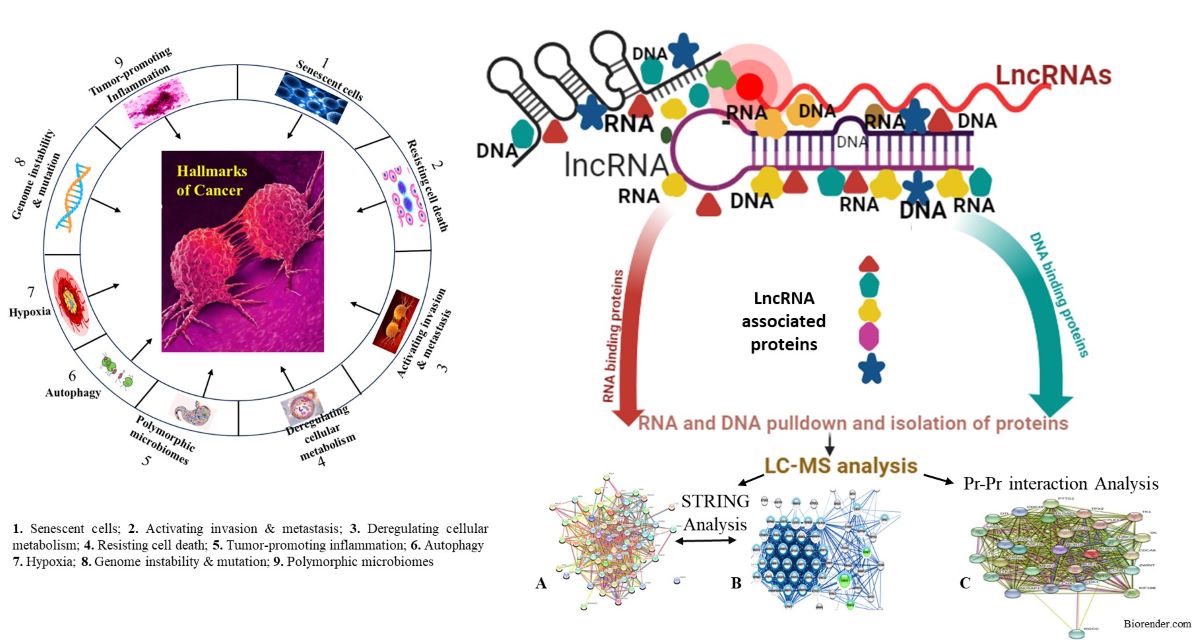
Keywords:
1. Introduction
2. Method:
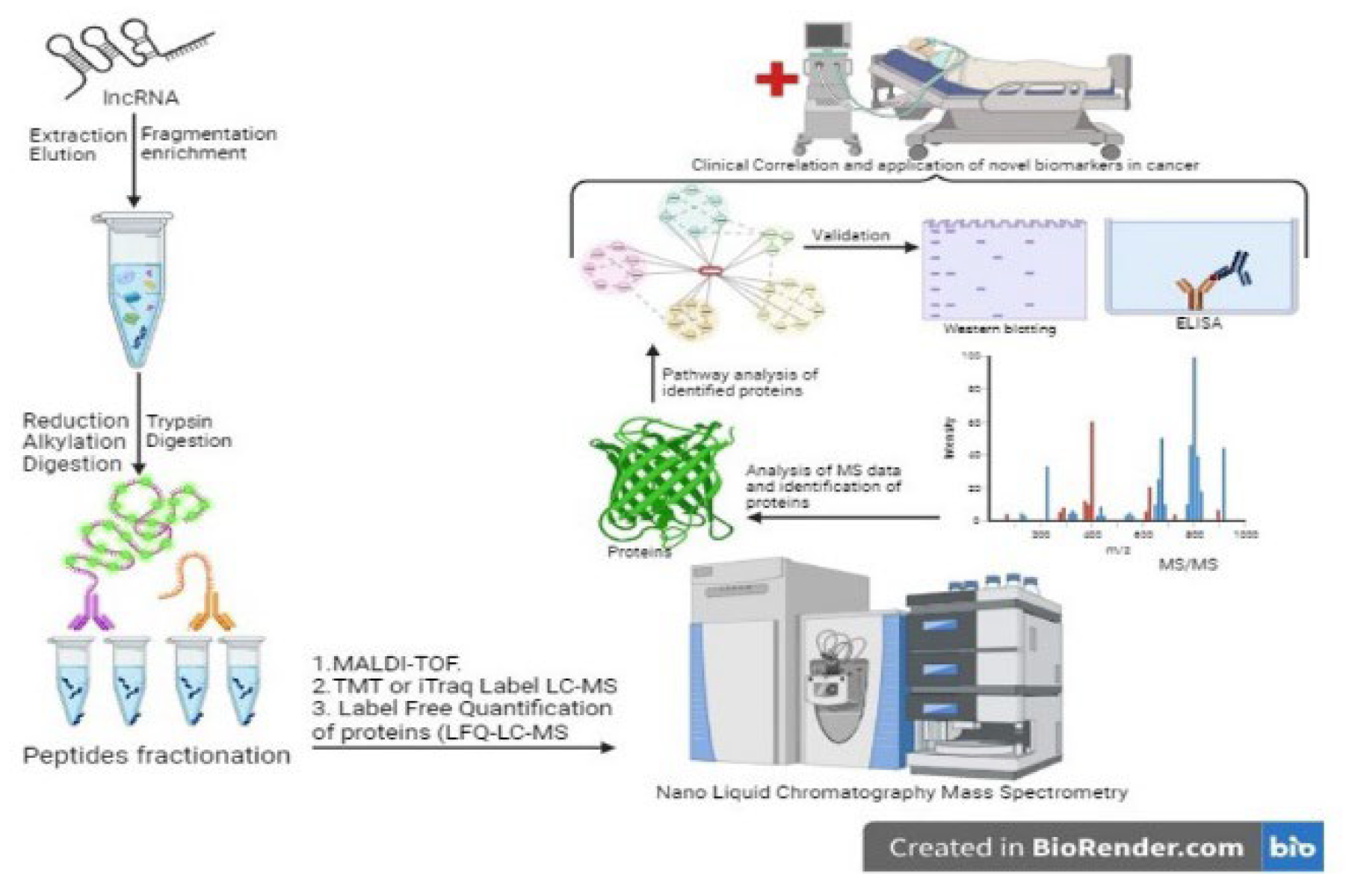
2.1. Isolation and Identification of LncRNAs Associated Proteins
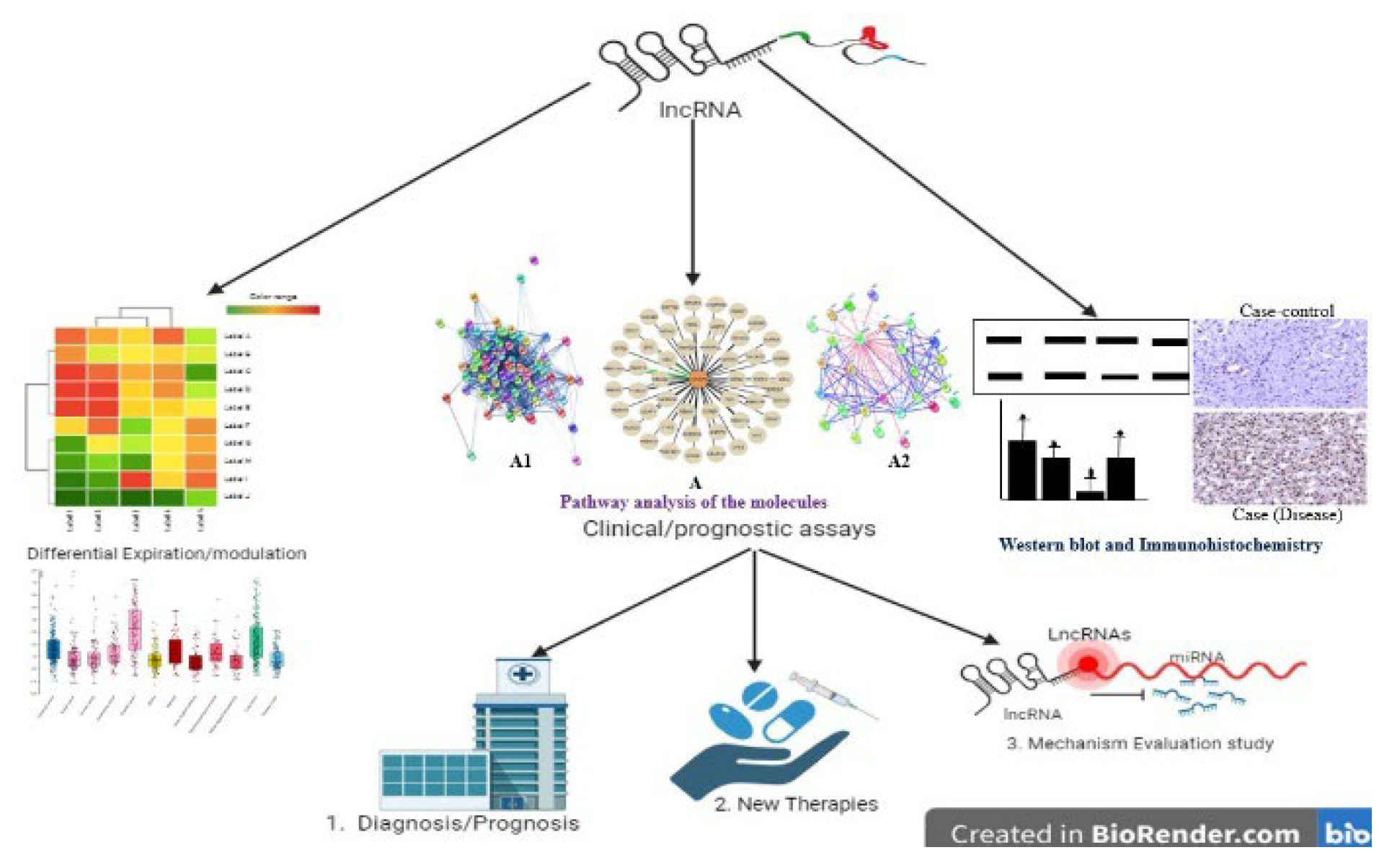
2.2. Identified Pathways Associated with lncRNAs:
3. Discussion:
4. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Balasundaram, P.; Anilkumar, A.C. Myoclonic Epilepsy of Infancy. In StatPearls; StatPearls Publishing Copyright © 2024, StatPearls Publishing LLC.: Treasure Island, FL, USA, 2024. [Google Scholar]
- Li, M.; Xue, Y.; He, R.; Huang, Q. Isolation of Protein Complexes Associated with Long Non-coding RNAs. Methods Mol Biol 2021, 2372, 27–33. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhang, T.; Pan, Y.; Hu, Z.; Yuan, J.; Hu, X.; Zhang, L.; Zhang, Y. Characterization of Long Non-coding RNA Associated Proteins by RNA-Immunoprecipitation. Methods Mol Biol 2021, 2372, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Nunez, R.T.; Sanford, J.R. Studying Isoform-Specific mRNA Recruitment to Polyribosomes with Frac-seq. Methods Mol Biol 2016, 1358, 99–108. [Google Scholar] [CrossRef]
- Roger, G.; Ducrocq, G.; Mesnier, J.; Sayah, N.; Abtan, J.; Ferrari, R.; Ford, I.; Fox, K.M.; Tardif, J.C.; Tendera, M.; et al. Chronic coronary syndromes without standard modifiable cardiovascular risk factors and outcomes: the CLARIFY registry. Eur Heart J 2024, 45, 2396–2406. [Google Scholar] [CrossRef]
- Qiu, B.; Simon, M.C. BODIPY 493/503 Staining of Neutral Lipid Droplets for Microscopy and Quantification by Flow Cytometry. Bio Protoc 2016, 6. [Google Scholar] [CrossRef]
- Lefaucheur, J.P.; Aleman, A.; Baeken, C.; Benninger, D.H.; Brunelin, J.; Di Lazzaro, V.; Filipović, S.R.; Grefkes, C.; Hasan, A.; Hummel, F.C.; et al. Evidence-based guidelines on the therapeutic use of repetitive transcranial magnetic stimulation (rTMS): An update (2014-2018). Clin Neurophysiol 2020, 131, 474–528. [Google Scholar] [CrossRef]
- Tran, T.H.; Hunger, S.P. The genomic landscape of pediatric acute lymphoblastic leukemia and precision medicine opportunities. Semin Cancer Biol 2022, 84, 144–152. [Google Scholar] [CrossRef]
- Genovese, G.; Carugo, A.; Tepper, J.; Robinson, F.S.; Li, L.; Svelto, M.; Nezi, L.; Corti, D.; Minelli, R.; Pettazzoni, P.; et al. Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 2017, 542, 362–366. [Google Scholar] [CrossRef]
- Breit, S.N.; Brown, D.A.; Tsai, V.W.W. GDF15 analogs as obesity therapeutics. Cell Metab 2023, 35, 227–228. [Google Scholar] [CrossRef]
- Imamura, S.; Morioka, T.; Yamazaki, Y.; Numaguchi, R.; Urata, H.; Motoyama, K.; Mori, K.; Fukumoto, S.; Shoji, T.; Emoto, M.; et al. Response to comment on Imamura et al. Plasma polyunsaturated fatty acid profile and delta-5 desaturase activity are altered in patients with type 2 diabetes. Metabolism 2014;63(11):1432-8. Metabolism 2015, 64, e3–e4. [Google Scholar] [CrossRef] [PubMed]
- Talamantes, S.; Lisjak, M.; Gilglioni, E.H.; Llamoza-Torres, C.J.; Ramos-Molina, B.; Gurzov, E.N. Non-alcoholic fatty liver disease and diabetes mellitus as growing aetiologies of hepatocellular carcinoma. JHEP Rep 2023, 5, 100811. [Google Scholar] [CrossRef] [PubMed]
- Characterization of Long Non-coding RNA Associated Proteins by RNA-Immunoprecipitation Junjie Jiang.
- Guo, Y.; Zhao, J.; Bi, J.; Wu, Q.; Wang, X.; Lai, Q. Heterogeneous nuclear ribonucleoprotein K (hnRNP K) is a tissue biomarker for detection of early hepatocellular carcinoma in patients with cirrhosis. J Hematol Oncol 2012, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Gutschner, T.; Haemmerle, M.; Genovese, G.; Draetta, G.F.; Chin, L. Post-translational Regulation of Cas9 during G1 Enhances Homology-Directed Repair. Cell Rep 2016, 14, 1555–1566. [Google Scholar] [CrossRef] [PubMed]
- Isolation of Protein Complexes Associated with Long Non-coding RNAs.
- Galamb, O.; Barták, B.K.; Kalmár, A.; Nagy, Z.B.; Szigeti, K.A.; Tulassay, Z.; Igaz, P.; Molnár, B. Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors. World J Gastroenterol 2019, 25, 5026–5048. [Google Scholar] [CrossRef]
- McHugh, C.A.; Chen, C.K.; Chow, A.; Surka, C.F.; Tran, C.; McDonel, P.; Pandya-Jones, A.; Blanco, M.; Burghard, C.; Moradian, A.; et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 2015, 521, 232–236. [Google Scholar] [CrossRef]
- Rodriguez-Frandsen, A.; Martin-Sancho, L.; Gounder, A.P.; Chang, M.W.; Liu, W.C.; De Jesus, P.D.; von Recum-Knepper, J.; Dutra, M.S.; Huffmaster, N.J.; Chavarria, M.; et al. Viral Determinants in H5N1 Influenza A Virus Enable Productive Infection of HeLa Cells. J Virol 2020, 94. [Google Scholar] [CrossRef]
- Spiniello, M.; Knoener, R.A.; Steinbrink, M.I.; Yang, B.; Cesnik, A.J.; Buxton, K.E.; Scalf, M.; Jarrard, D.F.; Smith, L.M. HyPR-MS for Multiplexed Discovery of MALAT1, NEAT1, and NORAD lncRNA Protein Interactomes. J Proteome Res 2018, 17, 3022–3038. [Google Scholar] [CrossRef]
- Razzaq, S.; Rauf, A.; Raza, A.; Akhtar, S.; Tabish, T.A.; Sandhu, M.A.; Zaman, M.; Ibrahim, I.M.; Shahnaz, G.; Rahdar, A.; et al. A Multifunctional Polymeric Micelle for Targeted Delivery of Paclitaxel by the Inhibition of the P-Glycoprotein Transporters. Nanomaterials (Basel) 2021, 11. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Sirokman, K.; McDonel, P.; Shishkin, A.A.; Surka, C.; Russell, P.; Grossman, S.R.; Chow, A.Y.; Guttman, M.; Lander, E.S. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 2014, 159, 188–199. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [PubMed]
- Torres, M.; Becquet, D.; Guillen, S.; Boyer, B.; Moreno, M.; Blanchard, M.P.; Franc, J.L.; François-Bellan, A.M. RNA Pull-down Procedure to Identify RNA Targets of a Long Non-coding RNA. J Vis Exp 2018. [Google Scholar] [CrossRef]
- Smith, A.L.; Friedman, D.B.; Yu, H.; Carnahan, R.H.; Reynolds, A.B. ReCLIP (reversible cross-link immuno-precipitation): an efficient method for interrogation of labile protein complexes. PLoS One 2011, 6, e16206. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.L.; Dohn, M.R.; Brown, M.V.; Reynolds, A.B. Association of Rho-associated protein kinase 1 with E-cadherin complexes is mediated by p120-catenin. Mol Biol Cell 2012, 23, 99–110. [Google Scholar] [CrossRef]
- Li, Y.; Wang, H.; Zhan, L.; Li, Q.; Li, Y.; Wu, G.; Wei, H.; Dong, X. LncRNA FER1L4 promotes differentiation and inhibits proliferation of NSCs via miR-874-3p/Ascl2. Am J Transl Res 2022, 14, 2256–2266. [Google Scholar]
- Rodriguez, P.D.; Paculova, H.; Kogut, S.; Heath, J.; Schjerven, H.; Frietze, S. Non-Coding RNA Signatures of B-Cell Acute Lymphoblastic Leukemia. Int J Mol Sci 2021, 22. [Google Scholar] [CrossRef]
- Bothos, E.; Hatzis, P.; Moulos, P. Interactive Analysis, Exploration, and Visualization of RNA-Seq Data with SeqCVIBE. Methods Protoc 2022, 5. [Google Scholar] [CrossRef]
- Liu, S.S.; Li, J.S.; Xue, M.; Wu, W.J.; Li, X.; Chen, W. LncRNA UCA1 Participates in De Novo Synthesis of Guanine Nucleotides in Bladder Cancer by Recruiting TWIST1 to Increase IMPDH1/2. Int J Biol Sci 2023, 19, 2599–2612. [Google Scholar] [CrossRef]
- Fu, S.; Wang, Y.; Li, H.; Chen, L.; Liu, Q. Regulatory Networks of LncRNA MALAT-1 in Cancer. Cancer Manag Res 2020, 12, 10181–10198. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Wang, C.; Li, Y.; Yan, S.; Wang, H.; Shao, X.; Xiao, M.; Yang, B.; Qin, G.; Kong, R.; Chen, R.; et al. Interactome analysis reveals that lncRNA HULC promotes aerobic glycolysis through LDHA and PKM2. Nat Commun 2020, 11, 3162. [Google Scholar] [CrossRef] [PubMed]
- Pei, C.; Gong, X.; Zhang, Y. LncRNA MALAT-1 promotes growth and metastasis of epithelial ovarian cancer via sponging microrna-22. Am J Transl Res 2020, 12, 6977–6987. [Google Scholar] [PubMed]
- Song, W.; Wang, K.; Zou, S.B. UCA1 lncRNA in metastases and prognosis. Panminerva Med 2017, 59, 278–279. [Google Scholar] [CrossRef] [PubMed]
- Ferrè, F.; Colantoni, A.; Helmer-Citterich, M. Revealing protein-lncRNA interaction. Brief Bioinform 2016, 17, 106–116. [Google Scholar] [CrossRef] [PubMed]
- Rastad, H.; Samimisedeh, P.; Alan, M.S.; Afshar, E.J.; Ghalami, J.; Hashemnejad, M.; Alan, M.S. The role of lncRNA CERS6-AS1 in cancer and its molecular mechanisms: A systematic review and meta-analysis. Pathol Res Pract 2023, 241, 154245. [Google Scholar] [CrossRef]
- Yu, X.; Zheng, H.; Chan, M.T.; Wu, W.K. HULC: an oncogenic long non-coding RNA in human cancer. J Cell Mol Med 2017, 21, 410–417. [Google Scholar] [CrossRef]
- Ou, C.; Sun, Z.; He, X.; Li, X.; Fan, S.; Zheng, X.; Peng, Q.; Li, G.; Li, X.; Ma, J. Targeting YAP1/LINC00152/FSCN1 Signaling Axis Prevents the Progression of Colorectal Cancer. Adv Sci (Weinh) 2020, 7, 1901380. [Google Scholar] [CrossRef]
- Sun, J.Y.; Ni, M.M. Long non-coding RNA HEIH: a novel tumor activator in multiple cancers. Cancer Cell Int 2021, 21, 558. [Google Scholar] [CrossRef]
- Jiang, H.; Zhou, L.; Shen, N.; Ning, X.; Wu, D.; Jiang, K.; Huang, X. M1 macrophage-derived exosomes and their key molecule lncRNA HOTTIP suppress head and neck squamous cell carcinoma progression by upregulating the TLR5/NF-κB pathway. Cell Death Dis 2022, 13, 183. [Google Scholar] [CrossRef]
- Rajagopal, T.; Talluri, S.; Akshaya, R.L.; Dunna, N.R. HOTAIR LncRNA: A novel oncogenic propellant in human cancer. Clin Chim Acta 2020, 503, 1–18. [Google Scholar] [CrossRef]
- Mu, Y.; Li, N.; Cui, Y.L. The lncRNA CCAT1 upregulates TGFβR1 via sponging miR-490-3p to promote TGFβ1-induced EMT of ovarian cancer cells. Cancer Cell Int 2018, 18, 145. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Sun, D.; Guo, H.; Wu, Z.; Chen, J. LncRNA CCAT2 promotes proliferation and suppresses apoptosis of colorectal cancer cells. J buon 2020, 25, 1840–1846. [Google Scholar] [PubMed]
- Hashemi, M.; Moosavi, M.S.; Abed, H.M.; Dehghani, M.; Aalipour, M.; Heydari, E.A.; Behroozaghdam, M.; Entezari, M.; Salimimoghadam, S.; Gunduz, E.S.; et al. Long non-coding RNA (lncRNA) H19 in human cancer: From proliferation and metastasis to therapy. Pharmacol Res 2022, 184, 106418. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Liu, H.M.; Wu, W.H.; Liu, H.; Pan, Y.; Li, W.J. Upregulation of long noncoding RNA CCAT1-L promotes epithelial-mesenchymal transition in gastric adenocarcinoma. Onco Targets Ther 2018, 11, 5647–5655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, H.; Yu, J.; Yao, X.; Yang, S.; Li, W.; Xu, L.; Zhao, L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol Cancer 2021, 20, 6. [Google Scholar] [CrossRef]
- Fan, Y.; Sheng, W.; Meng, Y.; Cao, Y.; Li, R. LncRNA PTENP1 inhibits cervical cancer progression by suppressing miR-106b. Artif Cells Nanomed Biotechnol 2020, 48, 393–407. [Google Scholar] [CrossRef]
- Jiang, J.; Azevedo-Pouly, A.C.; Redis, R.S.; Lee, E.J.; Gusev, Y.; Allard, D.; Sutaria, D.S.; Badawi, M.; Elgamal, O.A.; Lerner, M.R.; et al. Globally increased ultraconserved noncoding RNA expression in pancreatic adenocarcinoma. Oncotarget 2016, 7, 53165–53177. [Google Scholar] [CrossRef]
- Boyd, J.H.; Fjell, C.D.; Russell, J.A.; Sirounis, D.; Cirstea, M.S.; Walley, K.R. Increased Plasma PCSK9 Levels Are Associated with Reduced Endotoxin Clearance and the Development of Acute Organ Failures during Sepsis. J Innate Immun 2016, 8, 211–220. [Google Scholar] [CrossRef]
- Dhillon, R.S.; Yao, L.; Matey, V.; Chen, B.J.; Zhang, A.J.; Cao, Z.D.; Fu, S.J.; Brauner, C.J.; Wang, Y.S.; Richards, J.G. Interspecific differences in hypoxia-induced gill remodeling in carp. Physiol Biochem Zool 2013, 86, 727–739. [Google Scholar] [CrossRef]
- Compérat, E. Editorial for Cribriform architecture prostatic adenocarcinoma in needle biopsy is a strong independent predictor for lymph node metastases in radical prostatectomy (M. Downes et al.) and Ductal variant prostate carcinoma is associated with a significantly shorter metastasis-free survival (K. Chow et al.). Eur J Cancer 2021, 148, 430–431. [Google Scholar] [CrossRef]
- Lu, C.; Cai, R.; Grigg, J.C.; Ke, A. Using tRNA Scaffold to Assist RNA Crystallization. Methods Mol Biol 2021, 2323, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wu, L.; Zhong, Y.; Zhou, K.; Hou, Y.; Wang, Z.; Zhang, Z.; Xie, J.; Wang, C.; Chen, D.; et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell 2021, 184, 404–421.e416. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Sun, R.; Zhu, Y.; Li, Z.; She, X.; Jian, X.; Yu, F.; Deng, X.; Sai, B.; Wang, L.; et al. Lung microbiome alterations in NSCLC patients. Sci Rep 2021, 11, 11736. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, S.; Huang, L.; Zhao, R.; Dai, E.; Jiang, X.; He, Y.; Lu, J.; Peng, L.; Liu, W.; et al. CTNND1 variants cause familial exudative vitreoretinopathy through the Wnt/cadherin axis. JCI Insight 2022, 7. [Google Scholar] [CrossRef]
- Shang, S.; Liu, J.; Hua, F. Protein acylation: mechanisms, biological functions and therapeutic targets. Signal Transduct Target Ther 2022, 7, 396. [Google Scholar] [CrossRef]
- Li, Y.; Xu, J.; Chen, W.; Wang, X.; Zhao, Z.; Li, Y.; Zhang, L.; Jiao, J.; Yang, Q.; Ding, Q.; et al. Hepatocyte CD36 modulates UBQLN1-mediated proteasomal degradation of autophagic SNARE proteins contributing to septic liver injury. Autophagy 2023, 19, 2504–2519. [Google Scholar] [CrossRef]
- Li, Z.; Liu, X.; Luo, N.; Pang, Y.; Hou, Y.; Jiang, G. Long non-coding RNA CERS6-AS1 plays a prognostic role in promoting the progression of gastric cancer. Bioengineered 2021, 12, 12931–12939. [Google Scholar] [CrossRef]
- Lamsisi, M.; Wakrim, L.; Bouziyane, A.; Benhessou, M.; Oudghiri, M.; Laraqui, A.; Elkarroumi, M.; Ennachit, M.; El Mzibri, M.; Ennaji, M.M. The Biological Significance of Long noncoding RNAs Dysregulation and their Mechanism of Regulating Signaling Pathways in Cervical Cancer. Int J Mol Cell Med 2021, 10, 75–101. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Taheri, M.; Akbari Dilmaghani, N. A review on the role of PTENP1 in human disorders with an especial focus on tumor suppressor role of this lncRNA. Cancer Cell Int 2022, 22, 207. [Google Scholar] [CrossRef]
- Li, R.; Wang, X.; Zhu, C.; Wang, K. lncRNA PVT1: a novel oncogene in multiple cancers. Cell Mol Biol Lett 2022, 27, 84. [Google Scholar] [CrossRef]
- Loevenich, L.P.; Tschurtschenthaler, M.; Rokavec, M.; Silva, M.G.; Jesinghaus, M.; Kirchner, T.; Klauschen, F.; Saur, D.; Neumann, J.; Hermeking, H.; et al. SMAD4 Loss Induces c-MYC-Mediated NLE1 Upregulation to Support Protein Biosynthesis, Colorectal Cancer Growth, and Metastasis. Cancer Res 2022, 82, 4604–4623. [Google Scholar] [CrossRef] [PubMed]
- Frenkel-Pinter, M.; Haynes, J.W.; C, M.; Petrov, A.S.; Burcar, B.T.; Krishnamurthy, R.; Hud, N.V.; Leman, L.J.; Williams, L.D. Selective incorporation of proteinaceous over nonproteinaceous cationic amino acids in model prebiotic oligomerization reactions. Proc Natl Acad Sci U S A 2019, 116, 16338–16346. [Google Scholar] [CrossRef] [PubMed]

| S No. | Study | Target LncRNAs | Specific Method name (Cancer CRC & HCC) | Remarks/conclusions | Reference |
|---|---|---|---|---|---|
| 1 | Shijian Fu et al | MALAT-1 | Patient samples & ovarian cancer cell lines (SKOV3 & CAOV3) | MALAT-1 diagnostic or prognostic biomarker or therapeutic target in the treatment of many cancers. | [35] |
| 2 | An Yang et al | UCA1 | Via the miR-145/MYO6 axis | The UCA1/miR-145/MYO6 axis may serve as a potential therapeutic target for gastric cancer. | [36] |
| 3 | Enans M et al | T2D/ HCC | NAFLD/T2D-associated HCC | Metformin may reduce the risk of cancer in patients with T2D. The unadjusted odds ratio was 0.86 (95% CI 0.73 to 1.02). The unadjusted odds ratio for any exposure to metformin since 1993 was 0.79 (0.67 to 0.93) | [13,37] |
| 4 | Fabrizio Ferre et al | Revealing protein | RAPID-SELEX RNAcompete RNA Bin-n-Seq RNA-Ma | Better understanding of lncRNA cellular mechanisms and their disease-associated perturbations. | [37,38] |
| 5 | Fabrizio Ferre et al | LncRNA interaction | MS2 trapping SILAC-based Phage display Protein arrays | LncRNA-bound proteome, or if still uncharacterized protein domains and architectures are involved, network will be high | [37] |
| 6 | Arunoday Bhan et al | HULC | Tumorigenesis test in vitro and in vivo: RT-PCR, W. B | Potential implications in cancer diagnosis and therapy | [33,37] |
| 7 | Chunqing Wang et al | HULC | HULC interacts with the glycolytic enzyme LDHA | HULC promotes Warburg effect by orchestrating the enzymatic activities of glycolytic enzymes | [34,39] |
| 8 | Chunlin Ou et al | Linc00152 | Human Tissue Samples: | Targeting YAP1/LINC00152/FSCN1 Signaling Axis Prevents the Progression of Colorectal Cancer | [40] |
| 9 | Jie-yu Sun et al | HEIH | Non-coding RNAs | Nearly 8000 cancerspecifc lncRNAs have been nominated, PCA3 is prostate-specific, prognostic biomarker prostate cancer. | [41] |
| 10 | Huaili Jiang et al | HOTTIP | In silico analysis, Plasmid construction and transfection | Significantly, M1 exosomes and HOTTIP polarize circulating monocytes into the antitumor M1 phenotype, which may provide novel insight into HNSCC immunotherapy. | [42] |
| 11 | Taruna Rajagopal et al | HOTAIR | HOTAIR mediated gene silencing | It could be used in conjunction with current drugs to sensitize tumors to the existing therapies | [43] |
| 12 | Yang Mu et al | CCAT1 | RT-qPCR to level of miR-490-3p and CCAT1 | facilitate developing novel therapeutical therapies for treating ovarian cancer. | [44] |
| 13 | Peng Gao et al | CCAT2 | Dimethyl sulfoxide (DMSO) (Aldrich, St. Louis, | to explore genes co-expressed with lncRNA CCAT2 and functional molecular | [45] |
| 14 | Hashemi et al | H19 | Enhancing growth and cell cycle of cancers and by EMT induction | Increased proliferation Glycolysis induction miRNA-519d-3p down-regulation by H19 to increase LDHA expression | [46] |
| 15 | Hua Fang et al | CCAT1-L | Quantitative real-time PCR and Western blot, respectively. | inhibits epithelial–mesenchymal transition of gastric adenocarcinoma cells and thus suppresses the gastric adenocarcinoma metastasis. | [47] |
| 16 | Feifei Zhang1 | CRNDE | Chemosensitivity of GC in clinical samples and a PDX model. | Highlighting the significance of CRNDE as a potential prognostic marker and therapeutic target against chemoresistance in GC. | [48] |
| 17 | Yanping Li et al | FER1L4 | Cell was extracted from embryos of rat | FER1L4 modulates the proliferation and differentiation of NSCs via regulating Ascl2. | [28] |
| 18 | Yingrui Fan et al | PTENP | uciferase reporter assay and RNA-pulldown assay | Inhibit cell proliferation and EMT and induce cell apoptosisin cervical cancer cells. | [49] |
| 19 | Jinmai Jiang et al | T-UCRs | qPCR array to profile all 481 T-UCRs in pancreatic cancer specimens, pancreatic cancer cell lines | Expression of T-UCRs in both human and mouse PDAC and similar mechanism of upregulation in PDAC | [50] |
| 20 | Arunoday Bhan et al & John H Boyd | TUC338 | Plasma, treatment, and cell lines, MS2-MBP Protein Expression and Immobilization | The understanding of molecular mechanisms of lncRNAs. Inhibition of PCSK9 activity is an attractive target for treating the spectrum of sepsis and septic shock. |
[33,51] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





