Submitted:
24 October 2024
Posted:
25 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Participants and Biopsy Sampling
| Participant Number |
Age, years | Sex, M/F |
BMI, kg/m2 | Procedure Gastroscopy |
Procedure Colonoscopy |
Number of biopsy samples (total included) | |
| 1 | 24 | M | 20.1 | X | 10 | ||
| 2 | 55 | M | 28.0 | X | 10 | ||
| 3 | 49 | M | 27.0 | X | 10 | ||
| 4 | 18 | M | 28.1 | X | X | 20 | |
| 5 | 48 | F | 28.5 | X | 10 | ||
| 6 | 58 | F | 29.2 | X | 10(8) | ||
| 7 | 21 | F | 28.0 | X | 10 | ||
| 8 | 62 | F | 20.7 | X | X | 20 | |
| 9 | 35 | F | 23.1 | X | 10 | ||
| 10 | 60 | M | 29.3 | X | 10 | ||
| 11 | 21 | F | 23.3 | X | 10 | ||
| 12 | 35 | F | 23.8 | X | 10 | ||
| 13 | 44 | M | 28.4 | X | 10 | ||
| 14 | 56 | F | 24.1 | X | 10 | ||
| 15 | 56 | F | 21.3 | X | 10 | ||
| 16 | 56 | M | 24.4 | X | 10 | ||
| 17 | 57 | M | 24.7 | X | 10 | ||
| 18 | 44 | M | 24.0 | X | 10 | ||
| 19 | 61 | M | 23.3 | X | 10 | ||
| 20 | 64 | F | 26.6 | X | 10 | ||
| 21 | 65 | F | 23.5 | X | 10 | ||
| 22 | 55 | M | 25.3 | X | 10 | ||
| 23 | 44 | F | 29.8 | X | 10 | ||
| 24 | 52 | F | 24.5 | X | 10 | ||
| 25 | 30 | M | 30.0 | X | 10 | ||
| 26 | 41 | M | 29.9 | X | 10 | ||
| 27 | 63 | M | 28.2 | X | 10 | ||
| 28 | 55 | F | 27.2 | X | 10 | ||
| Average (mean, range) | 47.2, 18-65 |
25.8, 20.1-30 | |||||
| Total | 14/14 | 15 | 15 | 300 (298) | |||
2.2. TAS2R Transcription in the GI Tract
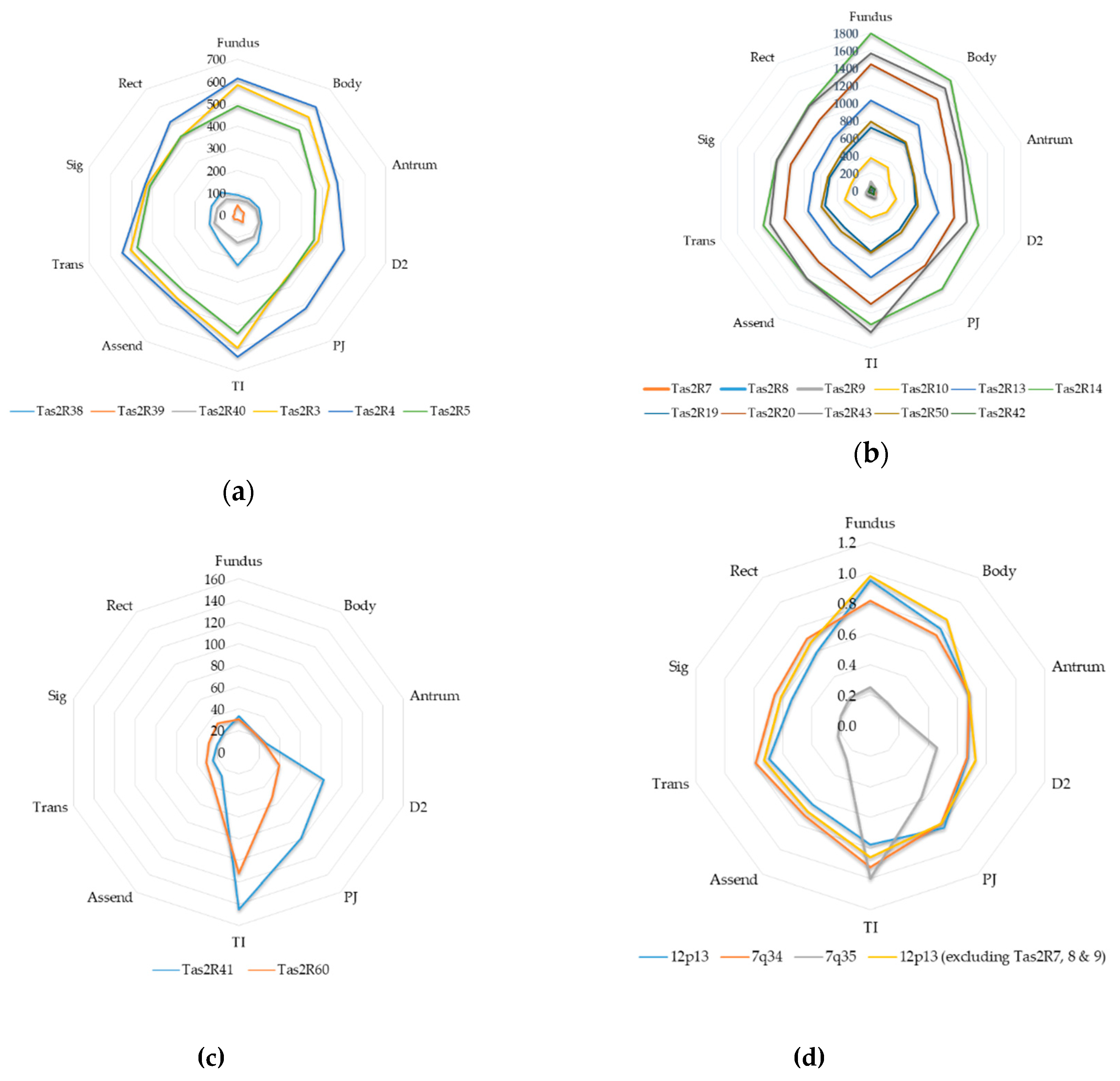
2.3. Inter-Regional Variation of GI TAS2Rs
| Region | Gastric Fundus |
Gastric Body |
Gastric Ant. |
D2 Duo. |
Prox. Jejunum |
Term. Ileum |
Assen. Colon |
Trans. Colon |
Sig. Colon |
Rectum |
Average |
| TAS2R1 | 23 | 14 | 20 | 16 | 29 | 11 | 9 | 7 | 5 | 9 | 14 |
| ±13a | ±8ab | ±15bc | ±9abc | ±17a | ±7bc | ±6c | ±7c | ±3c | ±5bc | ±3 | |
| TAS2R10 | 373 | 330 | 227 | 307 | 300 | 305 | 265 | 315 | 237 | 256 | 292 |
| ±68a | ±49ab | ±55c | ±56abc | ±88bc | ±69abc | ±48abc | ±73abc | ±42bc | ±48bc | ±19 | |
| TAS2R13 | 1031 | 925 | 649 | 810 | 811 | 988 | 751 | 752 | 681 | 736 | 813 |
| ±123a | ±92ab | ±68bc | ±105abc | ±147abc | ±146bc | ±92abc | ±84abc | ±72abc | ±108c | ±34 | |
| TAS2R14 | 1798 | 1553 | 1147 | 1291 | 1383 | 1527 | 1240 | 1291 | 1129 | 1206 | 1356 |
| ±218a | ±134ab | ±86c | ±146cd | ±211bcd | ±172abcd | ±143cd | ±172cd | ±110c | ±118c | ±51 | |
| TAS2R16 | 45 | 33 | 35 | 25 | 51 | 25 | 28 | 27 | 16 | 21 | 30 |
| ±26a | ±17ab | ±26bcd | ±15bcd | ±33abc | ±16d | ±18d | ±19d | ±10d | ±13cd | ±6 | |
| TAS2R19 | 718 | 668 | 511 | 533 | 554 | 691 | 514 | 555 | 497 | 505 | 575 |
| ±68a | ±48ab | ±48ab | ±60ab | ±106ab | ±78ab | ±63ab | ±66ab | ±53ab | ±59b | ±21 | |
| TAS2R20 | 1444 | 1292 | 955 | 1003 | 1052 | 1294 | 1010 | 1037 | 962 | 1001 | 1106 |
| ±110a | ±90a | ±72b | ±101b | ±149b | ±124ac | ±92bc | ±107b | ±85b | ±116b | ±35 | |
| TAS2R3 | 583 | 543 | 430 | 380 | 358 | 597 | 462 | 506 | 430 | 433 | 473 |
| ±30ab | ±26ab | ±29abc | ±21abc | ±37c | ±32a | ±26abc | ±35abc | ±34abc | ±54bc | ±12 | |
| TAS2R38 | 91 | 88 | 99 | 112 | 153 | 226 | 145 | 133 | 125 | 126 | 130 |
| ±29a | ±17b | ±16bc | ±13bc | ±20cd | ±28d | ±17cd | ±14cd | ±13bcd | ±17bc | ±7 | |
| TAS2R39 | 44 | 26 | 32 | 24 | 40 | 20 | 25 | 24 | 14 | 22 | 27 |
| ±23a | ±14ab | ±24bc | ±15bc | ±26bc | ±13c | ±16c | ±16c | ±9c | ±16bc | ±5 | |
| TAS2R4 | 615 | 598 | 469 | 503 | 520 | 637 | 480 | 545 | 439 | 515 | 532 |
| ±31 | ±26 | ±28 | ±13 | ±42 | ±28 | ±19 | ±38 | ±29 | ±77 | ±12 | |
| TAS2R40 | 68 | 71 | 86 | 100 | 122 | 124 | 98 | 112 | 98 | 88 | 97 |
| ±25a | ±14b | ±18bc | ±12bc | ±19c | ±15c | ±13bc | ±16bc | ±9bc | ±14bc | ±5 | |
| TAS2R41 | 33 | 24 | 27 | 83 | 98 | 145 | 27 | 25 | 21 | 23 | 50 |
| ±15a | ±8a | ±13a | ±9b | ±14b | ±30b | ±9a | ±10a | ±5a | ±6a | ±5 | |
| TAS2R42 | 49 | 35 | 47 | 34 | 58 | 27 | 30 | 31 | 14 | 24 | 35 |
| ±27a | ±20ab | ±36bc | ±21bc | ±38bc | ±18c | ±20c | ±20c | ±9c | ±15bc | ±7 | |
| TAS2R43* | 1567 | 1440 | 1095 | 1153 | 1082 | 1618 | 1241 | 1211 | 1132 | 1198 | 1274 |
| ±111ab | ±86ab | ±91ab | ±110ab | ±162a | ±122b | ±119ab | ±106ab | ±90ab | ±166ab | ±39 | |
| TAS2R5 | 490 | 469 | 365 | 360 | 364 | 531 | 418 | 475 | 415 | 437 | 433 |
| ±29ab | ±23abc | ±19d | ±20d | ±31d | ±37a | ±20abc | ±43bcd | ±38cd | ±48bcd | ±11 | |
| TAS2R50 | 789 | 683 | 519 | 567 | 591 | 707 | 572 | 598 | 513 | 548 | 609 |
| ±101a | ±77abc | ±56d | ±80bcd | ±123bd | ±86ac | ±80bcd | ±89bcd | ±65d | ±84d | ±27 | |
| TAS2R60 | 30 | 23 | 25 | 39 | 52 | 112 | 39 | 31 | 29 | 33 | 41 |
| ±13ab | ±9ab | ±14a | ±13bc | ±19cd | ±24d | ±18abc | ±12abc | ±9abc | ±8bc | ±5 | |
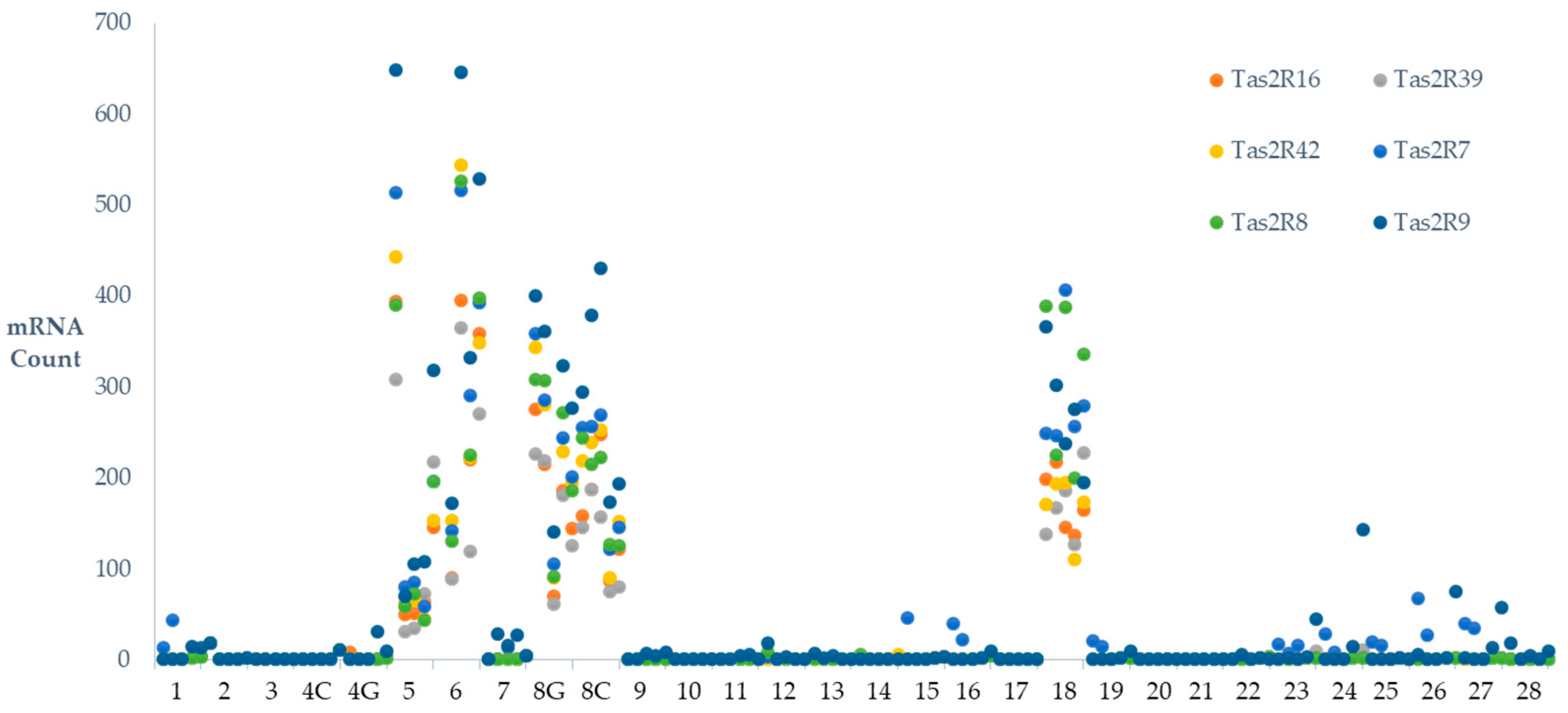
| TAS2R7 | 55 | 41 | 49 | 46 | 85 | 35 | 34 | 47 | 26 | 30 | 45 |
| ±30ab | ±24ac | ±34ac | ±20bd | ±41d | ±23c | ±23c | ±31c | ±18c | ±20ac | ±8 | |
| TAS2R8 | 54 | 37 | 47 | 34 | 52 | 44 | 30 | 42 | 23 | 33 | 40 |
| ±30a | ±22ab | ±35bc | ±22bc | ±34bc | ±29bc | ±20c | ±29c | ±15c | ±23bc | ±8 | |
| TAS2R9 | 100 | 57 | 60 | 41 | 78 | 48 | 48 | 48 | 33 | 29 | 54 |
| ±40a | ±29ab | ±43bc | ±26bc | ±52bc | ±30bc | ±31bc | ±31c | ±21bc | ±17bc | ±10 | |
| Average | 473 | 426 | 328 | 355 | 373 | 463 | 356 | 372 | 326 | 346 | |
| ±35 | ±30 | ±23 | ±25 | ±30 | ±32 | ±26 | ±27 | ±23 | ±26 |
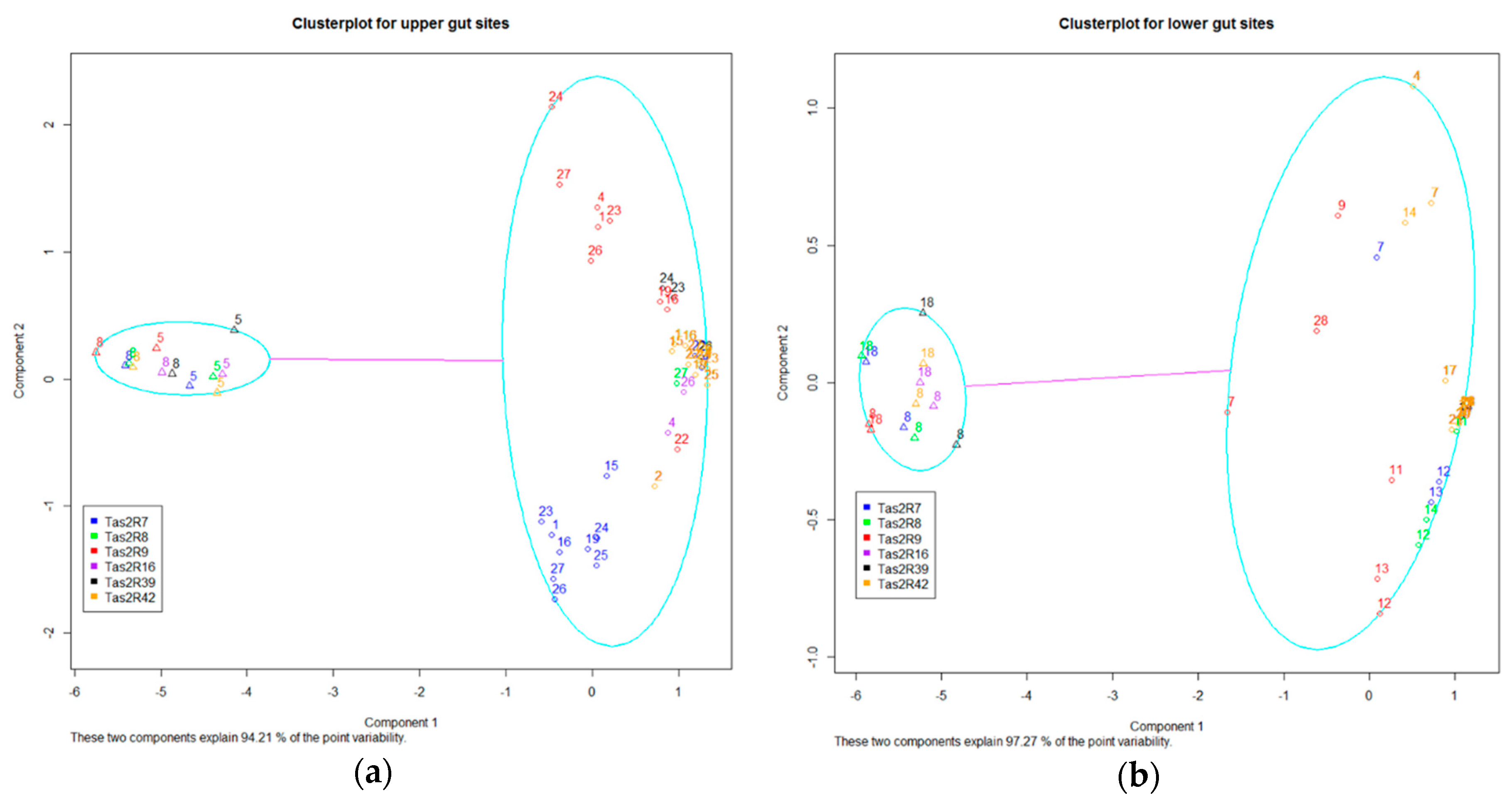
2.4. Inter-Individual Variation in TAS2Rs
2.5. Cytogeneic and Phylogenetic Variation of GI TAS2Rs
3. Discussion
4. Materials and Methods
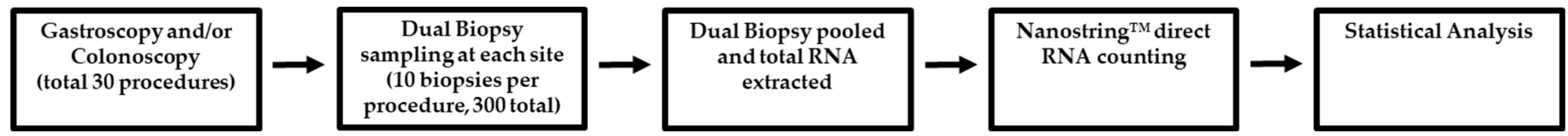
4.1. Ethics and Recruitment
4.2. Biopsy Tissue Sampling
| Sample Number | Procedure | Sample Name | Location Notes |
| 1 | Gastroscopy | Gastric Fundus |
|
| 2 | Gastroscopy | Gastric Body |
Approximately midpoint along the greater curvature |
| 3 | Gastroscopy | Gastric Antrum |
|
| 4 | Gastroscopy | D2 Duodenum |
|
| 5 | Gastroscopy | Proximal Jejunum |
Or as close to the Duodenojejunal Flexure as possible |
| 6 | Colonoscopy | Terminal Ileum |
Proximal to the ileocecal sphincter |
| 7 | Colonoscopy | Ascending Colon |
|
| 8 | Colonoscopy | Transverse Colon |
|
| 9 | Colonoscopy | Sigmoid Colon |
|
| 10 | Colonoscopy | Rectum |
4.3. NanoString Bitter Taste Receptor Analysis
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| TAS2R | Bitter Taste Receptor |
| LOD | Limit of Detection |
| LD | linear dichroism |
References
- Psichas, A.; Reimann, F.; Gribble, F.M. Gut chemosensing mechanisms. J. Clin. Investig. 2015, 125, 908–917. [CrossRef]
- Al Massadi, O.; Pardo, M.; Roca-Rivada, A.; Castelao, C.; Casanueva, F.F.; Camino, L.M.S. Macronutrients act directly on the stomach to regulate gastric ghrelin release. J. Endocrinol. Investig. 2010, 33, 599–602. [CrossRef]
- Janssen, S.; Laermans, J.; Verhulst, P.-J.; Thijs, T.; Tack, J.; Depoortere, I. Bitter taste receptors and α-gustducin regulate the secretion of ghrelin with functional effects on food intake and gastric emptying. Proc. Natl. Acad. Sci. 2011, 108, 2094–2099. [CrossRef]
- Feng, X.; Zhong, S.; Yang, J.; Wang, Y.; Liu, J. Effects on Glucagon-Like Peptide-1 Secretion by Distal Ileal Administration of Nutrients. Obes. Surg. 2013, 23, 1774–1782. [CrossRef]
- Díez-Sampedro, A.; Hirayama, B.A.; Osswald, C.; Gorboulev, V.; Baumgarten, K.; Volk, C.; Wright, E.M.; Koepsell, H. A glucose sensor hiding in a family of transporters. Proc. Natl. Acad. Sci. USA 2003, 100, 11753–11758. [CrossRef]
- Jang, H.-J.; Kokrashvili, Z.; Theodorakis, M.J.; Carlson, O.D.; Kim, B.-J.; Zhou, J.; Kim, H.H.; Xu, X.; Chan, S.L.; Juhaszova, M.; et al. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc. Natl. Acad. Sci. USA 2007, 104, 15069–15074. [CrossRef]
- Tolhurst, G.; Heffron, H.; Lam, Y.S.; Parker, H.E.; Habib, A.M.; Diakogiannaki, E.; Cameron, J.; Grosse, J.; Reimann, F.; Gribble, F.M. Short-Chain Fatty Acids Stimulate Glucagon-Like Peptide-1 Secretion via the G-Protein-Coupled Receptor FFAR2. Diabetes 2012, 61, 364–371. [CrossRef]
- Avau, B.; Depoortere, I. The bitter truth about bitter taste receptors: beyond sensing bitter in the oral cavity. Acta Physiol. 2015, 216, 407–420. [CrossRef]
- Meyerhof, W.; Batram, C.; Kuhn, C.; Brockhoff, A.; Chudoba, E.; Bufe, B.; Appendino, G.; Behrens, M. The Molecular Receptive Ranges of Human TAS2R Bitter Taste Receptors. Chem. Senses 2010, 35, 157–170. [CrossRef]
- Beauchamp, G.K.; Mennella, J.A. Flavor Perception in Human Infants: Development and Functional Significance. Digestion 2011, 83, 1–6. [CrossRef]
- Rozin, P.; A Vollmecke, T. Food Likes and Dislikes. Annu. Rev. Nutr. 1986, 6, 433–456. [CrossRef]
- Latorre, R.; Huynh, J.; Mazzoni, M.; Gupta, A.; Bonora, E.; Clavenzani, P.; Chang, L.; Mayer, E.A.; De Giorgio, R.; Sternini, C. Expression of the Bitter Taste Receptor, T2R38, in Enteroendocrine Cells of the Colonic Mucosa of Overweight/Obese vs. Lean Subjects. PLOS ONE 2016, 11, e0147468. [CrossRef]
- Gunawardene, A.R.; Corfe, B.M.; Staton, C.A. Classification and functions of enteroendocrine cells of the lower gastrointestinal tract. Int. J. Exp. Pathol. 2011, 92, 219–231. [CrossRef]
- Sternini, C., L. Anselmi, and E. Rozengurt, Enteroendocrine cells: a site of 'taste' in gastrointestinal chemosensing. Curr Opin Endocrinol Diabetes Obes, 2008. 15(1): p. 73-8.
- Moran, G.W., et al., Enteroendocrine cells: neglected players in gastrointestinal disorders? Therap Adv Gastroenterol, 2008. 1(1): p. 51-60.
- Posovszky, C.; Wabitsch, M. Regulation of Appetite, Satiation, and Body Weight by Enteroendocrine Cells. Part 1: Characteristics of Enteroendocrine Cells and Their Capability of Weight Regulation. Horm. Res. Paediatr. 2015, 83, 1–10. [CrossRef]
- Walker, E.; Lo, K.; Tham, S.; Pahl, M.; Lomiwes, D.; Cooney, J.; Wohlers, M.; Gopal, P. New Zealand Bitter Hops Extract Reduces Hunger During a 24 h Water Only Fast. Nutrients 2019, 11, 2754. [CrossRef]
- Walker, E.G.; Lo, K.R.; Pahl, M.C.; Shin, H.S.; Lang, C.; Wohlers, M.W.; Poppitt, S.D.; Sutton, K.H.; Ingram, J.R. An extract of hops (Humulus lupulus L.) modulates gut peptide hormone secretion and reduces energy intake in healthy-weight men: a randomized, crossover clinical trial. Am. J. Clin. Nutr. 2022, 115, 925–940. [CrossRef]
- Walker, E.; Lo, K.; Gopal, P. Gastrointestinal delivery of bitter hop extract reduces appetite and food cravings in healthy adult women undergoing acute fasting. Obes. Pillars 2024, 11, 100117. [CrossRef]
- Sakai, H.; Sato, K.; Kai, Y.; Chiba, Y.; Narita, M. Denatonium and 6-n-Propyl-2-thiouracil, Agonists of Bitter Taste Receptor, Inhibit Contraction of Various Types of Smooth Muscles in the Rat and Mouse. Biol. Pharm. Bull. 2016, 39, 33–41. [CrossRef]
- Clark, A.A.; Liggett, S.B.; Munger, S.D. Extraoral bitter taste receptors as mediators of off-target drug effects. FASEB J. 2012, 26, 4827–4831. [CrossRef]
- Jeon, T.-I.; Seo, Y.-K.; Osborne, T.F. Gut bitter taste receptor signalling induces ABCB1 through a mechanism involving CCK. Biochem. J. 2011, 438, 33–37. [CrossRef]
- Liszt, K.I.; Wang, Q.; Farhadipour, M.; Segers, A.; Thijs, T.; Nys, L.; Deleus, E.; Van der Schueren, B.; Gerner, C.; Neuditschko, B.; et al. Human intestinal bitter taste receptors regulate innate immune responses and metabolic regulators in obesity. J. Clin. Investig. 2022, 132. [CrossRef]
- Lei, H.; Yu, D.; Xue, Y.-B.; Li, Y.-H.; Gong, S.-M.; Peng, Y.-Y.; Liu, K.-F.; Buratto, D.; Yang, Y.; Zhang, S.-S.; et al. Tuft cells utilize taste signaling molecules to respond to the pathobiont microbe Ruminococcus gnavus in the proximal colon. Front. Immunol. 2023, 14, 1259521. [CrossRef]
- Schneider, C.; O’leary, C.E.; Locksley, R.M. Regulation of immune responses by tuft cells. Nat. Rev. Immunol. 2019, 19, 584–593. [CrossRef]
- Gustafsson, J.K.; Johansson, M.E.V. The role of goblet cells and mucus in intestinal homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 785–803. [CrossRef]
- Gassler, N. Paneth cells in intestinal physiology and pathophysiology. World J. Gastrointest. Pathophysiol. 2017, 8, 150–160. [CrossRef]
- Wu, S.V.; Rozengurt, N.; Yang, M.; Young, S.H.; Sinnett-Smith, J.; Rozengurt, E. Expression of bitter taste receptors of the T2R family in the gastrointestinal tract and enteroendocrine STC-1 cells. Proc. Natl. Acad. Sci. USA 2002, 99, 2392–2397. [CrossRef]
- Rozengurt, N.; Wu, S.V.; Chen, M.C.; Huang, C.; Sternini, C.; Rozengurt, E. Colocalization of the α-subunit of gustducin with PYY and GLP-1 in L cells of human colon. Am. J. Physiol. Liver Physiol. 2006, 291, G792–G802. [CrossRef]
- Kaji, I.; Karaki, S.-I.; Fukami, Y.; Terasaki, M.; Kuwahara, A. Secretory effects of a luminal bitter tastant and expressions of bitter taste receptors, T2Rs, in the human and rat large intestine. Am. J. Physiol. Liver Physiol. 2009, 296, G971–G981. [CrossRef]
- Le Nevé, B.; Foltz, M.; Daniel, H.; Gouka, R. The steroid glycoside H.g.-12 from Hoodia gordonii activates the human bitter receptor TAS2R14 and induces CCK release from HuTu-80 cells. Am. J. Physiol. Liver Physiol. 2010, 299, G1368–G1375. [CrossRef]
- Descamps-Solà, M.; Vilalta, A.; Jalsevac, F.; Blay, M.T.; Rodríguez-Gallego, E.; Pinent, M.; Beltrán-Debón, R.; Terra, X.; Ardévol, A. Bitter taste receptors along the gastrointestinal tract: comparison between humans and rodents. Front. Nutr. 2023, 10, 1215889. [CrossRef]
- Nolden, A.A.; McGeary, J.E.; Hayes, J.E. Differential bitterness in capsaicin, piperine, and ethanol associates with polymorphisms in multiple bitter taste receptor genes. Physiol. Behav. 2016, 156, 117–127. [CrossRef]
- Kauer, J., et al., Adult picky eating. Phenomenology, taste sensitivity, and psychological correlates. Appetite, 2015. 90: p. 219-28.
- Green, E.; Jacobson, A.; Haase, L.; Murphy, C. Neural correlates of taste and pleasantness evaluation in the metabolic syndrome. Brain Res. 2015, 1620, 57–71. [CrossRef]
- A Mennella, J. Ontogeny of taste preferences: basic biology and implications for health. Am. J. Clin. Nutr. 2014, 99, 704S–711S. [CrossRef]
- Dotson, C.D.; Zhang, L.; Xu, H.; Shin, Y.-K.; Vigues, S.; Ott, S.H.; Elson, A.E.T.; Choi, H.J.; Shaw, H.; Egan, J.M.; et al. Bitter Taste Receptors Influence Glucose Homeostasis. PLOS ONE 2008, 3, e3974. [CrossRef]
- Kok, B.P., et al., Intestinal bitter taste receptor activation alters hormone secretion and imparts metabolic benefits. Mol Metab, 2018. 16: p. 76-87.
- Liszt, K.I.; Ley, J.P.; Lieder, B.; Behrens, M.; Stöger, V.; Reiner, A.; Hochkogler, C.M.; Köck, E.; Marchiori, A.; Hans, J.; et al. Caffeine induces gastric acid secretion via bitter taste signaling in gastric parietal cells. Proc. Natl. Acad. Sci. USA 2017, 114, E6260–E6269. [CrossRef]
- Lee, R.J.; Xiong, G.; Kofonow, J.M.; Chen, B.; Lysenko, A.; Jiang, P.; Abraham, V.; Doghramji, L.; Adappa, N.D.; Palmer, J.N.; et al. T2R38 taste receptor polymorphisms underlie susceptibility to upper respiratory infection. J. Clin. Investig. 2012, 122, 4145–4159. [CrossRef]
- Verbeurgt, C.; Veithen, A.; Carlot, S.; Tarabichi, M.; Dumont, J.E.; Hassid, S.; Chatelain, P. The human bitter taste receptor T2R38 is broadly tuned for bacterial compounds. PLOS ONE 2017, 12, e0181302. [CrossRef]
- Egerod, K.L.; Engelstoft, M.S.; Grunddal, K.V.; Nøhr, M.K.; Secher, A.; Sakata, I.; Pedersen, J.; Windeløv, J.A.; Füchtbauer, E.-M.; Olsen, J.; et al. A Major Lineage of Enteroendocrine Cells Coexpress CCK, Secretin, GIP, GLP-1, PYY, and Neurotensin but Not Somatostatin. Endocrinology 2012, 153, 5782–5795. [CrossRef]
- Harada, Y.; Koseki, J.; Sekine, H.; Fujitsuka, N.; Kobayashi, H. Role of Bitter Taste Receptors in Regulating Gastric Accommodation in Guinea Pigs. J. Pharmacol. Exp. Ther. 2019, 369, 466–472. [CrossRef]
- Kuwahara, A. Involvement of the Gut Chemosensory System in the Regulation of Colonic Anion Secretion. BioMed Res. Int. 2015, 2015, 1–9. [CrossRef]
- Lossow, K.; Hübner, S.; Roudnitzky, N.; Slack, J.P.; Pollastro, F.; Behrens, M.; Meyerhof, W. Comprehensive Analysis of Mouse Bitter Taste Receptors Reveals Different Molecular Receptive Ranges for Orthologous Receptors in Mice and Humans. J. Biol. Chem. 2016, 291, 15358–15377. [CrossRef]
- Thalmann, S.; Behrens, M.; Meyerhof, W. Major haplotypes of the human bitter taste receptor TAS2R41 encode functional receptors for chloramphenicol. Biochem. Biophys. Res. Commun. 2013, 435, 267–273. [CrossRef]
- Fiebich, B.L.; Appel, K. Anti-inflammatory effects of willow bark extract. Clin. Pharmacol. Ther. 2003, 74, 96–96. [CrossRef]
- Saller, R., J. Melzer, and M. Felder, Pain Relief with a Proprietary Extract of Willow Bark in Rheumatology. An Open Trial. Swiss Journal of Integrative Medicine, 2008. 20(3): p. 156-162.
- Widmayer, P.; Goldschmid, H.; Henkel, H.; Kã¼Per, M.; Kã¶Nigsrainer, A.; Breer, H.; Küper, M. High fat feeding affects the number of GPR120 cells and enteroendocrine cells in the mouse stomach. Front. Physiol. 2015, 6, 53–53. [CrossRef]
- Roudnitzky, N.; Behrens, M.; Engel, A.; Kohl, S.; Thalmann, S.; Hübner, S.; Lossow, K.; Wooding, S.P.; Meyerhof, W. Receptor Polymorphism and Genomic Structure Interact to Shape Bitter Taste Perception. PLOS Genet. 2015, 11, e1005530. [CrossRef]
- Prandi, S.; Voigt, A.; Meyerhof, W.; Behrens, M. Expression profiling of Tas2r genes reveals a complex pattern along the mouse GI tract and the presence of Tas2r131 in a subset of intestinal Paneth cells. Cell. Mol. Life Sci. 2018, 75, 49–65. [CrossRef]
- Khan, M.; Vaes, E.; Mombaerts, P. Regulation of the Probability of Mouse Odorant Receptor Gene Choice. Cell 2011, 147, 907–921. [CrossRef]
- Behrens, M.; Born, S.; Redel, U.; Voigt, N.; Schuh, V.; Raguse, J.-D.; Meyerhof, W. Immunohistochemical Detection of TAS2R38 Protein in Human Taste Cells. PLOS ONE 2012, 7, e40304. [CrossRef]
- Veldman-Jones, M.H., et al., Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res, 2015. 75(13): p. 2587-93.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





