Submitted:
22 October 2024
Posted:
23 October 2024
You are already at the latest version
Abstract
One of the most pressing environmental problems contemporary civilizations confront is the ever-increasing amount of plastic waste. Because of their impact on every living thing, these wastes are seen as a major issue on a global scale. To counteract the harmful environmental effects caused by conventional disposal methods, it is critical to show that eco-friendly alternatives are viable. Biodegradation is one of the best eco-friendly methods for removing plastic waste. In this study, we aimed to identify bacteria from sewage wastewater treatment plants (SWW) that could degrade polyethylene. The bacterial strains isolated from sewerage wastewater were incubated for 120 days in 50 ml of minimal salt media (MSM) containing 60mg polyethylene. After four months, our research revealed that Bacillus tropicus (SH4) demonstrated significant potential, degrading poly-ethylene up to 21.6%. We observed the changes after biodegradation using FTIR, GC-MS, and SEM analysis. In conclusion, microorganisms extracted from sewage wastewater possess the ability to mitigate plastic contamination in aquatic ecosystems. Future proteomics and genome investiga-tions are necessary to elucidate the enzymes and metabolic processes implicated in plastic breakdown.
Keywords:
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Applying Stress to Bacteria to Use Plastic as a Carbon Source
2.2.1. Minimal Salt Media (MSM) Preparation
2.3. Molecular Identification of Bacteria
2.4. Weight Loss Experiment
2.5. Scanning Electron Microscopy (SEM) of Degraded Plastic Pieces
2.6. Gas Chromatography-Mass Spectrometry Analysis (GC-MS)
2.7. Fourier Transform Infrared Spectroscopy (FTIR)
3. Results
3.1. Molecular Identification
3.2. Weight Reduction
3.3. GC_MS Analysis
3.4. FTIR Analysis
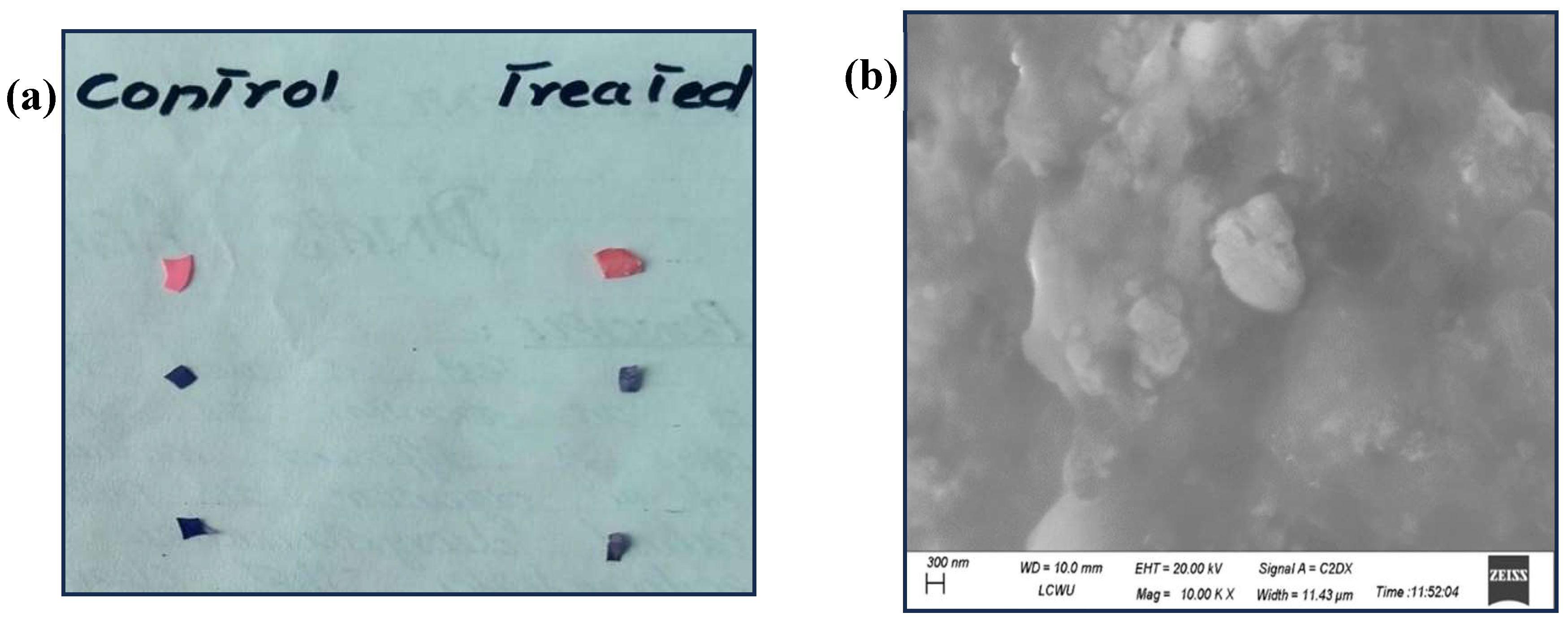
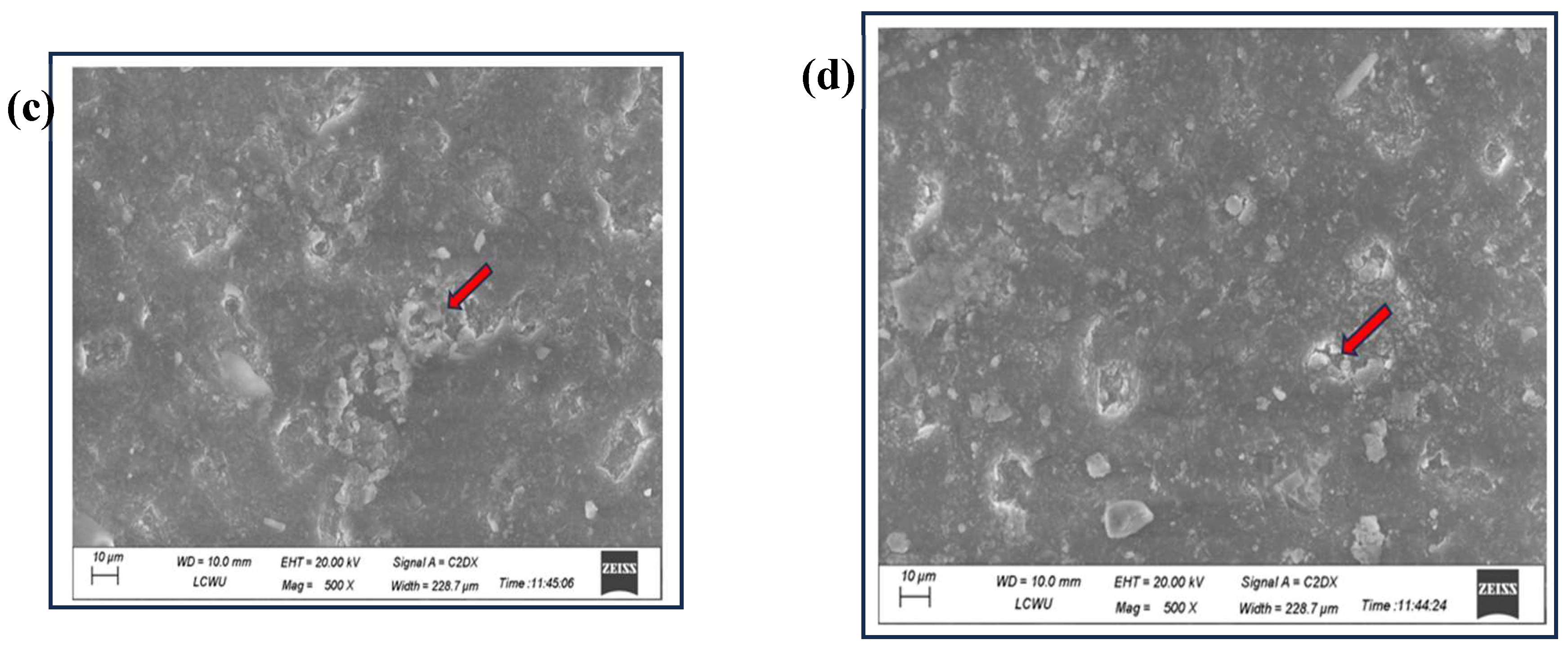
3.5. SEM Analysis for Physical Change in Plastic Pieces
4. Discussion
4. Conclusions
Author Contributions
Data Availability Statement
Conflicts of Interest
References
- Sonke, J. E., Koenig, A. M., Yakovenko, N., Hagelskjær, O., Margenat, H., Hansson, S. V., ... & Thomas, J. L. (2022). A mass budget and box model of global plastics cycling, degradation and dispersal in the land-ocean-atmosphere system. Microplastics and Nanoplastics, 2(1), 28. [CrossRef]
- Schirmeister, C. G., & Mülhaupt, R. (2022). Closing the carbon loop in the circular plastics economy. Macromolecular rapid communications, 43(13), 2200247. [CrossRef]
- Mohamadi, M. (2023). Plastic Types and Applications. Plastic Waste Treatment and Management: Gasification Processes, 1-19. [CrossRef]
- Cudjoe, D., Brahim, T., & Zhu, B. (2023). Assessing the economic and ecological viability of generating electricity from oil derived from pyrolysis of plastic waste in China. Waste Management, 168, 354-365. [CrossRef]
- Arisman, & Fatimah, Y. A. (2023). Waste management in indonesia: strategies and implementation of sustainable development goals (sdgs) and circular economy. In Circular Economy Adoption: Catalysing Decarbonisation Through Policy Instruments (pp. 131-157). Singapore: Springer Nature Singapore. [CrossRef]
- Drzyzga, O., & Prieto, A. (2019). Plastic waste management, a matter for the ‘community’. Microbial biotechnology, 12(1), 66. [CrossRef]
- Ali, S., Isha, & Chang, Y. C. (2023). Ecotoxicological Impact of Bioplastics Biodegradation: A Comprehensive Review. Processes, 11(12), 3445. [CrossRef]
- Kibria, M. G., Masuk, N. I., Safayet, R., Nguyen, H. Q., & Mourshed, M. (2023). Plastic waste: challenges and opportunities to mitigate pollution and effective management. International Journal of Environmental Research, 17(1), 20. [CrossRef]
- Bakht, A., Rasool, N., & Iftikhar, S. (2020). Characterization of plastic degrading bacteria isolated from landfill sites. International journal of clinical microbiology and biochemical technology, 3(1), 030-035. [CrossRef]
- Korotkov, E. V., Suvorova, Y. M., Kostenko, D. O., & Korotkova, M. A. (2021). Multiple alignment of promoter sequences from the Arabidopsis thaliana l. Genome. Genes, 12(2), 135. [CrossRef]
- Saitou, N., & Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular biology and evolution, 4(4), 406-425. [CrossRef]
- Felsenstein, J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. evolution, 39(4), 783-791. [CrossRef]
- Tamura, K., Nei, M., & Kumar, S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proceedings of the National Academy of Sciences, 101(30), 11030-11035. [CrossRef]
- Tamura, K., Stecher, G., & Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Molecular biology and evolution, 38(7), 3022-3027. [CrossRef]
- Gao, L., & Gu, J. D. (2021). A new unified conceptual framework involving maintenance energy, metabolism and toxicity for research on degradation of organic pollutants. International Biodeterioration & Biodegradation, 162, 105253. [CrossRef]
- Tamnou, E. B. M., Arfao, A. T., Nougang, M. E., Metsopkeng, C. S., Ewoti, O. V. N., Moungang, L. M., ... & Nola, M. (2021). Biodegradation of polyethylene by the bacterium Pseudomonas aeruginosa in acidic aquatic microcosm and effect of the environmental temperature. Environmental Challenges, 3, 100056. [CrossRef]
- Yao, Z., Seong, H. J., & Jang, Y. S. (2022). Degradation of low density polyethylene by Bacillus species. Applied Biological Chemistry, 65(1), 84. [CrossRef]
- Sun, X., Chen, Z., Kong, T., Chen, Z., Dong, Y., Kolton, M., ... & Sun, W. (2022). Mycobacteriaceae mineralizes micropolyethylene in riverine ecosystems. Environmental Science & Technology, 56(22), 15705-15717. [CrossRef]
- Abraham, J., Ghosh, E., Mukherjee, P., & Gajendiran, A. (2017). Microbial degradation of low density polyethylene. Environmental Progress & Sustainable Energy, 36(1), 147-154. [CrossRef]
- Hou, L., Xi, J., Liu, J., Wang, P., Xu, T., Liu, T., ... & Lin, Y. B. (2022). Biodegradability of polyethylene mulching film by two Pseudomonas bacteria and their potential degradation mechanism. Chemosphere, 286, 131758. [CrossRef]
- Kavitha, R., & Bhuvaneswari, V. (2021). Assessment of polyethylene degradation by biosurfactant producing ligninolytic bacterium. Biodegradation, 32(5), 531-549. [CrossRef]
- Samanta, S., Datta, D., & Halder, G. (2020). Biodegradation efficacy of soil inherent novel sp. Bacillus tropicus (MK318648) onto low-density polyethylene matrix. Journal of Polymer Research, 27(10), 1-16. [CrossRef]
- Arkatkar, A., Juwarkar, A. A., Bhaduri, S., Uppara, P. V., & Doble, M. (2010). Growth of Pseudomonas and Bacillus biofilms on pretreated polypropylene surface. International Biodeterioration & Biodegradation, 64(6), 530-536. [CrossRef]
- Gupta, K. K., & Devi, D. (2020). Characteristics investigation on biofilm formation and biodegradation activities of Pseudomonas aeruginosa strain ISJ14 colonizing low density polyethylene (LDPE) surface. Heliyon, 6(7). [CrossRef]
- MacLeod, M., Arp, H. P. H., Tekman, M. B., & Jahnke, A. (2021). The global threat from plastic pollution. Science, 373(6550), 61-65. [CrossRef]
- Li, X., Tian, Y., Xu, C., & Cheng, B. (2019). The impact of marine pollution control on the output value of marine fisheries based on the spatial econometric model. Journal of Coastal Research, 98(SI), 381-384. [CrossRef]
- Velis, C. A. (2014). Plastic waste in marine litter: Action now and at the source. Waste Management & Research, 32(4), 251-253. [CrossRef]
- Mukhaifi, E. A., Al-Atbi, H. S., & Ali, S. F. (2023). Isolation and Identification of Polyethylene Terephthalate Degrading Bacteria from Shatt Al-Arab and Sewage Water of Basrah City. Baghdad Science Journal, 20(5 (Suppl.)). [CrossRef]
- Meng, Q., Yi, X., Zhou, H., Song, H., Liu, Y., Zhan, J., & Pan, H. (2024). Isolation of marine polyethylene (PE)-degrading bacteria and its potential degradation mechanisms. Marine Pollution Bulletin, 207, 116875. [CrossRef]
- Skariyachan, S., Patil, A. A., Shankar, A., Manjunath, M., Bachappanavar, N., & Kiran, S. (2018). Enhanced polymer degradation of polyethylene and polypropylene by novel thermophilic consortia of Brevibacillus sps. and Aneurinibacillus sp. screened from waste management landfills and sewage treatment plants. Polymer Degradation and Stability, 149, 52-68. [CrossRef]
- Shahnawaz, M., Sangale, M. K., & Ade, A. B. (2016). Bacteria-based polythene degradation products: GC-MS analysis and toxicity testing. Environmental Science and Pollution Research, 23(11), 10733-10741. [CrossRef]
- Roy, P. K., Titus, S., Surekha, P., Tulsi, E., Deshmukh, C., & Rajagopal, C. (2008). Degradation of abiotically aged LDPE films containing pro-oxidant by bacterial consortium. Polymer degradation and stability, 93(10), 1917-1922. [CrossRef]
- Khandare, S. D., Chaudhary, D. R., & Jha, B. (2021). Marine bacterial biodegradation of low-density polyethylene (LDPE) plastic. Biodegradation, 32(2), 127-143. [CrossRef]
- Nadeem, H., Alia, K. B., Muneer, F., Rasul, I., Siddique, M. H., Azeem, F., & Zubair, M. (2021). Isolation and identification of low-density polyethylene degrading novel bacterial strains. Archives of Microbiology, 203(9), 5417-5423. [CrossRef]
- Selke, S., Auras, R., Nguyen, T. A., Castro Aguirre, E., Cheruvathur, R., & Liu, Y. (2015). Evaluation of biodegradation-promoting additives for plastics. Environmental Science & Technology, 49(6), 3769-3777. [CrossRef]
- Shah, A. A., Hasan, F., Akhter, J. I., Hameed, A., & Ahmed, S. (2008). Degradation of polyurethane by novel bacterial consortium isolated from soil. Annals of microbiology, 58, 381-386. [CrossRef]
- Dwicania, E., Rinanti, A., & Fachrul, M. F. (2019, December). Biodegradation of LLDPE plastic by mixed bacteria culture of Pseudomonas aeruginosa and Brevibacterium sp. In Journal of Physics: Conference Series (Vol. 1402, No. 2, p. 022105). IOP Publishing. [CrossRef]
- Sanin, S. L., Sanin, F. D., & Bryers, J. D. (2003). Effect of starvation on the adhesive properties of xenobiotic degrading bacteria. Process biochemistry, 38(6), 909-914. [CrossRef]
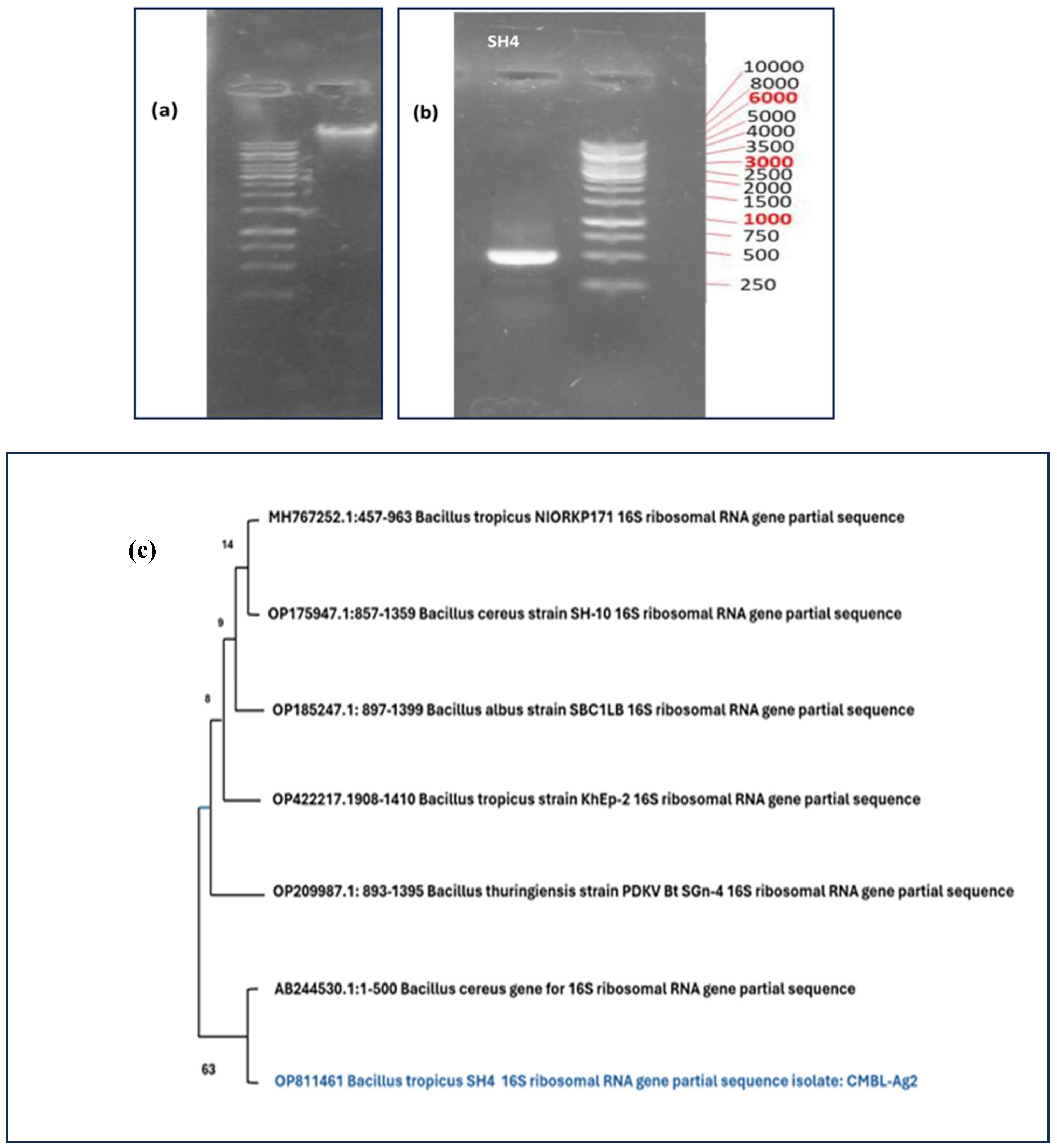
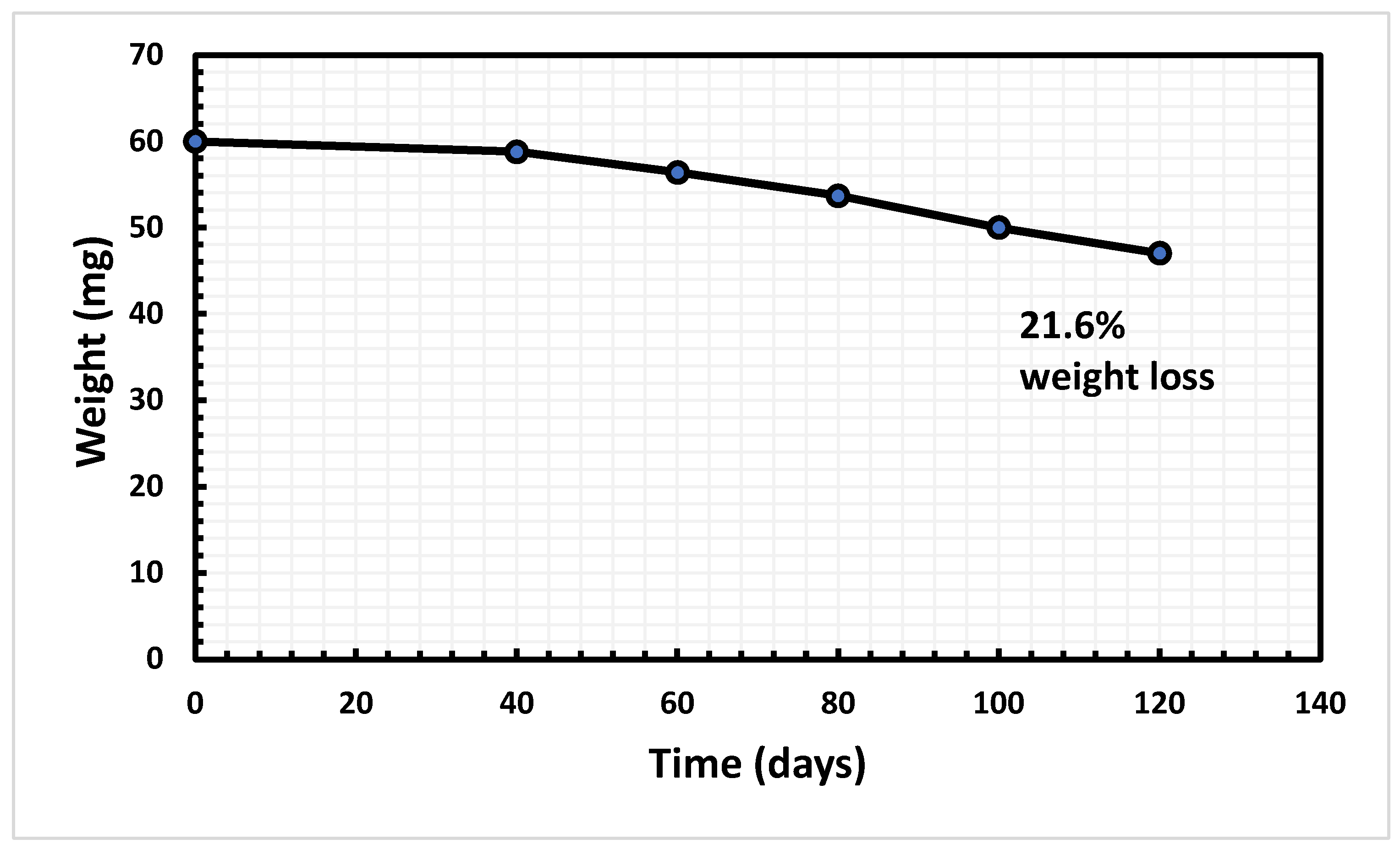
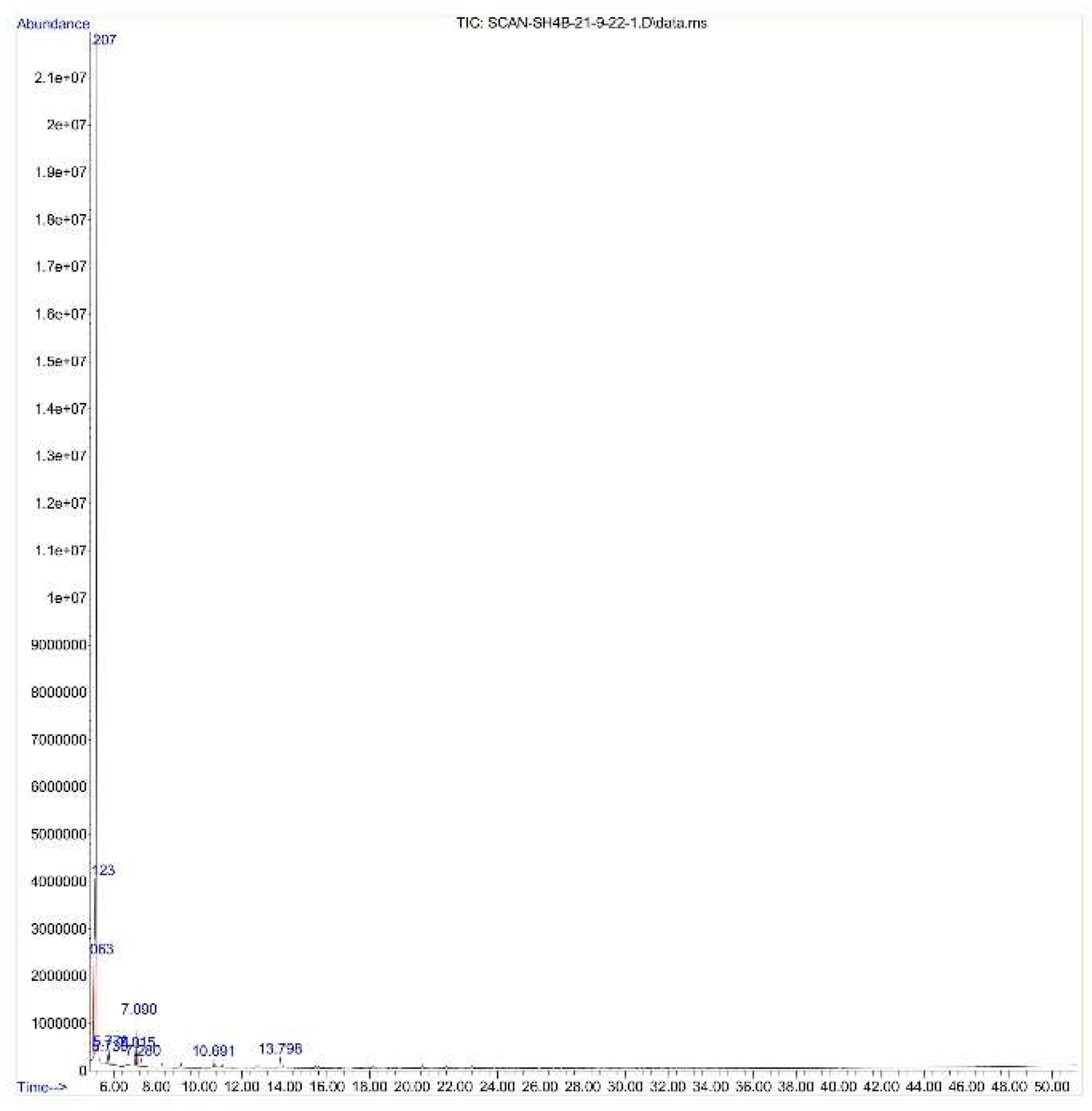
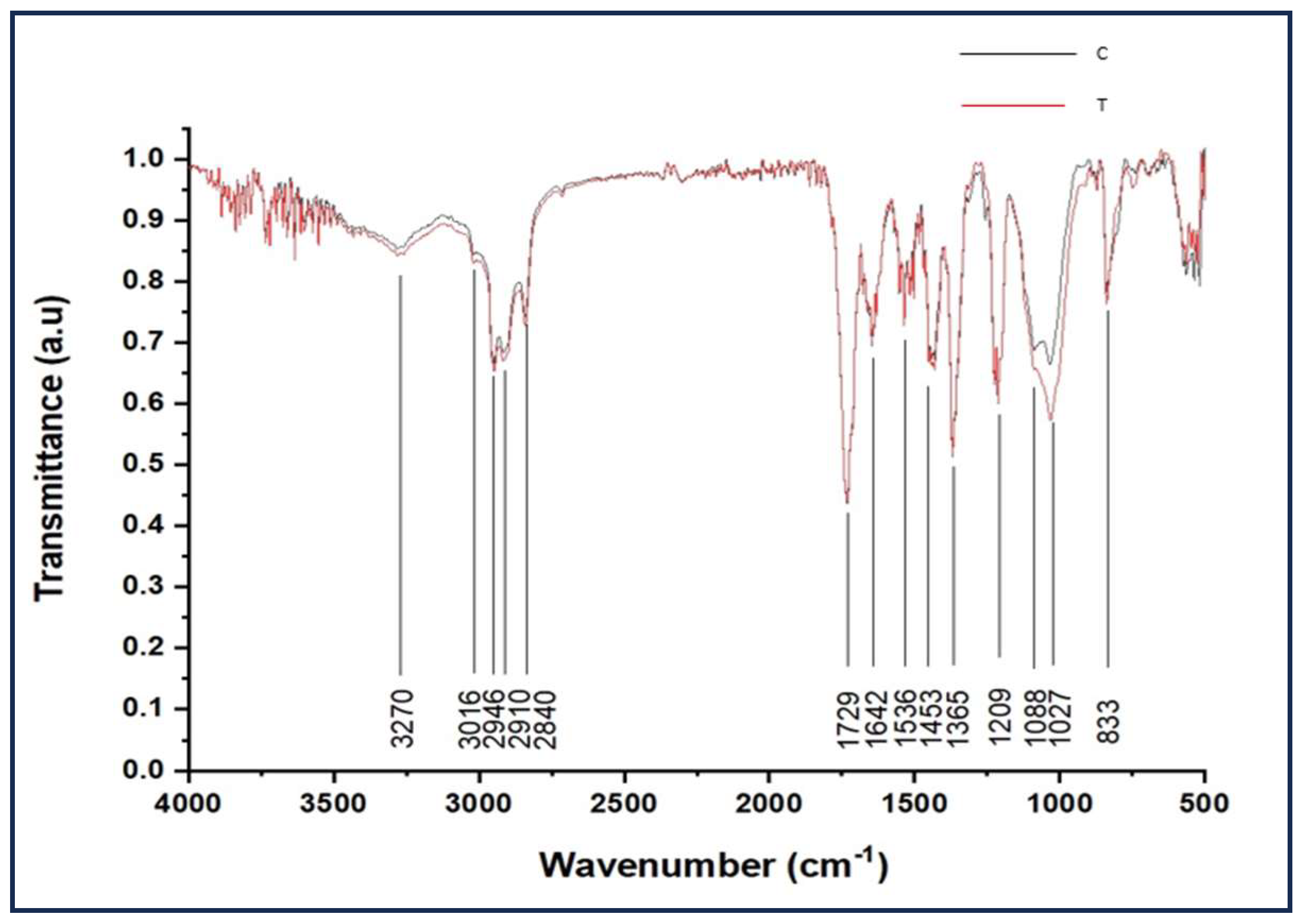
| Serial No. | Compound | Quantity/500ml |
| 1 | K2HPO4 | 2.27g |
| 2 | Na2HPO4 | 5.97g |
| 3 | NH4Cl | 0.5g |
| 4 | MgSO4 | 0.25g |
| 5 | CaCl2 | 0.0025g |
| 6 | FeSO4 | 0.001g |
| 7 | MnSO4 | 0.0005g |
| 8 | ZnSO4 | 0.001g |
| Bacteria | Compounds |
|---|---|
| Bacillus tropicus | Pentasiloxane, 1,1,3,3,5,5,7,7,9,9-decamethyl |
| 1,3,5-Benzetriol, 3TMS derivative | |
| Cyclotrisiloxane, hexamethyl | |
| Arsenous acid, tris(trimethylsilyl) ester | |
| Tris(tert-butyldimethylsilyloxy)arsane | |
| Cyclopentasiloxane, decamethyl- | |
| Cyclohexasiloxane, dodecamethyl | |
| 3-Amino-2-phenazinol ditms | |
| Benzeneethanamine, N-[(pentafluorophenyl)methylene]-.beta.,3,4-tris[(trimethylsilyl)oxy] | |
| Benzamide, 4-ethyl-N-benzyl-N-propyl | |
| trans-(2-Chlorovinyl)dimethylethoxysilane | |
| Ethanone, 2-(4-hydroxy-5,6-dimethylthieno [2,3-d]pyrimidin-2-ylthio)-1-(4-ethylphenyl) | |
| Cyclotetrasiloxane, octamethyl | |
| 1,4-Benzenedimethanethiol, 2TBDM | |
| 2,6-Dihydroxybenzoic acid, 3TMS | |
| Octasiloxane, 1,1,3,3,5,5,7,7,9,9,11,11,13,13,15,15-hexadecamethyl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





