Biographical notes: Aswin Mohan is a dynamic and diligent Research Assistant at Accubits Invent Pvt Ltd. He pursued his Masters degree in Computational Biology from Dept. of Computational Biology and Bioinformatics, University of Kerala, and a degree in Chemistry from DB Pampa College, Parumala. He has one year of experience in molecular biology and CADD at Accubits Invent. He is proficient in handling computational and biological databases and tools. His area of expertise includes all the CADD-related aspects such as Molecular modeling (Target and small molecules), Molecular docking (Discovery Studio), Molecular dynamics simulation (GROMACS), Phylogenetic analysis ( MEGA), Visualization tools(Argus lab), Sequence alignment and analysis and multiple sequence alignment(BLAST, ClustalW). He has exposure to programming languages such as python and R and experience conducting wet lab experiments.
Gayathri S S is an efficient Computational biologist, working as a Research Assistant at Accubits Invent Pvt Ltd. She is a Post Graduate in Computational Biology and pursued her graduation in Physics from the University of Kerala. She has two years of experience at Accubits Invent Pvt Ltd. She is an expert in the field of CADD, particularly on Molecular docking tools(Discovery Studio, MOE, Autodock, ClusPro), Molecular Visualization tools( PyMOL, Argus Lab, VMD), Dynamic simulation tools(Discovery studio, NAMD), Various Sequence Alignment tools( MEGA, CLUSTALW, BLAST) and published two journals in this field. She is well-trained in sequence retrieval from biological databases and their processing using bioinformatic tools. She has exposure to python, Perl, and R programming and has good hands-on experience with Bio-perl, Bio-python, and R Bioconductor packages. She also has exposure to wet lab experiments.
Mr. Sidharth Selvin, a Biotechnology and Biochemical Engineer from Sree Chithira Thirunal College of Engineering, currently serves as a Research Assistant at Accubits Invent for the past one and a half years. With expertise in Computer-Aided Drug Design (CADD) and molecular docking tools like Hex, Autodock, DiffDock, and ClusPro, he has also delved into advanced technologies such as Transformer models and neural networks. Sidharth’s proficiency extends to molecular visualization tools like PyMOL, Argus Lab, and VMD, as well as sequence alignment tools such as MEGA, CLUSTALW, and BLAST. He actively engages in data analysis, employing techniques like Knowledge Graphs to extract insights from complex biological datasets. His multidisciplinary skill set and commitment to staying updated on emerging technologies make him a valuable asset in bioinformatics and computational biology.
Shahanas Naisam is a Research Associate at Accubits Invent Pvt. Ltd. She completed her post-graduation in Bioinformatics, graduated in Botany, and has an ITI diploma in Computer Operator Programming Assistant from the University of Kerala. She has worked as a Project Trainee at the Department of Computational Biology and Bioinformatics at the University of Kerala, Karyavattom campus. Being a bioinformatic post-graduate and having a history as a research personnel, she has meticulous attention to detail, ready to investigate and manipulate complex material for biological and cellular study. She has expertise in data mining, collection, management, data integration, and data visualization. Proficient in utilizing standard software for Bioinformatics and computational biology applications, she is adept in Molecular modeling, Molecular docking interaction studies, Molecular dynamics simulation, Visualization tools, Phylogenetic Tree Analysis, Sequence analysis, alignment tools, Structure characterization, prediction tools, etc. She is also well-versed in handling microbes and phytocompounds.
Nidhin Sreekumar is a Director and Chief Research Scientist at Accubits Invent Pvt. Ltd. He completed his postgraduate studies with an MHRD Scholarship (GATE) in Energy Engineering (Chemical Engineering) from the National Institute of Technology, Trichy, with a research project in Bioenergy in collaboration with the CSIR-National Institute of Interdisciplinary Science and Technology (NIIST), Trivandrum. After which he joined Sree Chitra Thirunal College of Engineering (SCTCE) as a Faculty in Biotechnology. He went on to pursue his doctoral research with MHRD Scholarship at the National Institute of Technology Calicut, in the area of bioenergy production from wastewater in collaboration with the Ex-Chairman of Kerala State Pollution Control Board and NIIST. After that, he went back to SCTCE followed by Kalasalingam University, Tamilnadu as an Associate Professor in the Department of Biotechnology. He has many peer-reviewed publications in various areas like bioprocess, non-conventional energy resources, biomedical/healthcare, and environmental engineering.
Introduction
In the continuous hounding to develop therapeutic practices along with reducing the systemic side effects, the drug delivery field has dealt with a significant transition towards precision and selectivity. Poor drug delivery and low bioavailability of lead molecules are two major hurdles in drug development. This is due to the selectively permeable barrier plasma membrane that separates the interior of the cell from the outside environment, thereby protecting the cell from harmful substances which acts as a major challenge for the delivery of therapeutic molecules[
1]. Over the years, researchers have developed numerous delivery systems to overcome this barrier. Despite the tremendous progress, the existing delivery methods result in high toxicity, immunogenicity, and low delivery yield. Targeted drug delivery systems have emerged as a transformative approach, aiming to deliver therapeutic payloads exclusively to diseased cells or tissues[
2]. Among the myriad strategies employed, the utilization of cell-penetrating peptides (CPPs) has gained much recognition as efficient, promising, and versatile mediums or carriers for achieving precise drug delivery[
3,
4].
CPPs exhibit a remarkable ability to traverse eukaryotic membranes without causing substantial membrane damage and can carry diverse cargoes like proteins, peptides, drugs, nucleic acids, siRNAs, nanoparticles, etc [
5]. The process starts with the interaction between the negatively charged cell-surface phospholipids and the positively charged residues in CPPs[
6]. Moreover, other factors including secondary structure, charge distribution, and hydrophobicity also play significant roles in effective translocation. Some studies report the application of CPPs in delivering the natural ligands to the target sites[
7]. The CPPs conjugated with natural ligands including neuropeptide Y, melanocortin, bombesin, and vasoactive intestinal peptide selectively recognize and bind to the target receptors in various types of cancer such as prostate, colorectal, melanoma, and breast cancer[
4,
8].
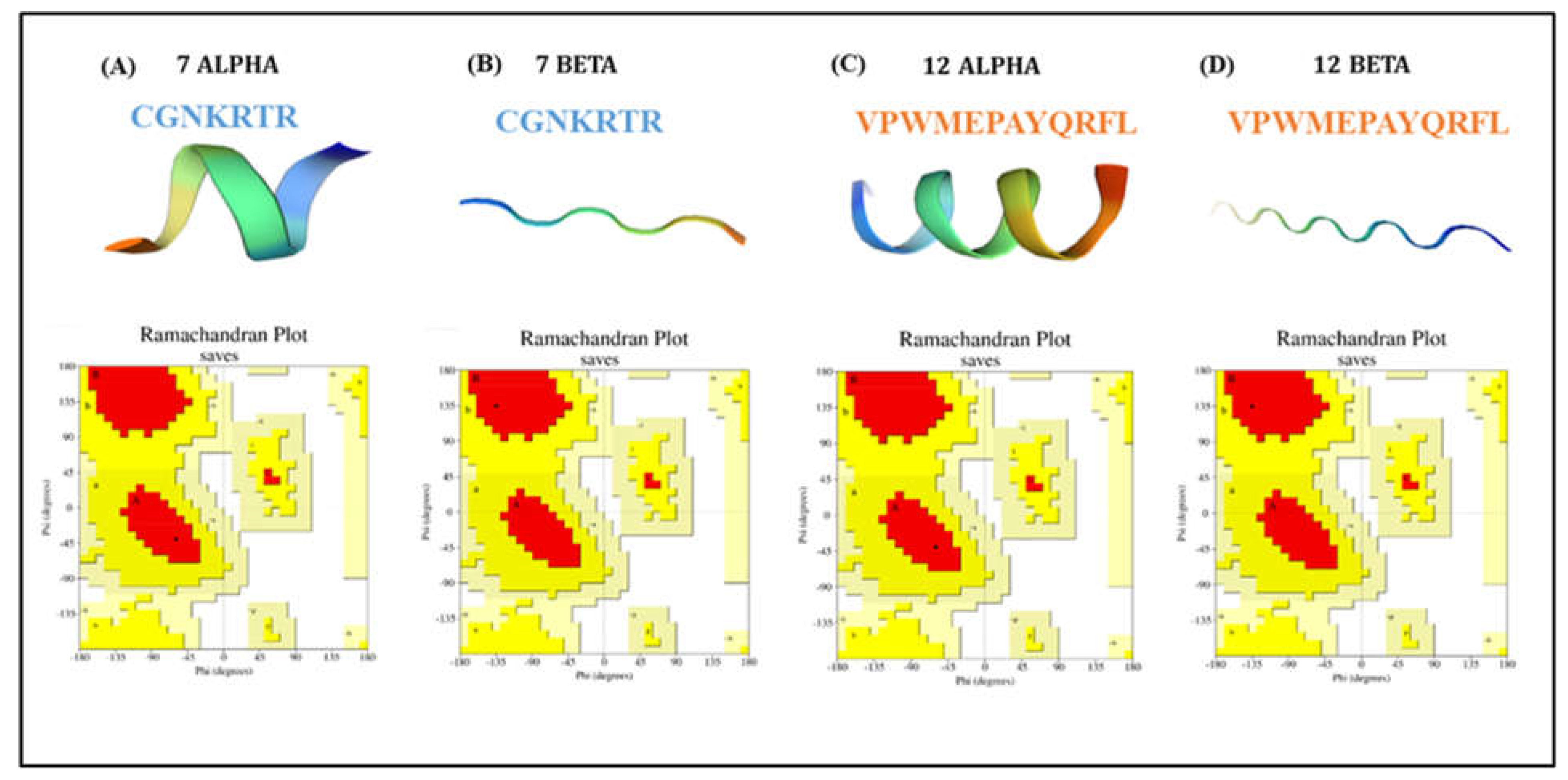
In this work, we considered both the alpha and beta conformation of two CPPs: a 7-amino acid CPP (CGNKRTR) and a 12-amino acid CPP (VPWMEPAYQRFL). The selection of these peptides was based on their ability to cell-penetrate breast cancer cells. The 7-amino acid CPP is a tumor-homing and penetrating peptide that contains a CendR motif (R/K-XX-R/K)[
9]. This motif facilitates the internalization of the peptide and its cargo into the cells. The potential of tLyp-1 in cancer diagnosis and therapy was reported in some studies. The 12-amino acid CPP, also known as p160 was identified by phage display technology as a peptide that has an affinity for neuroblastoma cells, a type of cancer cell2. It can be used as a carrier for drugs or radiopharmaceuticals that target neuroblastoma cells, offering potential applications in the development of therapeutic interventions for this particular form of cancer[
10].
As the continuation of an in-house study, we employed
in silico analysis to validate the cell-penetrating ability of 7 amino acid peptides and 12 amino acid peptides. We conjugated the CPPs with four in-house lead molecules (phytocompounds) - Phytocompound A, Phytocompound B, Phytocompound C, and Phytocompound D. They possessed a terminal hydroxyl group suitable for ester linkage with CPPs with target-specific cancer-related proteins - Cyclin-Dependent Kinase (CDK1, CDK2, CDK4, CDK6), tumor protein p53 (P53), Epidermal Growth Factor Receptor (EGFR), and Proteasomal ubiquitin receptor (PSMD4, ADRM1) that are involved in cell growth and cell signaling pathways. CDKs are a family of serine/threonine kinases that regulate the cell cycle progression by interacting with various cyclins and are specifically involved in different stages of the cell cycle[
11]. Their dysregulation has led to cancer cell development. p53 is a tumor suppressor protein that regulates cell growth and apoptosis[
12]. EGFR is a transmembrane receptor that binds to the epidermal growth factor and triggers downstream signaling pathways involved in cell growth, proliferation, and differentiation[
13]. PSMD4 and ADRM1 are proteasomal ubiquitin receptors implicated in cancer development and progression[
14].
The cell-penetrating activity validation of peptides (CPPs) was performed through the in-silico analysis of the CPP conjugated ligands and receptors incorporating molecular interaction studies in multiple software.
Materials and Methods
Target
The 3D structure of eight target proteins including Cyclin-Dependent Kinase (CDK1, CDK2, CDK4, CDK6), tumor protein p53 (P53), Epidermal Growth Factor Receptor (EGFR), and Proteasomal ubiquitin receptor (PSMD4, ADRM1) were retrieved from the Protein Data Bank in .pdb format. The retrieved structures were saved in .pdbqt format. The protein preparation steps were carried out using different software and tools. The structure was purified by using Discovery Studio Visualizer, and missing residues were added using the Swiss PDB viewer[
15]. Followed by the addition of polar hydrogen and charges using Autodock tools in the MGL tools package.
Ligands and CPPS
For this study, we considered in-house lead molecules (phytocompounds) as ligands named Phytocompound A, Phytocompound B, Phytocompound C, and Phytocompound D. The 2D structure of these ligands were retrieved from the PubChem database in .sdf format. Then the structure format was converted to .pdbqt format using Chemical Toolbox OpenBabel after the ligand preparation[
16].
CPPs were listed based on a literature review, emphasizing CPPs having good penetration in breast cancer cells. We screened the listed CPPs based on their properties using an in silico method CELLPPD, which is developed to predict and design efficient CPPs and a sequence-based tool C2Pred for identifying cell-penetrating peptides. After screening we selected the alpha and beta chains of two CPPs, the 7-amino acid CPP (CGNKRTR) and the 12-amino acid CPP (VPWMEPAYQRFL), that is 7-alpha, 7-beta, 12-alpha, and 12-beta chains (
Figure 1). The predicted physicochemical properties of selected CPPs are included in
Table 1. These CPPs were modeled using the advanced molecular editing and visualization tool Avogadro and optimized the structure for maintaining stability[
17].
Finally, the four lead molecules, Phytocompounds A, B, C, and D. were conjugated to these two CPPs in both alpha helix and beta sheet conformations, resulting in a total of 16 drug-CPP conjugates.
Molecular Docking
Molecular docking was performed using the open-source program Autodock vina (v.1.2.0.)[
18]. In this study blind docking was carried out to determine the protein-ligand interactions of 16 drug-CPP conjugates against eight target proteins. The grid box was generated with a grid spacing of 1Å for eight proteins and
Table 2 shows the corresponding grid box value of considered proteins.
After docking the result was analyzed based on the binding affinity, and H bond interaction. The H bond interaction of the complex was visualized with the help of
Discovery Studio Visualizer 2021. For a comparison study, we considered the web server Hpep dock 2.0 (
http://huanglab.phys.hust.edu.cn/hpepdock/) for molecular docking of CPP-conjugated lead molecules with eight cancer target proteins[
19]. The results were analyzed and screened based on their binding affinity and H-bond interaction.
Thus, the dock results from the two software Autodock vina and Hpep dock were compared. The best complexes with better interaction and good binding affinity in both tools were selected and subjected to further analysis.
Result and Discussion
CPPs have widely been used in anticancer drug delivery to tumor sites and many preclinical-clinical trials have proven it as a potential strategy in the field of oncology. It can facilitate the transport of therapeutic drugs across various physiological drug barriers. Even though CPPs have certain drawbacks such as short duration of action, lack of cell specificity, poor stability in vivo, poor therapeutic efficacy, compatibility problems (i.e. immunogenicity), and formation of unwanted metabolites in cancer treatment, they can be improved by optimizing the CPPs by chemical modification or by clever design. There are continuous advancements in the field of targeted drug delivery using CPPs for enhancing efficacy, especially for the transportation of anti-cancer drugs and imaging reagents. Therefore, the development of CPPs conjugated with potential lead molecules can serve as an excellent approach for the enhancement of target drug delivery in target-specific cancer-related proteins. For this study, four different Cpp chains namely 7 alpha, 7 beta, 12 Alpha & 12 beta were modeled using the Avogadro tool and validated with the help of Ramachandran Plot using SAVES v6.0 (PROCHECK) server[
20]. It was found that the number of residues found in the favored region was ~100.0% (
Figure 1.). So, the 3D structure of these CPPs was found to be acceptable for
in silico interaction analysis.
Targeting proteins involved in cell growth and signaling pathways holds immense significance in cancer treatment[
21]. The malfunctioning of the proteins leads to the dysregulation of the pathways often pilots to uncontrolled cell growth. The selected eight targets - ADRM1, PSMD4, CDK1, CDK2, CDK4, CDK6, P53, and EGFR, involved in cell growth and signaling pathways and hold notable significance. The significance lies in these proteins being targets for numerous drugs, indicating their potential as a therapeutic intervention. The CPP-conjugated drug delivery to these protein sites will help to deliver the drug to the accurate target or site where these proteins are located or function. This targeted drug delivery will reduce the off-target effects, minimize toxicity, and enhance the overall safety profile of the treatment[
22].
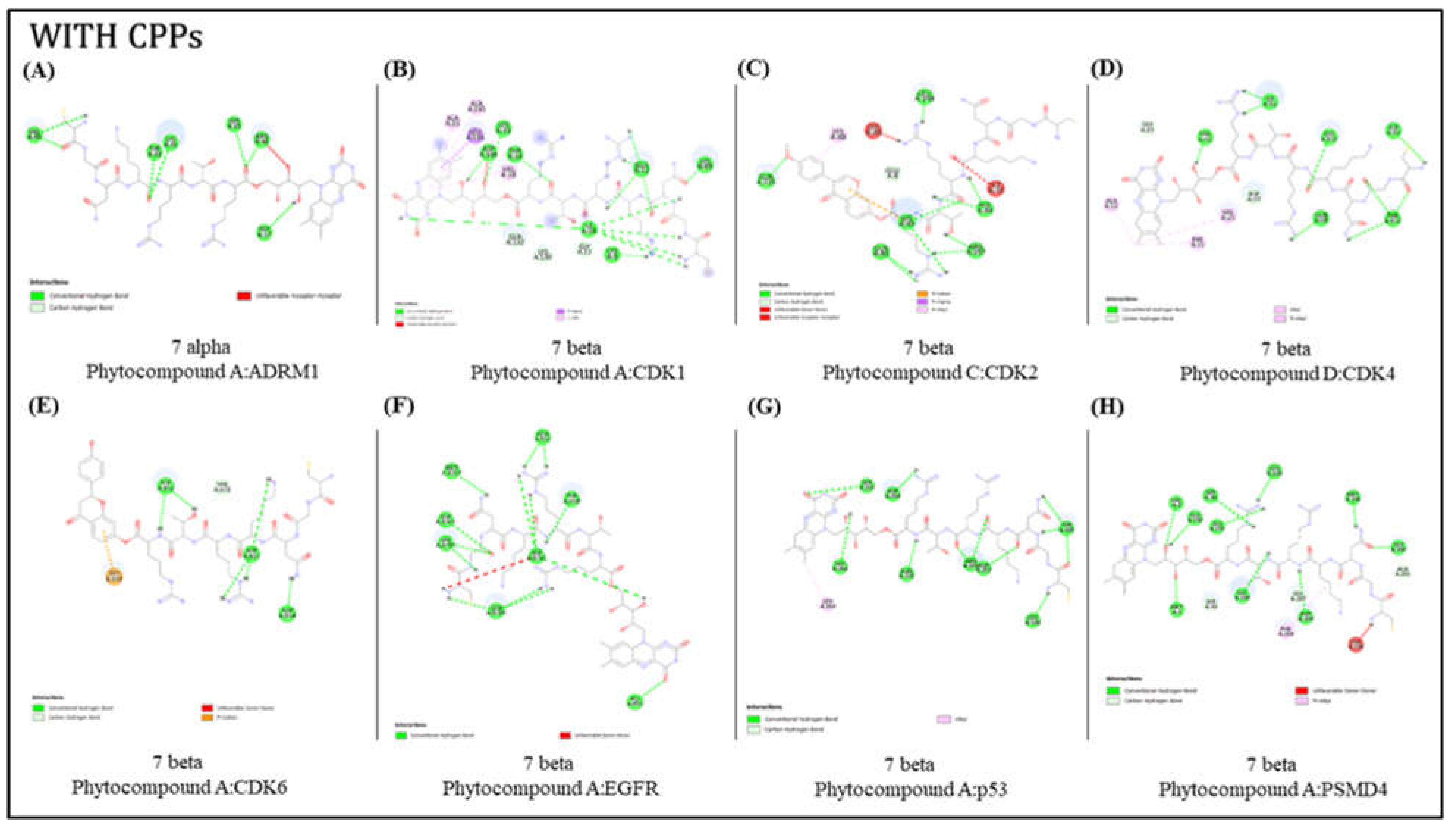
The interaction validated four in-house lead molecules (phytocompounds) Phytocompound A, Phytocompound B, Phytocompound C, and Phytocompound D were considered to conjugate with the CPPs for the targeted drug delivery study.
The in-silico interaction analysis was executed in two parts: i) Molecular docking of the lead molecules with CPPs against targets and ii) without CPPs.
- i).
Molecular docking of CPP conjugated lead molecule
A comprehensive analysis was conducted using both Autodock Vina and Hpep Dock, totaling 192 docking assessments. The outcomes were screened based on better interaction and binding affinities (
Table 3). It was observed that among the considered CPPs chains, the beta-confirmation 12 and 7 peptide chains exhibited more favorable interactions compared to the alpha-confirmation 12 and 7 chains (
Figure 2).
The Hpep dock results reveal favorable binding affinities between 12 beta chains and proteins ADRM1, CDK4, CDK6, and P53, while 7 beta chains exhibit strong affinities with EGFR and PSMD4. Additionally, 12 alpha chains demonstrate good binding affinity with CDK1 and CDK2. In contrast, the Autodock Vina results indicate that 7 beta chains exhibit enhanced binding affinities with CDK1, CDK2, CDK4, CDK6, EGFR, P53, and PSMD4, whereas 7 alpha chains exhibit strong binding affinities with ADRM1.
When assessing and comparing the dock outputs of Hpep docking and Autodock Vina, it was observed that the ligand molecules conjugated with 12 beta chains and 7 beta chains, exhibited favorable binding affinities.
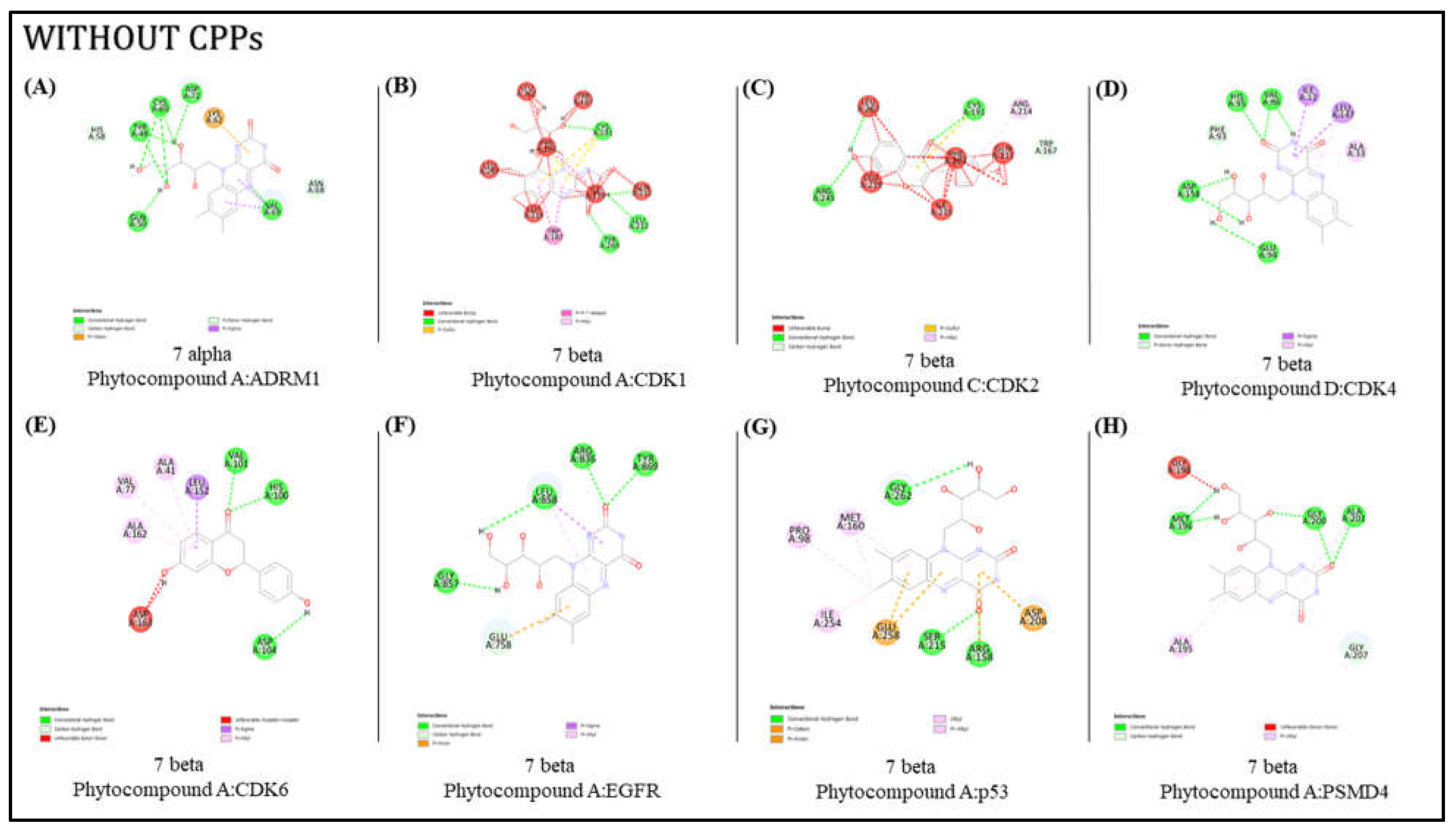
- ii).
Molecular docking of the lead molecule without CPPs
The lead molecules were subjected to docking individually using Autodock Vina software, and a comparative analysis was conducted between the Autodock results. The evaluation focused on the docking outcomes derived from the protein-ligand complex with CPPs (cell-penetrating peptides) conjugated and the complex without CPPs. The binding affinity and the count of hydrogen bonds for both scenarios, i.e., the complex with CPPs and the one without CPPs, are presented in
Table 4 (
Figure 3.).
Upon scrutinizing the docking result complexes, it is evident that there is no substantial difference in the binding affinity between complexes conjugated with CPPs and those without CPPs. However, when considering hydrogen bonds, there is a noteworthy increase in the number of hydrogen bonds in the eight complexes with CPPs compared to the ones without CPPs. This indicates that the conjugation of CPPs does not compromise the ligands’ potentiality. The results illustrate a delicate equilibrium between binding affinity and the number of interactions, emphasizing the role of CPP conjugation in augmenting protein-ligand interactions. Furthermore, it implies that the effectiveness of CPPs may be contingent on the specific target proteins; thus, further optimization and experimental validation are imperative for a comprehensive understanding of the potential advantages of CPPs in targeted drug delivery.
Conclusion
The study delves into the promising realm of cell-penetrating peptides (CPPs) for target-based drug delivery, employing an integrated approach involving data mining, molecular modeling, property calculation, and comprehensive docking analyses using multiple software tools (Autodock Vina and Hpep Dock). The meticulous screening process led to the selection of two distinct types of CPPs, namely alpha and beta conformations, each represented by 7 and 12 amino acid chains. These CPPs were then conjugated with in-house lead molecules possessing anticancer potential, specifically targeting crucial proteins involved in cell growth and signaling pathways. The in-silico analysis demonstrated the successful validation of cell-penetrating ability for both 7 and 12 amino acid peptides, with noteworthy findings on binding affinity and hydrogen bond interactions. While no substantial differences in binding affinity were observed between CPP-conjugated and non-conjugated complexes, the significant increase in hydrogen bond formation in CPP-conjugated complexes underscores the augmentative role of CPPs in protein-ligand interactions. This delicate equilibrium emphasizes the potential of CPPs in targeted drug delivery, suggesting a nuanced effectiveness dependent on specific target proteins. As a continuation of our in-house study, further optimization and experimental validation are crucial to unravel the full spectrum of advantages that CPPs may offer in the realm of targeted drug delivery for cancer therapy.
Author Contributions
Shahanas Naisam initiated outlining the computational framework and supervised the research, Aswin Mohan crafted the idea and conducted computations. Gayathri S.S executed the data collection, conducted computations and article writing activities. Shahanas Naisam took responsibility for result validation and interpretation. Sidharth Selvin assisted in the computational analysis. Dr. Nidhin Sreekumar provided invaluable support, encouraging the investigation, and overseeing the project’s progression. All authors engaged in constructive discussions concerning the outcomes and jointly contributed to shaping the final manuscript.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Statements and Declarations
Data Availability
The datasets generated during and/or analyzed during the current study are not publicly available due to the confidential nature of the work but are available from the corresponding author on reasonable request.
Acknowledgments
This research was supported by Accubits Invent Pvt. Ltd. and supervised by Dr. Nidhin Sreekumar, Chief Research Scientist. We thank all our colleagues who provided insight and expertise that greatly assisted the research.
Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Ethics approval
Ethics approval is not required for this study as it solely involves computational work.
References
- M. Stielow, A. Witczyńska, N. Kubryń, Ł. Fijałkowski, J. Nowaczyk, and A. Nowaczyk, “The Bioavailability of Drugs—The Current State of Knowledge,” Molecules, vol. 28, no. 24, 2023. [CrossRef]
- T. C. Ezike et al., “Advances in drug delivery systems, challenges and future directions,” Heliyon, vol. 9, no. 6, p. e17488, 2023. [CrossRef]
- N. Khairkhah, A. Namvar, and A. Bolhassani, “Application of Cell Penetrating Peptides as a Promising Drug Carrier to Combat Viral Infections,” Mol. Biotechnol., vol. 65, no. 9, pp. 1387–1402, 2023. [CrossRef]
- M. Zhou et al., “The role of cell-penetrating peptides in potential anti-cancer therapy,” Clin. Transl. Med., vol. 12, no. 5, 2022. [CrossRef]
- A. Gautam et al., “In silico approaches for designing highly effective cell penetrating peptides,” J. Transl. Med., vol. 11, no. 1, pp. 1–12, 2013. [CrossRef]
- I. Ruseska and A. Zimmer, “Internalization mechanisms of cell-penetrating peptides,” Beilstein J. Nanotechnol., vol. 11, pp. 101–123, 2020. [CrossRef]
- J. Xie et al., “Cell-Penetrating Peptides in Diagnosis and Treatment of Human Diseases: From Preclinical Research to Clinical Application,” Front. Pharmacol., vol. 11, no. May, pp. 1–23, 2020. [CrossRef]
- P. Hoppenz, S. Els-Heindl, and A. G. Beck-Sickinger, “Peptide-Drug Conjugates and Their Targets in Advanced Cancer Therapies,” Front. Chem., vol. 8, no. July, pp. 1–24, 2020. [CrossRef]
- L. Larue, B. Kenzhebayeva, M. G. Al-thiabat, and V. Jouan, “tLyp – 1 : a peptide suitable to target NRP – 1 receptor,” 2022.
- V. Askoxylakis et al., “Characterization and development of a peptide (p160) with affinity for neuroblastoma cells,” J. Nucl. Med., vol. 47, no. 6, pp. 981–988, 2006.
- P. Łukasik, M. Załuski, and I. Gutowska, “Cyclin-dependent kinases (Cdk) and their role in diseases development–review,” Int. J. Mol. Sci., vol. 22, no. 6, pp. 1–33, 2021. [CrossRef]
- T. Ozaki and A. Nakagawara, “Role of p53 in cell death and human cancers,” Cancers (Basel)., vol. 3, no. 1, pp. 994–1013, 2011. [CrossRef]
- P. Wee and Z. Wang, “Epidermal growth factor receptor cell proliferation signaling pathways,” Cancers (Basel)., vol. 9, no. 5, pp. 1–45, 2017. [CrossRef]
- I. A. Voutsadakis, “Proteasome expression and activity in cancer and cancer stem cells,” Tumor Biol., vol. 39, no. 3, pp. 1–17, 2017. [CrossRef]
- M.U. Johansson, V. Zoete, O. Michielin, and N. Guex, “Defining and searching for structural motifs using DeepView/Swiss-PdbViewer,” BMC Bioinformatics, vol. 13, no. 1, 2012. [CrossRef]
- N.M. O’Boyle, M. Banck, C. A. James, C. Morley, T. Vandermeersch, and G. R. Hutchison, “Open Babel,” J. Cheminform., vol. 3, no. 33, pp. 1–14, 2011.
- R. López, “Capillary surfaces with free boundary in a wedge,” Adv. Math. (N. Y)., vol. 262, pp. 476–483, 2014. [CrossRef]
- J.Eberhardt, D. Santos-Martins, A. F. Tillack, and S. Forli, “AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings,” J. Chem. Inf. Model., vol. 61, no. 8, pp. 3891–3898, Aug. 2021.
- P. Zhou, B. Jin, H. Li, and S. Y. Huang, “HPEPDOCK: A web server for blind peptide-protein docking based on a hierarchical algorithm,” Nucleic Acids Res., vol. 46, no. W1, pp. W443–W450, 2018. [CrossRef]
- H. A. Sawal, S. Nighat, T. Safdar, and L. Anees, “Comparative In Silico Analysis and Functional Characterization of TANK-Binding Kinase 1–Binding Protein 1,” Bioinform. Biol. Insights, vol. 17, pp. 1–5, 2023. [CrossRef]
- H. Y. K. Yip and A. Papa, “Signaling pathways in cancer: Therapeutic targets, combinatorial treatments, and new developments,” Cells, vol. 10, no. 3, pp. 1–30, 2021. [CrossRef]
- R. A. Bottens and T. Yamada, “Cell-Penetrating Peptides (CPPs) as Therapeutic and Diagnostic Agents for Cancer,” Cancers (Basel)., vol. 14, no. 22, pp. 1–17, 2022. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).