Submitted:
14 October 2024
Posted:
16 October 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Fractal Analysis of Vertebrate Respiratory Systems
- Quantify the fractal dimensions of respiratory systems across diverse vertebrate species.
- Analyze how these fractal properties correlate with metabolic rates, body size, and environmental adaptations.
- Explore the potential of fractal analysis as a tool for understanding evolutionary trends in respiratory system development.
- Investigate the implications of these findings for biomimetic design and medical applications.
2. Methodology
2.1. Mathematics
2.1.1. Fractal Dimension Calculation
- is the number of boxes of side length required to cover the fractal,
- is the box size.
- is the number of self-similar pieces,
- is the scale factor.
2.1.2. L-System Modeling
- represents a branch,
- [and ] denote the start and end of a branch,
2.1.3. Branch Coordinate Calculation
- are the coordinates of the branch’s starting point,
- is the angle of the branch,
- is the initial branch length,
- is the scale factor,
- is the depth (iteration number).
2.1.4. Branching Angle Calculation
- is the angle of the parent branch,
- is the species-specific branching angle.
2.1.5. Scale Factor Application
- is the length of the parent branch,
- is the species-specific scale factor.
2.1.6. Surface Area to Volume Ratio Analysis
- is the surface area to volume ratio,
- is the fractal dimension.
2.1.7. Metabolic Rate Correlation
2.1.8. Lacunarity Analysis
- is the lacunarity at box size ,
- is the number of occupied sites in a box of size at location ,
- is the total number of boxes.
2.1.9. Fractal Iteration Depth
- is the smallest desired branch size,
- is the scale factor.
2.2. Implementation and Analysis
3. Results
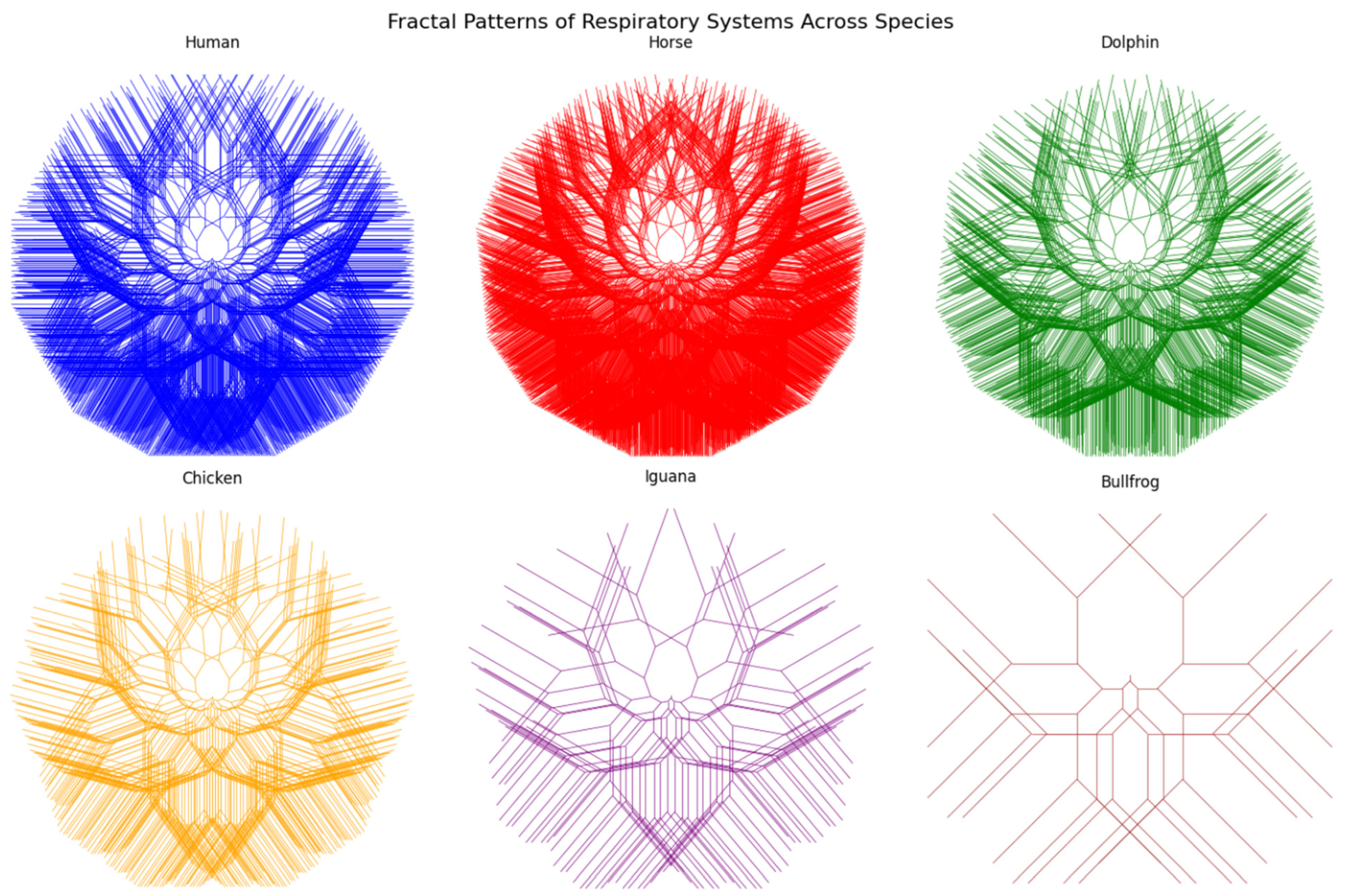
3.1. Overview
3.2. Human
3.3. Horse
3.4. Dolphin
3.5. Chicken
3.6. Iguana
3.7. Bullfrog
3.8. Summary
4. Discussion
4.1. Gradient of Complexity in Respiratory Structures
4.2. Correlation with Metabolic Rates and Body Size
4.3. Environmental Adaptations and Respiratory Efficiency
4.4. Evolutionary Implications
4.5. Methodological Insights and Limitations
4.6. Implications for Biomimetic Design and Medicine
4.7. Future Directions
5. Conclusion
- The identification of a complexity gradient in respiratory structures, from the highly intricate lungs of mammals to the simpler structures of amphibians, reflecting a spectrum of metabolic needs and environmental adaptations.
- The demonstration of how fractal dimension correlates with metabolic rates, providing a mathematical basis for understanding the relationship between form and function in respiratory systems.
- Insights into unique adaptations, such as the efficient avian respiratory system and the specialized lungs of marine mammals, showcasing how fractal analysis can reveal functional adaptations to diverse environments.
- A new perspective on the evolution of respiratory systems, suggesting that increasing fractal complexity has been a key factor in enabling the physiological innovations observed across vertebrate taxa.
- The development of a robust methodological framework combining fractal dimension calculations, L-system modeling, and lacunarity analysis, which can be applied to other branching biological structures.
6. Atachment
-
Python Code:import numpy as npimport matplotlib.pyplot as pltfrom matplotlib.collections import LineCollection
-
def create_fractal_tree(n, angle, scale, initial_length=1.0):def generate_tree(x, y, angle, depth):if depth == 0:return []
-
nx = x + np.cos(angle) * initial_length * scale**depthny = y + np.sin(angle) * initial_length * scale**depth
-
left_branch = generate_tree(nx, ny, angle—np.radians(branching_angle), depth—1)right_branch = generate_tree(nx, ny, angle + np.radians(branching_angle), depth—1)
- return [[(x, y), (nx, ny)]] + left_branch + right_branch
- return generate_tree(0, 0, -np.pi/2, n)
-
def plot_fractal_tree(ax, tree, color):lines = LineCollection(tree, colors=color, linewidths=0.5)ax.add_collection(lines)ax.autoscale()ax.set_aspect(‘equal’)
-
# Species parametersspecies = {‘Human’: {‘iterations’: 12, ‘angle’: 30, ‘scale’: 0.7, ‘color’: ‘blue’},‘Horse’: {‘iterations’: 13, ‘angle’: 28, ‘scale’: 0.72, ‘color’: ‘red’},‘Dolphin’: {‘iterations’: 11, ‘angle’: 32, ‘scale’: 0.68, ‘color’: ‘green’},‘Chicken’: {‘iterations’: 10, ‘angle’: 35, ‘scale’: 0.65, ‘color’: ‘orange’},‘Iguana’: {‘iterations’: 8, ‘angle’: 40, ‘scale’: 0.6, ‘color’: ‘purple’},‘Bullfrog’: {‘iterations’: 6, ‘angle’: 45, ‘scale’: 0.55, ‘color’: ‘brown’}}
-
# Create plotfig, axs = plt.subplots(2, 3, figsize=(15, 10))fig.suptitle(‘Fractal Patterns of Respiratory Systems Across Species’, fontsize=16)
-
for (species_name, params), ax in zip(species.items(), axs.flatten()):branching_angle = params[‘angle’]tree = create_fractal_tree(params[‘iterations’], params[‘angle’], params[‘scale’])plot_fractal_tree(ax, tree, params[‘color’])ax.set_title(species_name)ax.axis(‘off’)
-
plt.tight_layout()plt.show()
Conflicts of Interest
References
- Alencar, A. M., Arold, S. P., Buldyrev, S. V., Majumdar, A., Stamenović, D., Stanley, H. E., & Suki, B. (2003). Physiology: Dynamic instabilities in the inflated lung. Nature, 417(6891), 809-811. [CrossRef]
- Altemeier, W. A., McKinney, S., & Glenny, R. W. (2000). Fractal nature of regional ventilation distribution. Journal of Applied Physiology, 88(5), 1551-1557. [CrossRef]
- 3 Boser, S. R., Park, H., Perry, S. F., Menache, M. G., & Green, F. H. (2005). Fractal geometry of airway remodeling in human asthma. American Journal of Respiratory and Critical Care Medicine, 172(7), 817-823. [CrossRef]
- Farmer, C. G. (1997). Did lungs and the intracardiac shunt evolve to oxygenate the heart in vertebrates? Paleobiology, 23(3), 358-372.
- Feder, M. E., & Burggren, W. W. (1985). Cutaneous gas exchange in vertebrates: design, patterns, control and implications. Biological Reviews, 60(1), 1-45. [CrossRef]
- Glenny, R. W. (2011). Emergence of matched airway and vascular trees from fractal rules. Journal of Applied Physiology, 110(4), 1119-1129. [CrossRef]
- Goldberger, A. L., Rigney, D. R., & West, B. J. (1990). Chaos and fractals in human physiology. Scientific American, 262(2), 42-49. [CrossRef]
- Goniakowska-Witalińska, L. (1978). Ultrastructural and morphometric studies of the lung of the European salamander, Salamandra salamandra L. Cell and Tissue Research, 191(2), 343-356. [CrossRef]
- Hicks, J. W., & Wang, T. (1999). Hypoxic hypometabolism in the anesthetized turtle, Trachemys scripta. American Journal of Physiology-Regulatory, Integrative and Comparative Physiology, 277(1), R18-R23. [CrossRef]
- Horsfield, K. (1990). Diameters, generations, and orders of branches in the bronchial tree. Journal of Applied Physiology, 68(2), 457-461. [CrossRef]
- Maina, J. N. (2000). What it takes to fly: the structural and functional respiratory refinements in birds and bats. Journal of Experimental Biology, 203(20), 3045-3064. [CrossRef]
- Mandelbrot, B. B. (1982). The fractal geometry of nature. W.H. Freeman and Company.
- Marlin, D., & Nankervis, K. (2002). Equine exercise physiology. John Wiley & Sons.
- Pabst, D. A., Rommel, S. A., & McLellan, W. A. (1999). The functional morphology of marine mammals. In Biology of marine mammals (pp. 15-72). Smithsonian Institution Press.
- Perry, S. F. (1998). Lungs: comparative anatomy, functional morphology, and evolution. In Biology of the Reptilia (Vol. 19, pp. 1-92). Academic Press.
- Piscitelli, M. A., McLellan, W. A., Rommel, S. A., Blum, J. E., Barco, S. G., & Pabst, D. A. (2013). Lung size and thoracic morphology in shallow-and deep-diving cetaceans. Journal of Morphology, 274(10), 1066-1079. [CrossRef]
- Prusinkiewicz, P., & Lindenmayer, A. (1990). The algorithmic beauty of plants. Springer-Verlag.
- Weibel, E. R. (1991). Fractal geometry: a design principle for living organisms. American Journal of Physiology-Lung Cellular and Molecular Physiology, 261(6), L361-L369. [CrossRef]
- Weibel, E. R., & Gomez, D. M. (1962). Architecture of the human lung: use of quantitative methods establishes fundamental relations between size and number of lung structures. Science, 137(3530), 577-585. [CrossRef]
- West, G. B., Brown, J. H., & Enquist, B. J. (1997). A general model for the origin of allometric scaling laws in biology. Science, 276(5309), 122-126. [CrossRef]
- West, G. B., Brown, J. H., & Enquist, B. J. (1999). The fourth dimension of life: fractal geometry and allometric scaling of organisms. Science, 284(5420), 1677-1679. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



