Submitted:
03 October 2024
Posted:
04 October 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Materials and Methods
Results
Discussion
Data Availability Statement
Acknowledgments
References
- Dos Santos, N. B. et al. Thimet oligopeptidase (EC 3.4.24.15) key functions suggested by knockout mice phenotype characterization. Biomolecules 9, 382 (2019).
- Fontenele-Neto, J. D., Massarelli, E. E., Garrido, P. A. G., Beaudet, A. & Ferro, E. S. Comparative fine structural distribution of endopeptidase 24.15 (EC3.4.24.15) and 24.16 (EC3.4.24.16) in rat brain. J. Comp. Neurol. 438, 399–410 (2001).
- Ferro, E. S. et al. Secretion of a neuropeptide-metabolizing enzyme similar to endopeptidase 22.19 by glioma C6 cells. Biochem. Biophys. Res. Commun. 191, 275–281 (1993). [CrossRef]
- Carreño, F. R., Goñi, C. N., Castro, L. M. & Ferro, E. S. 14-3-3 epsilon modulates the stimulated secretion of endopeptidase 24.15. J. Neurochem. 93, 10–25 (2005).
- Garrido, P. A. G. et al. Confocal Microscopy Reveals Thimet Oligopeptidase (EC 3.4.24.15) and Neurolysin (EC 3.4.24.16) in the Classical Secretory Pathway. DNA Cell Biol. 18, 323–331 (1999). [CrossRef]
- Ferro, E. S., Tullai, J. W., Glucksman, M. J. & Roberts, J. L. Secretion of Metalloendopeptidase 24.15 (EC 3.4.24.15). DNA Cell Biol. 18, 781–789 (1999). [CrossRef]
- Ferro, E. S., Gewehr, M. C. F. & Navon, A. Thimet oligopeptidase biochemical and biological significances: Past, present, and future directions. Biomolecules 10, 1–30 (2020).
- Ray, K., Hines, C. S., Coll-Rodriguez, J. & Rodgers, D. W. Crystal Structure of Human Thimet Oligopeptidase Provides Insight into Substrate Recognition, Regulation, and Localization. J. Biol. Chem. 279, 20480–20489 (2004).
- Healy, D. P. & Orlowski, M. Immunocytochemical localization of endopeptidase 24.15 in rat brain. Brain Res. 571, 121–128 (1992). [CrossRef]
- Rioli, V. et al. Neuropeptide specificity and inhibition of recombinant isoforms of the endopeptidase 3.4.24.16 family: Comparison with the related recombinant endopeptidase 3.4.24.15. Biochem. Biophys. Res. Commun. 250, 5–11 (1998).
- Acker, G. R., Molineaux, C. & Orlowski, M. Synaptosomal Membrane-Bound Form of Endopeptidase-24.15 Generates Leu-Enkephalin from Dynorphin 1-8, α- and β-Neoendorphin, and Met-Enkephalin from Met-Enkephalin-Arg6-Gly7-Leu. J. Neurochem. 48, 284–292 (1987).
- Molineaux, C. J., Lasdun, A., Michaud, C. & Orlowski, M. Endopeptidase-24.15 Is the Primary Enzyme that Degrades Luteinizing Hormone Releasing Hormone Both In Vitro and In Vivo. J. Neurochem. 51, 624–633 (1988).
- Visniauskas, B. et al. Sleep deprivation changes thimet oligopeptidase (THOP1) expression and activity in rat brain. Heliyon 5, (2019).
- Thompson, A., Grueninger-Leitch, F., Huber, G. & Malherbe, P. Expression and characterization of human β-secretase candidates metalloendopeptidase MP78 and cathepsin D in βAPP-overexpressing cells. Mol. Brain Res. 48, 206–214 (1997).
- McDermott, J. R., Biggins, J. A. & Gibson, A. M. Human brain peptidase activity with the specificity to generate the N-terminus of the Alzheimer β-amyloid protein from its precursor. Biochem. Biophys. Res. Commun. 185, 746–752 (1992).
- Papastoitsis, G., Abraham, C. R., Siman, R. & Scott, R. Identification of a Metalloprotease from Alzheimer’s Disease Brain Able To Degrade the β-Amyloid Precursor Protein and Generate Amyloidogenic Fragments. Biochemistry 33, 192–199 (1994).
- Yamin, R., Malgeri, E. G., Sloane, J. A., McGraw, W. T. & Abraham, C. R. Metalloendopeptidase EC 3.4.24.15 is necessary for Alzheimer’s amyloid- β peptide degradation. J. Biol. Chem. 274, 18777–18784 (1999).
- Meckelein, B. et al. Human endopeptidase (THOP1) is localized on chromosome 19 within the linkage region for the late-onset Alzheimer disease AD2 locus. Genomics 31, 246–249 (1996).
- Torres, M. P., Prange, C. & Lennon, G. Human endopeptidase 24.15 (THOP1) is localized on chromosome 19p13.3 and is excluded from the linkage region for late-onset Alzheimer disease. Genomics 53, 239–240 (1998).
- Pollio, G. et al. Increased expression of the oligopeptidase THOP1 is a neuroprotective response to Aβ toxicity. Neurobiol. Dis. 31, 145–158 (2008).
- Shi, Y. et al. Transcriptomic Analyses for Identification and Prioritization of Genes Associated With Alzheimer’s Disease in Humans. Front. Bioeng. Biotechnol. 8, 504172 (2020).
- del Campo, M. et al. CSF proteome profiling across the Alzheimer’s disease spectrum reflects the multifactorial nature of the disease and identifies specific biomarker panels. Nat. Aging 2, 1040–1053 (2022).
- del Campo, M. et al. CSF proteome profiling reveals biomarkers to discriminate dementia with Lewy bodies from Alzheimer´s disease. Nat. Commun. 14, 1–14 (2023).
- Hok-A-Hin, Y. S. et al. Thimet oligopeptidase as a potential CSF biomarker for Alzheimer’s disease: A cross-platform validation study. Alzheimer’s Dement. Diagnosis, Assess. Dis. Monit. 15, e12456 (2023).
- Hok-A-Hin, Y. S. et al. Thimet Oligopeptidase is a potential CSF biomarker for Alzheimer’s Disease. Alzheimer’s Dement. 18, e065528 (2022).
- Climer, S., Templeton, A. R. A. R. & Zhang, W. Allele-specific network reveals combinatorial interaction that transcends small effects in psoriasis GWAS. PLoS Comput. Biol. 10, e1003766 (2014).
- Climer, S., Yang, W., de las Fuentes, L., Dávila-Román, V. G. V. G. & Gu, C. C. C. A custom correlation coefficient (CCC) approach for fast identification of multi-SNP association patterns in genome-wide SNPs data. Genet. Epidemiol. 38, 610–621 (2014).
- Climer, S. Connecting the dots: The boons and banes of network modeling. Patterns 2, 100374 (2021).
- Altshuler, D. M. et al. Integrating common and rare genetic variation in diverse human populations. Nature 467, 52–8 (2010).
- Climer, S., Templeton, A. R. A. R. & Zhang, W. Human gephyrin is encompassed within giant functional noncoding yin-yang sequences. Nat. Commun. 6, (2015).
- Bogdanovic, M. et al. The ZIP3 Zinc Transporter Is Localized to Mossy Fiber Terminals and Is Required for Kainate-Induced Degeneration of CA3 Neurons. J. Neurosci. 42, 2824–2834 (2022).
- Qian, J. et al. Knockout of Zn transporters Zip-1 and Zip-3 attenuates seizure-induced CA1 neurodegeneration. J. Neurosci. 31, 97–104 (2011).
- Philp, L. K. et al. SGTA: A New Player in the Molecular Co-Chaperone Game. Hormones and Cancer 4, 343–357 (2013).
- Benarroch, R., Austin, J. M., Ahmed, F. & Isaacson, R. L. The roles of cytosolic quality control proteins, SGTA and the BAG6 complex, in disease. in Advances in Protein Chemistry and Structural Biology 114, 265–313 (Academic Press Inc., 2019).
- Kubota, S. et al. SGTA associates with intracellular aggregates in neurodegenerative diseases. Mol. Brain 14, 1–10 (2021).
- Winnefeld, M., Rommelaere, J. & Cziepluch, C. The human small glutamine-rich TPR-containing protein is required for progress through cell division. Exp. Cell Res. 293, 43–57 (2004).
- Rothhammer-Hampl, T. et al. Frequent Epigenetic Inactivation of DIRAS-1 and DIRAS-2 Contributes to Chemo-Resistance in Gliomas. Cancers (Basel). 13, 5113 (2021).
- Sommerer, Y. et al. Entorhinal cortex epigenome-wide association study highlights four novel loci showing differential methylation in Alzheimer’s disease. Alzheimer’s Res. Ther. 15, 1–13 (2023).
- Oliveira, V. et al. Calcium modulates endopeptidase 24.15 (EC 3.4.24.15) membrane association, secondary structure and substrate specificity. FEBS J. 272, 2978–2992 (2005).
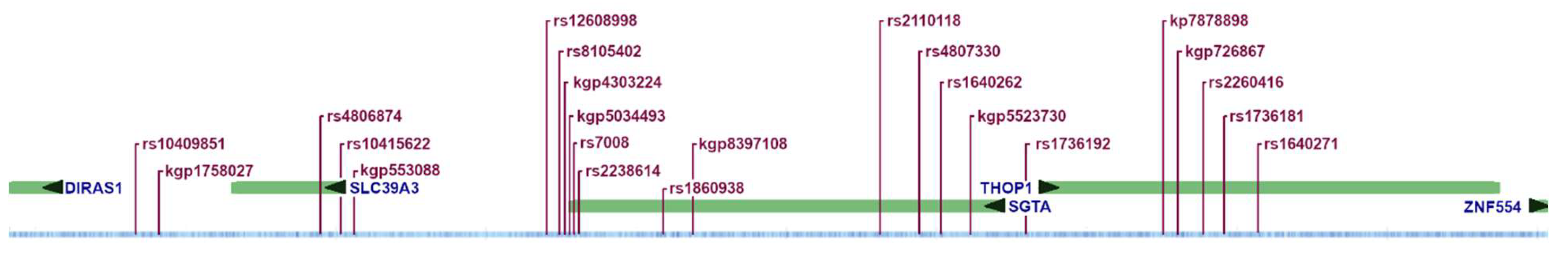
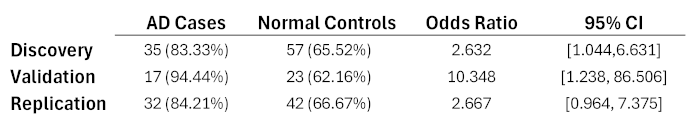 |
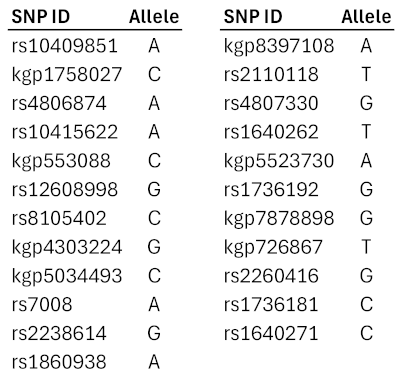 |
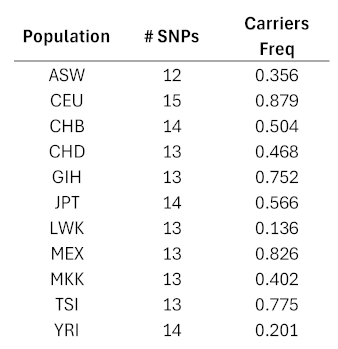 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





