Submitted:
27 September 2024
Posted:
30 September 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction: Who Are You, Mr. IDP?
2. Roles of Intrinsic Disorder in Origin of Life
2.1. Prebiotic Life on the Earth: Intrinsic Disorder of the Extraterrestrial Peptides
2.2. Prebiotic Life on the Earth: Intrinsic Disorder of the Primordial Proteins
3. Roles of Intrinsic Disorder in Evolution
3.1. Wavy Evolution of Intrinsic Disorder: Back to the Future or Blast from the Past
3.2. Intrinsic Disorder and LLPS: From Prebiotic Life to Origin of Cellular Life and Evolution
3.3. Intrinsic Disorder in Nucleic Acid-Binding Proteins
4. Intrinsic Disorder as Means for Increasing the Proteome Complexity
4.1. Alternative Splicing
4.2. Posttranslational Modifications
4.3. Intrinsic Disorder, Structural Heterogeneity, Multifunctionality, and Binding Promiscuity
5. Protein Intrinsic Disorder and Evolution of Multicellularity
5.1. Intrinsic Disorder and Proteoforms
- The agreement between phylogeny and biogeography;
- The correspondence between phylogeny and the paleontological record;
- The existence of numerous predicted transitional fossils;
- The hierarchical classification of morphological characteristics;
- The marked similarities of biological structures with different functions (that is, homologies); and
- The congruence of morphological and molecular phylogenies.
5.2. Casual Emergence
- The constituents of a complex system are interdependent;
- A complex system possesses a structure spanning several scales and may be nested; i.e., the components of a complex system may themselves be complex systems;
- A complex system is capable of emergent behavior, which is unanticipated behavior shown by the system, for example the arising of novel and coherent structures, patterns and properties during the process of self-organization;
- Complexity involves an interplay between chaos (disorder) and order;
- Complexity involves an interplay between cooperation and competition, and complex systems contain both positive (amplifying) and negative (damping) feedbacks;
- Complex systems may have a memory. In other words, the history of a complex system may be important, since due to their dynamic nature, complex systems change over time, and prior states may have an influence on present states (for example, no two genetically identical mice or even two single cells that share the exact same DNA sequence are absolutely identical because of environmental influences, random variations in gene expression, and epigenetic modifications).
5.3. Intrinsic Disorder, Noise/Stochasticity of Transcriptional Regulation, and Development
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fischer, E. Einfluss der configuration auf die wirkung der enzyme. Ber. Dt. Chem. Ges. 1894, 27, 2985-2993.
- Uversky, V.N. Natively unfolded proteins: a point where biology waits for physics. Protein Sci 2002, 11, 739-756. [CrossRef]
- Uversky, V.N. A decade and a half of protein intrinsic disorder: biology still waits for physics. Protein Sci 2013, 22, 693-724. [CrossRef]
- Petsko, G.A.; Ringe, D. Primers in Biology. Protein Structure and Function.; New Science Press Ltd., Sinauer Associates, Inc. Publishers, Blackwell Publishing: London, 2004.
- Uversky, V.N.; Dunker, A.K. Understanding protein non-folding. Biochim Biophys Acta 2010, 1804, 1231-1264. [CrossRef]
- Bernstein, F.C.; Koetzle, T.F.; Williams, G.J.; Meyer, E.F., Jr.; Brice, M.D.; Rodgers, J.R.; Kennard, O.; Shimanouchi, T.; Tasumi, M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol 1977, 112, 535-542.
- Bloomer, A.C.; Champness, J.N.; Bricogne, G.; Staden, R.; Klug, A. Protein disk of tobacco mosaic virus at 2.8 A resolution showing the interactions within and between subunits. Nature 1978, 276, 362-368.
- Bode, W.; Schwager, P.; Huber, R. The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding. The refined crystal structures of the bovine trypsinogen-pancreatic trypsin inhibitor complex and of its ternary complex with Ile-Val at 1.9 A resolution. J Mol Biol 1978, 118, 99-112.
- Le Gall, T.; Romero, P.R.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn 2007, 24, 325-342. [CrossRef]
- Dunker, A.K.; Garner, E.; Guilliot, S.; Romero, P.; Albrecht, K.; Hart, J.; Obradovic, Z.; Kissinger, C.; Villafranca, J.E. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput 1998, 473-484.
- Wright, P.E.; Dyson, H.J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 1999, 293, 321-331.
- Uversky, V.N.; Gillespie, J.R.; Fink, A.L. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins 2000, 41, 415-427.
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J Mol Graph Model 2001, 19, 26-59.
- Tompa, P. Intrinsically unstructured proteins. Trends Biochem Sci 2002, 27, 527-533.
- Daughdrill, G.W.; Pielak, G.J.; Uversky, V.N.; Cortese, M.S.; Dunker, A.K. Natively disordered proteins. In Handbook of Protein Folding, Buchner, J., Kiefhaber, T., Eds.; Wiley-VCH, Verlag GmbH & Co. KGaA: Weinheim, Germany, 2005; pp. 271-353.
- Dunker, A.K.; Babu, M.M.; Barbar, E.; Blackledge, M.; Bondos, S.E.; Dosztányi, Z.; Dyson, H.J.; Forman-Kay, J.; Fuxreiter, M.; Gsponer, J.; et al. What’s in a name? Why these proteins are intrinsically disordered. Intrinsically Disordered Proteins 2013, 1, e24157.
- Dunker, A.K.; Obradovic, Z.; Romero, P.; Garner, E.C.; Brown, C.J. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform 2000, 11, 161-171.
- Ward, J.J.; Sodhi, J.S.; McGuffin, L.J.; Buxton, B.F.; Jones, D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 2004, 337, 635-645. [CrossRef]
- Uversky, V.N. The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J Biomed Biotechnol 2010, 2010, 568068. [CrossRef]
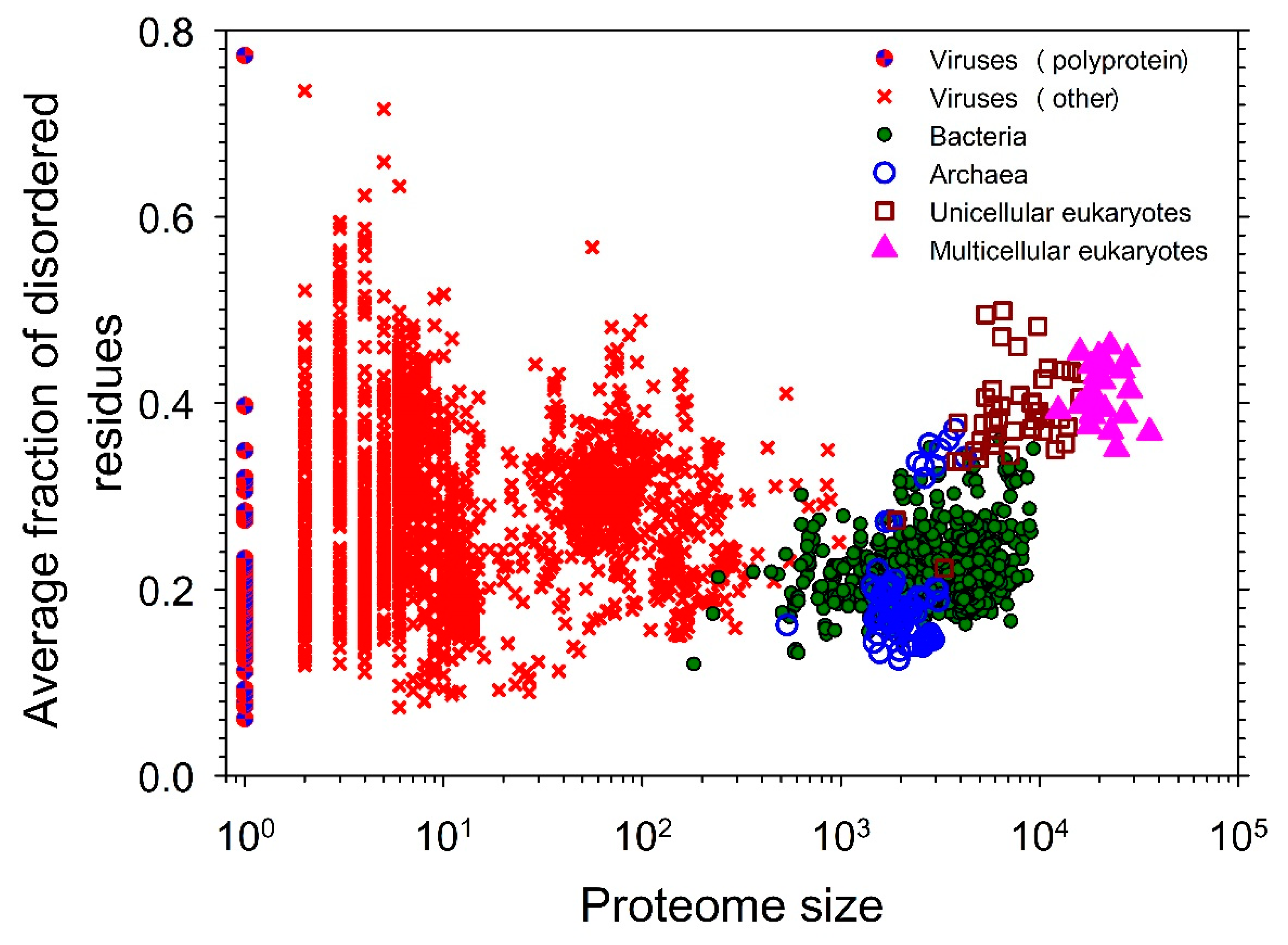
- Xue, B.; Dunker, A.K.; Uversky, V.N. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn 2012, 30, 137-149. [CrossRef]
- Wathen, B.; Jia, Z. Folding by numbers: primary sequence statistics and their use in studying protein folding. Int J Mol Sci 2009, 10, 1567-1589. [CrossRef]
- Uversky, V.N. Unusual biophysics of intrinsically disordered proteins. Biochim Biophys Acta 2013, 1834 932-951. [CrossRef]
- Campen, A.; Williams, R.M.; Brown, C.J.; Meng, J.; Uversky, V.N.; Dunker, A.K. TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept Lett 2008, 15, 956-963.
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder and functional proteomics. Biophys J 2007, 92, 1439-1456. [CrossRef]
- Romero, P.; Obradovic, Z.; Li, X.; Garner, E.C.; Brown, C.J.; Dunker, A.K. Sequence complexity of disordered protein. Proteins 2001, 42, 38-48.
- Garner, E.; Cannon, P.; Romero, P.; Obradovic, Z.; Dunker, A.K. Predicting Disordered Regions from Amino Acid Sequence: Common Themes Despite Differing Structural Characterization. Genome Inform Ser Workshop Genome Inform 1998, 9, 201-213.
- Williams, R.M.; Obradovi, Z.; Mathura, V.; Braun, W.; Garner, E.C.; Young, J.; Takayama, S.; Brown, C.J.; Dunker, A.K. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput 2001, 89-100.
- Vacic, V.; Uversky, V.N.; Dunker, A.K.; Lonardi, S. Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics 2007, 8, 211. [CrossRef]
- Ferron, F.; Longhi, S.; Canard, B.; Karlin, D. A practical overview of protein disorder prediction methods. Proteins 2006, 65, 1-14. [CrossRef]
- Bourhis, J.M.; Canard, B.; Longhi, S. Predicting protein disorder and induced folding: from theoretical principles to practical applications. Curr Protein Pept Sci 2007, 8, 135-149.
- Dosztanyi, Z.; Sandor, M.; Tompa, P.; Simon, I. Prediction of protein disorder at the domain level. Curr Protein Pept Sci 2007, 8, 161-171.
- Dosztanyi, Z.; Tompa, P. Prediction of protein disorder. Methods Mol Biol 2008, 426, 103-115. [CrossRef]
- He, B.; Wang, K.; Liu, Y.; Xue, B.; Uversky, V.N.; Dunker, A.K. Predicting intrinsic disorder in proteins: an overview. Cell Res 2009, 19, 929-949. [CrossRef]
- Jin, F.; Liu, Z. Inherent Relationships among Different Biophysical Prediction Methods for Intrinsically Disordered Proteins. Biophys J 2013, 104, 488-495. [CrossRef]
- Romero, P.; Obradovic, Z.; Kissinger, C.R.; Villafranca, J.E.; Garner, E.; Guilliot, S.; Dunker, A.K. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput 1998, 437-448.
- Feng, Z.P.; Zhang, X.; Han, P.; Arora, N.; Anders, R.F.; Norton, R.S. Abundance of intrinsically unstructured proteins in P. falciparum and other apicomplexan parasite proteomes. Mol Biochem Parasitol 2006, 150, 256-267. [CrossRef]
- Tompa, P.; Dosztanyi, Z.; Simon, I. Prevalent structural disorder in E. coli and S. cerevisiae proteomes. J Proteome Res 2006, 5, 1996-2000. [CrossRef]
- Galea, C.A.; High, A.A.; Obenauer, J.C.; Mishra, A.; Park, C.G.; Punta, M.; Schlessinger, A.; Ma, J.; Rost, B.; Slaughter, C.A.; et al. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. Journal of proteome research 2009, 8, 211-226. [CrossRef]
- Xue, B.; Williams, R.W.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N. Archaic chaos: intrinsically disordered proteins in Archaea. BMC Syst Biol 2010, 4 Suppl 1, S1. [CrossRef]
- Burra, P.V.; Kalmar, L.; Tompa, P. Reduction in structural disorder and functional complexity in the thermal adaptation of prokaryotes. PLoS One 2010, 5, e12069. [CrossRef]
- Dunker, A.K.; Cortese, M.S.; Romero, P.; Iakoucheva, L.M.; Uversky, V.N. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J 2005, 272, 5129-5148. [CrossRef]
- Uversky, V.N.; Oldfield, C.J.; Dunker, A.K. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit 2005, 18, 343-384. [CrossRef]
- Iakoucheva, L.M.; Brown, C.J.; Lawson, J.D.; Obradovic, Z.; Dunker, A.K. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 2002, 323, 573-584.
- Vucetic, S.; Xie, H.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res 2007, 6, 1899-1916. [CrossRef]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Obradovic, Z.; Uversky, V.N. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res 2007, 6, 1917-1932. [CrossRef]
- Xie, H.; Vucetic, S.; Iakoucheva, L.M.; Oldfield, C.J.; Dunker, A.K.; Uversky, V.N.; Obradovic, Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res 2007, 6, 1882-1898. [CrossRef]
- Wickramasinghe, N.C.; Wickramasinghe, J.; Napier, W. Comets and the Origin of Life; World Scientific: 2009.
- Nakashima, S.; Kebukawa, Y.; Kitadai, N.; Igisu, M.; Matsuoka, N. Geochemistry and the Origin of Life: From Extraterrestrial Processes, Chemical Evolution on Earth, Fossilized Life's Records, to Natures of the Extant Life. Life (Basel) 2018, 8. [CrossRef]
- Rimola, A.; Balucani, N.; Ceccarelli, C.; Ugliengo, P. Tracing the Primordial Chemical Life of Glycine: A Review from Quantum Chemical Simulations. Int J Mol Sci 2022, 23. [CrossRef]
- Irvine, W.M. Extraterrestrial organic matter: a review. Orig Life Evol Biosph 1998, 28, 365-383. [CrossRef]
- Krasnokutski, S.; Chuang, K.-J.; Jäger, C.; Ueberschaar, N.; Henning, T. A pathway to peptides in space through the condensation of atomic carbon. Nature Astronomy 2022, 6, 381-386.
- Kulkarni, P.; Salgia, R.; Uversky, V.N. Intrinsic disorder, extraterrestrial peptides, and prebiotic life on the earth. J Biomol Struct Dyn 2023, 41, 5481-5485. [CrossRef]
- Rivera-Valentin, E.G.; Filiberto, J.; Lynch, K.L.; Mamajanov, I.; Lyons, T.W.; Schulte, M.; Mendez, A. Introduction-First Billion Years: Habitability. Astrobiology 2021, 21, 893-905. [CrossRef]
- Cronin, J.R.; Pizzarello, S. Amino acids in meteorites. Adv Space Res 1983, 3, 5-18. [CrossRef]
- McGeoch, J.E.; McGeoch, M.W. A 4641Da polymer of amino acids in Acfer 086 and Allende meteorites. arXiv preprint arXiv:1707.09080 2017.
- McGeoch, M.; Dikler, S.; McGeoch, J.E. Hemolithin: a meteoritic protein containing iron and lithium. arXiv preprint arXiv:2002.11688 2020.
- Radzicka, A.; Wolfenden, R. Rates of uncatalyzed peptide bond hydrolysis in neutral solution and the transition state affinities of proteases. Journal of the American Chemical Society 1996, 118, 6105-6109.
- Oparin, A.I. The Origin of Life (in Russian); Moscow Worker publisher: Moscow, 1924.
- Haldane, J.B.S. The origin of life. In The Rationalist Annual for the Year 1929, Watts, C.A., Ed.; Watts & Co: London 1929; pp. 3-10.
- Miller, S.L. A production of amino acids under possible primitive earth conditions. Science 1953, 117, 528-529.
- Miller, S.L.; Urey, H.C. Organic compound synthesis on the primitive earth. Science 1959, 130, 245-251.
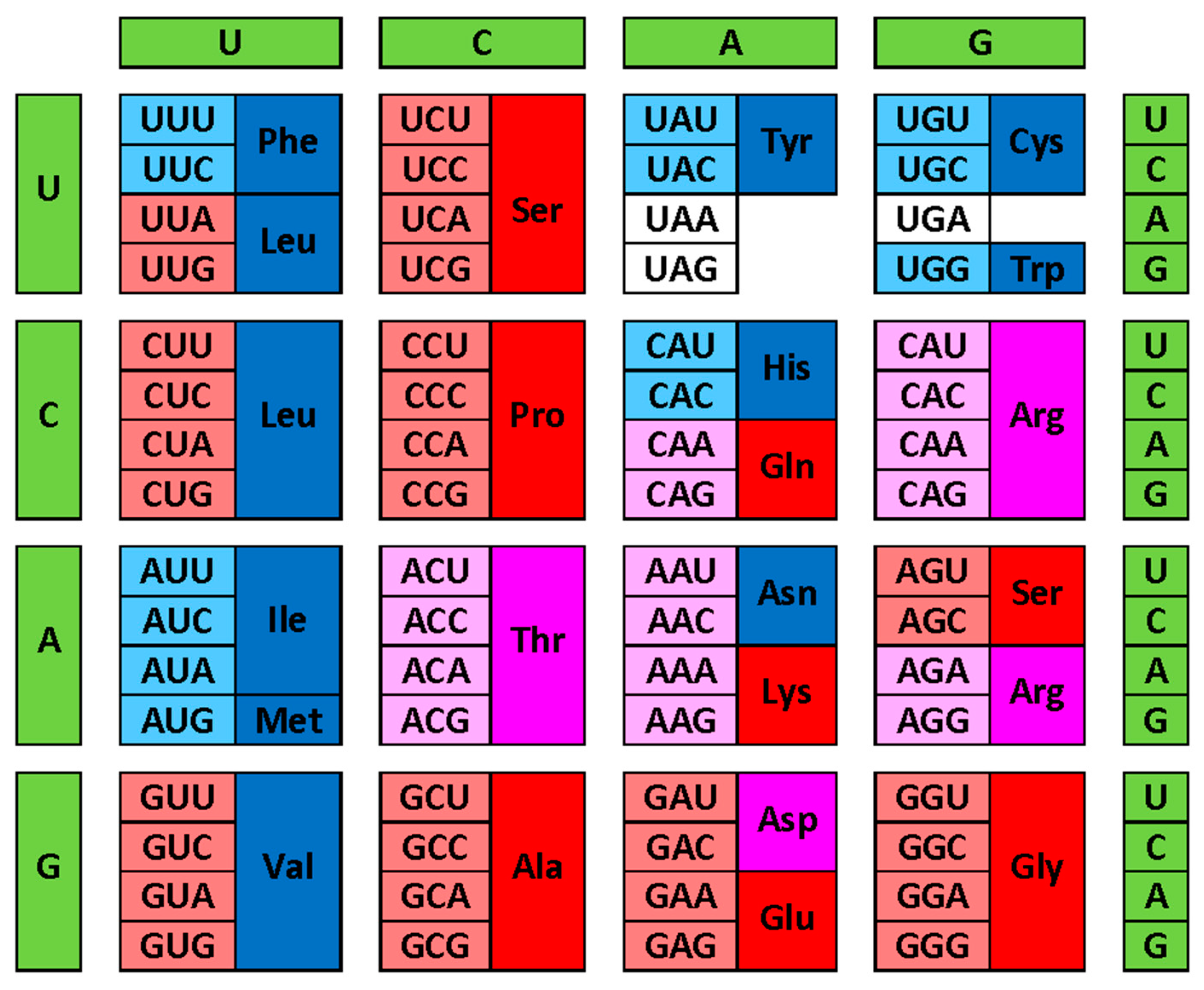
- Crick, F.H. The origin of the genetic code. J Mol Biol 1968, 38, 367-379, doi:0022-2836(68)90392-6 [pii].
- Wong, J.T. A co-evolution theory of the genetic code. Proc Natl Acad Sci U S A 1975, 72, 1909-1912.
- Jukes, T.H. Possibilities for the evolution of the genetic code from a preceding form. Nature 1973, 246, 22-26.
- Trifonov, E.N. Consensus temporal order of amino acids and evolution of the triplet code. Gene 2000, 261, 139-151, doi:S0378-1119(00)00476-5 [pii].
- Sickmeier, M.; Hamilton, J.A.; LeGall, T.; Vacic, V.; Cortese, M.S.; Tantos, A.; Szabo, B.; Tompa, P.; Chen, J.; Uversky, V.N.; et al. DisProt: the Database of Disordered Proteins. Nucleic Acids Res 2007, 35, D786-793. [CrossRef]
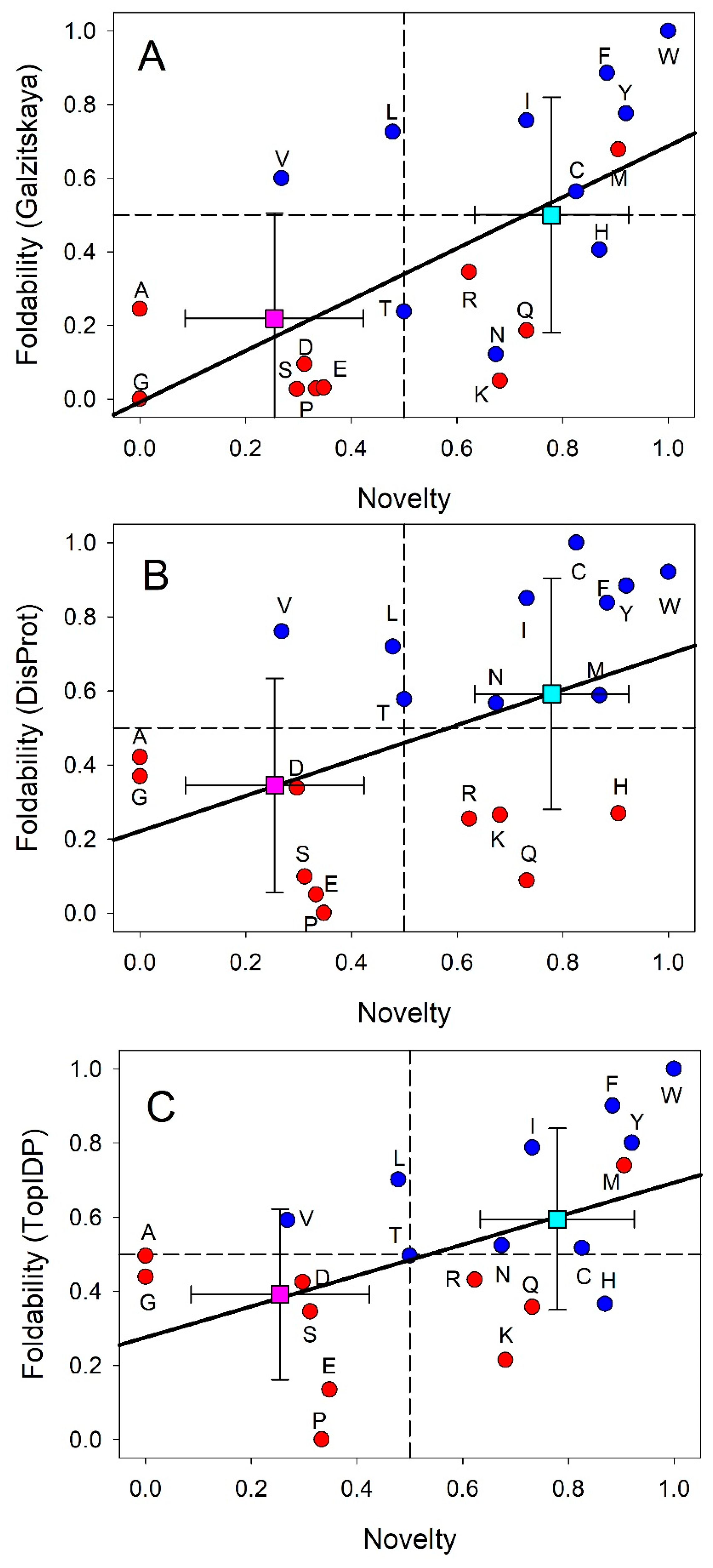
- Garbuzynskiy, S.O.; Lobanov, M.Y.; Galzitskaya, O.V. To be folded or to be unfolded? Protein Sci 2004, 13, 2871-2877. [CrossRef]
- Xia, T.; SantaLucia, J., Jr.; Burkard, M.E.; Kierzek, R.; Schroeder, S.J.; Jiao, X.; Cox, C.; Turner, D.H. Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs. Biochemistry 1998, 37, 14719-14735. [CrossRef]
- Zhou, H.; Zhou, Y. Quantifying the effect of burial of amino acid residues on protein stability. Proteins 2004, 54, 315-322. [CrossRef]
- Poole, A.M.; Jeffares, D.C.; Penny, D. The path from the RNA world. J Mol Evol 1998, 46, 1-17.
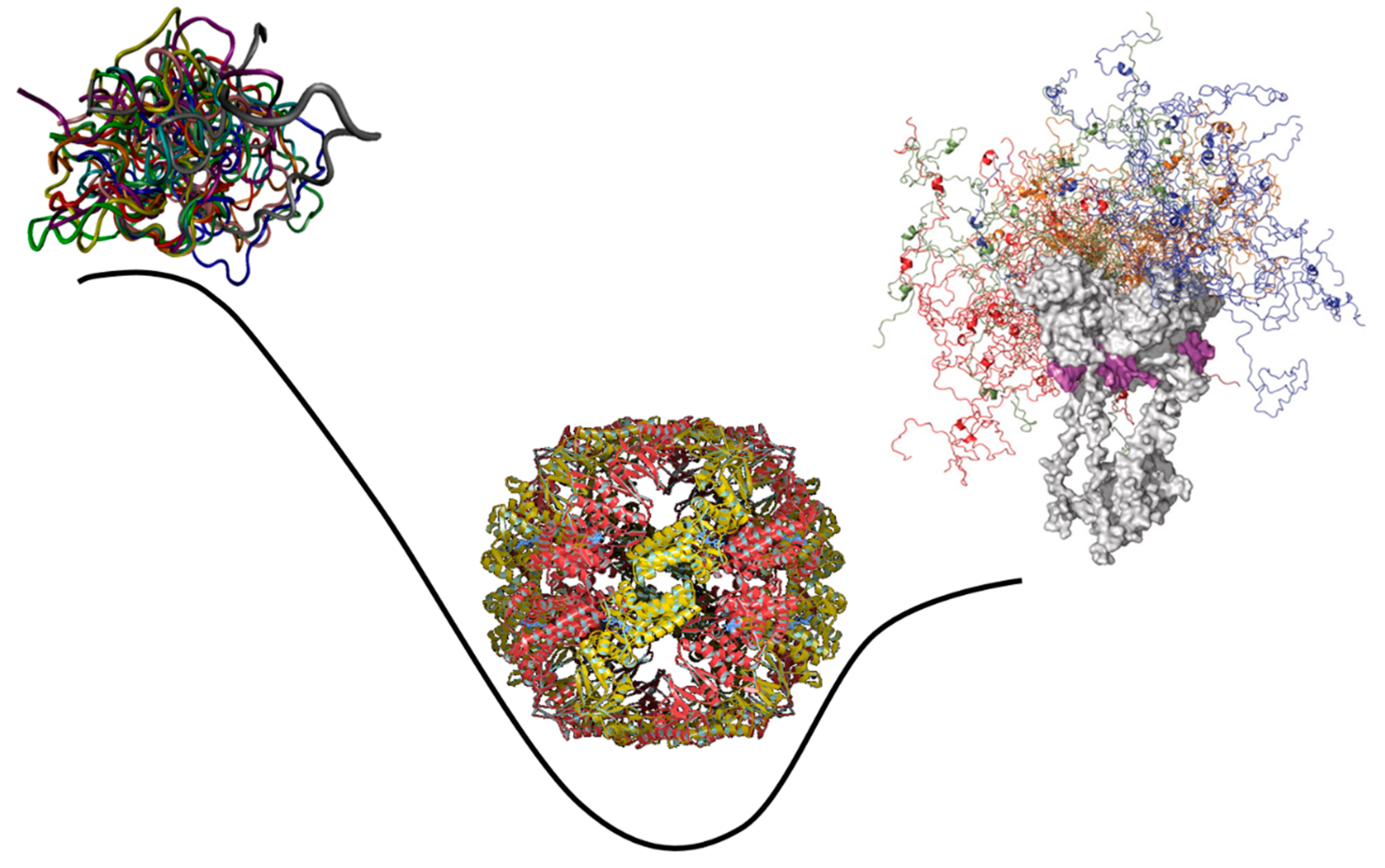
- Peng, Z.; Oldfield, C.J.; Xue, B.; Mizianty, M.J.; Dunker, A.K.; Kurgan, L.; Uversky, V.N. A creature with a hundred waggly tails: intrinsically disordered proteins in the ribosome. Cell Mol Life Sci 2014, 71, 1477-1504. [CrossRef]
- Jeffares, D.C.; Poole, A.M.; Penny, D. Relics from the RNA world. J Mol Evol 1998, 46, 18-36.
- Tompa, P.; Csermely, P. The role of structural disorder in the function of RNA and protein chaperones. Faseb J 2004, 18, 1169-1175. [CrossRef]
- Treiber, D.K.; Williamson, J.R. Beyond kinetic traps in RNA folding. Curr Opin Struct Biol 2001, 11, 309-314, doi:S0959-440X(00)00206-2 [pii].
- Cristofari, G.; Darlix, J.L. The ubiquitous nature of RNA chaperone proteins. Prog Nucleic Acid Res Mol Biol 2002, 72, 223-268.
- Gilbert, W. Origin of life - the RNA world. Nature 1986, 319, 618-618, doi:Doi 10.1038/319618a0.
- Csermely, P. Proteins, RNAs and chaperones in enzyme evolution: A folding perspective. Trends in Biochemical Sciences 1997, 22, 147-149, doi:Doi 10.1016/S0968-0004(97)01026-8.
- Doolittle, W.F. Uprooting the tree of life. Sci Am 2000, 282, 90-95.
- Theobald, D.L. A formal test of the theory of universal common ancestry. Nature 2010, 465, 219-222. [CrossRef]
- Lahav, N.; White, D.; Chang, S. Peptide formation in the prebiotic era: thermal condensation of glycine in fluctuating clay environments. Science 1978, 201, 67-69. [CrossRef]
- Rodriguez-Garcia, M.; Surman, A.J.; Cooper, G.J.T.; Suarez-Marina, I.; Hosni, Z.; Lee, M.P.; Cronin, L. Formation of oligopeptides in high yield under simple programmable conditions. Nat Commun 2015, 6, 8385. [CrossRef]
- Campbell, T.D.; Febrian, R.; McCarthy, J.T.; Kleinschmidt, H.E.; Forsythe, J.G.; Bracher, P.J. Prebiotic condensation through wet-dry cycling regulated by deliquescence. Nat Commun 2019, 10, 4508. [CrossRef]
- Sakata, K.; Kitadai, N.; Yokoyama, T. Effects of pH and temperature on dimerization rate of glycine: evaluation of favorable environmental conditions for chemical evolution of life. Geochimica et Cosmochimica Acta 2010, 74, 6841-6851.
- Imai, E.; Honda, H.; Hatori, K.; Brack, A.; Matsuno, K. Elongation of oligopeptides in a simulated submarine hydrothermal system. Science 1999, 283, 831-833. [CrossRef]
- Ohara, S.; Kakegawa, T.; Nakazawa, H. Pressure effects on the abiotic polymerization of glycine. Orig Life Evol Biosph 2007, 37, 215-223. [CrossRef]
- Muller, F.; Escobar, L.; Xu, F.; Wegrzyn, E.; Nainyte, M.; Amatov, T.; Chan, C.Y.; Pichler, A.; Carell, T. A prebiotically plausible scenario of an RNA-peptide world. Nature 2022, 605, 279-284. [CrossRef]
- Sumie, Y.; Sato, K.; Kakegawa, T.; Furukawa, Y. Boron-assisted abiotic polypeptide synthesis. Commun Chem 2023, 6, 89. [CrossRef]
- Lazcano, A. Historical development of origins research. Cold Spring Harb Perspect Biol 2010, 2, a002089. [CrossRef]
- Lei, L.; Burton, Z.F. Chaos, order and systematics in evolution of the genetic code. 2020.
- Kauffman, S.A. The origins of order: Self-organization and selection in evolution; Oxford University Press, USA: 1993.
- Kacar, B.; Garcia, A.K.; Anbar, A.D. Evolutionary History of Bioessential Elements Can Guide the Search for Life in the Universe. Chembiochem 2021, 22, 114-119. [CrossRef]
- Matveev, V.V. Cell theory, intrinsically disordered proteins, and the physics of the origin of life. Prog Biophys Mol Biol 2019, 149, 114-130. [CrossRef]
- Matsuo, M.; Kurihara, K. Proliferating coacervate droplets as the missing link between chemistry and biology in the origins of life. Nat Commun 2021, 12, 5487. [CrossRef]
- Brangwynne, C.P.; Eckmann, C.R.; Courson, D.S.; Rybarska, A.; Hoege, C.; Gharakhani, J.; Julicher, F.; Hyman, A.A. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009, 324, 1729-1732. [CrossRef]
- Darling, A.L.; Uversky, V.N. Known types of membrane-less organelles and biomolecular condensates. In Droplets of Life: Membrane-Less Organelles, Biomolecular Condensates, and Biological Liquid-Liquid Phase Separation, 1st ed.; Uversky, V.N., Ed.; Elsevier: Amsterdam, Netherlands, 2023; pp. 271-335.
- Brangwynne, C.P. Phase transitions and size scaling of membrane-less organelles. The Journal of cell biology 2013, 203, 875-881. [CrossRef]
- Brangwynne, C.P.; Tompa, P.; Pappu, R.V. Polymer physics of intracellular phase transitions. Nat Physics 2015, 11, 899–904. [CrossRef]
- Uversky, V.N.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B. Intrinsically disordered proteins as crucial constituents of cellular aqueous two phase systems and coacervates. FEBS Lett 2015, 589, 15-22. [CrossRef]
- Dundr, M.; Misteli, T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2010, 2, a000711. [CrossRef]
- Zhu, L.; Brangwynne, C.P. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 2015, 34, 23-30. [CrossRef]
- Uversky, V.N. Intrinsically disordered proteins in overcrowded milieu: Membrane-less organelles, phase separation, and intrinsic disorder. Curr Opin Struct Biol 2017, 44, 18-30. [CrossRef]
- Uversky, V.N. Protein intrinsic disorder-based liquid-liquid phase transitions in biological systems: Complex coacervates and membrane-less organelles. Adv Colloid Interface Sci 2017, 239, 97-114. [CrossRef]
- Feric, M.; Vaidya, N.; Harmon, T.S.; Mitrea, D.M.; Zhu, L.; Richardson, T.M.; Kriwacki, R.W.; Pappu, R.V.; Brangwynne, C.P. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell 2016, 165, 1686-1697. [CrossRef]
- Mitrea, D.M.; Kriwacki, R.W. Phase separation in biology; functional organization of a higher order. Cell Commun Signal 2016, 14, 1. [CrossRef]
- Martin, E.W.; Holehouse, A.S. Intrinsically disordered protein regions and phase separation: sequence determinants of assembly or lack thereof. Emerg Top Life Sci 2020, 4, 307-329. [CrossRef]
- Antifeeva, I.A.; Fonin, A.V.; Fefilova, A.S.; Stepanenko, O.V.; Povarova, O.I.; Silonov, S.A.; Kuznetsova, I.M.; Uversky, V.N.; Turoverov, K.K. Liquid-liquid phase separation as an organizing principle of intracellular space: overview of the evolution of the cell compartmentalization concept. Cell Mol Life Sci 2022, 79, 251. [CrossRef]
- Turoverov, K.K.; Kuznetsova, I.M.; Fonin, A.V.; Darling, A.L.; Zaslavsky, B.Y.; Uversky, V.N. Stochasticity of Biological Soft Matter: Emerging Concepts in Intrinsically Disordered Proteins and Biological Phase Separation. Trends Biochem Sci 2019, 44, 716-728. [CrossRef]
- Darling, A.L.; Liu, Y.; Oldfield, C.J.; Uversky, V.N. Intrinsically Disordered Proteome of Human Membrane-Less Organelles. Proteomics 2018, 18, e1700193. [CrossRef]
- Uversky, V.N. Protein intrinsic disorder and structure-function continuum. Prog Mol Biol Transl Sci 2019, 166, 1-17. [CrossRef]
- Uversky, V.N. Recent Developments in the Field of Intrinsically Disordered Proteins: Intrinsic Disorder–Based Emergence in Cellular Biology in Light of the Physiological and Pathological Liquid–Liquid Phase Transitions. Annual Review of Biophysics 2021, 50, 135-156. [CrossRef]
- Meng, F.; Na, I.; Kurgan, L.; Uversky, V.N. Compartmentalization and Functionality of Nuclear Disorder: Intrinsic Disorder and Protein-Protein Interactions in Intra-Nuclear Compartments. Int J Mol Sci 2015, 17. [CrossRef]
- Uversky, V.N. The roles of intrinsic disorder-based liquid-liquid phase transitions in the "Dr. Jekyll-Mr. Hyde" behavior of proteins involved in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Autophagy 2017, 13, 2115-2162. [CrossRef]
- Brangwynne, C.P.; Mitchison, T.J.; Hyman, A.A. Active liquid-like behavior of nucleoli determines their size and shape in Xenopus laevis oocytes. Proc Natl Acad Sci U S A 2011, 108, 4334-4339. [CrossRef]
- Li, P.; Banjade, S.; Cheng, H.C.; Kim, S.; Chen, B.; Guo, L.; Llaguno, M.; Hollingsworth, J.V.; King, D.S.; Banani, S.F.; et al. Phase transitions in the assembly of multivalent signalling proteins. Nature 2012, 483, 336-340. [CrossRef]
- Aggarwal, S.; Snaidero, N.; Pahler, G.; Frey, S.; Sanchez, P.; Zweckstetter, M.; Janshoff, A.; Schneider, A.; Weil, M.T.; Schaap, I.A.; et al. Myelin membrane assembly is driven by a phase transition of myelin basic proteins into a cohesive protein meshwork. PLoS biology 2013, 11, e1001577. [CrossRef]
- Feric, M.; Brangwynne, C.P. A nuclear F-actin scaffold stabilizes ribonucleoprotein droplets against gravity in large cells. Nature cell biology 2013, 15, 1253-1259. [CrossRef]
- Wippich, F.; Bodenmiller, B.; Trajkovska, M.G.; Wanka, S.; Aebersold, R.; Pelkmans, L. Dual specificity kinase DYRK3 couples stress granule condensation/dissolution to mTORC1 signaling. Cell 2013, 152, 791-805. [CrossRef]
- Nesterov, S.V.; Ilyinsky, N.S.; Uversky, V.N. Liquid-liquid phase separation as a common organizing principle of intracellular space and biomembranes providing dynamic adaptive responses. Biochim Biophys Acta Mol Cell Res 2021, 1868, 119102. [CrossRef]
- Fonin, A.V.; Antifeeva, I.A.; Kuznetsova, I.M.; Turoverov, K.K.; Zaslavsky, B.Y.; Kulkarni, P.; Uversky, V.N. Biological soft matter: intrinsically disordered proteins in liquid-liquid phase separation and biomolecular condensates. Essays Biochem 2022, 66, 831-847. [CrossRef]
- Nott, T.J.; Petsalaki, E.; Farber, P.; Jervis, D.; Fussner, E.; Plochowietz, A.; Craggs, T.D.; Bazett-Jones, D.P.; Pawson, T.; Forman-Kay, J.D.; et al. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell 2015, 57, 936-947. [CrossRef]
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.B.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. Elife 2016, 5. [CrossRef]
- Elbaum-Garfinkle, S.; Kim, Y.; Szczepaniak, K.; Chen, C.C.; Eckmann, C.R.; Myong, S.; Brangwynne, C.P. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc Natl Acad Sci U S A 2015, 112, 7189-7194. [CrossRef]
- Lin, Y.; Protter, D.S.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 2015, 60, 208-219. [CrossRef]
- Toretsky, J.A.; Wright, P.E. Assemblages: functional units formed by cellular phase separation. The Journal of cell biology 2014, 206, 579-588. [CrossRef]
- Csizmok, V.; Follis, A.V.; Kriwacki, R.W.; Forman-Kay, J.D. Dynamic Protein Interaction Networks and New Structural Paradigms in Signaling. Chem Rev 2016. [CrossRef]
- Rigau, M.; Juan, D.; Valencia, A.; Rico, D. Intronic CNVs and gene expression variation in human populations. PLoS Genet 2019, 15, e1007902. [CrossRef]
- Berget, S.M.; Moore, C.; Sharp, P.A. Spliced segments at the 5' terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci U S A 1977, 74, 3171-3175. [CrossRef]
- Chow, L.T.; Gelinas, R.E.; Broker, T.R.; Roberts, R.J. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell 1977, 12, 1-8. [CrossRef]
- Williamson, B. DNA insertions and gene structure. Nature 1977, 270, 295-297.
- Eddy, S.R. The C-value paradox, junk DNA and ENCODE. Curr Biol 2012, 22, R898-899. [CrossRef]
- Palazzo, A.F.; Gregory, T.R. The case for junk DNA. PLoS Genet 2014, 10, e1004351. [CrossRef]
- Consortium, T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57-74. [CrossRef]
- Taft, R.J.; Pheasant, M.; Mattick, J.S. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 2007, 29, 288-299. [CrossRef]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat Rev Genet 2014, 15, 423-437. [CrossRef]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat Rev Genet 2014, 15, 829-845. [CrossRef]
- Van Nostrand, E.L.; Freese, P.; Pratt, G.A.; Wang, X.; Wei, X.; Xiao, R.; Blue, S.M.; Chen, J.Y.; Cody, N.A.L.; Dominguez, D.; et al. A large-scale binding and functional map of human RNA-binding proteins. Nature 2020, 583, 711-719. [CrossRef]
- Wang, C.; Uversky, V.N.; Kurgan, L. Disordered nucleiome: Abundance of intrinsic disorder in the DNA- and RNA-binding proteins in 1121 species from Eukaryota, Bacteria and Archaea. Proteomics 2016, 16, 1486-1498. [CrossRef]
- Peng, Z.; Mizianty, M.J.; Xue, B.; Kurgan, L.; Uversky, V.N. More than just tails: intrinsic disorder in histone proteins. Mol Biosyst 2012, 8, 1886-1901. [CrossRef]
- Bhalla, J.; Storchan, G.B.; MacCarthy, C.M.; Uversky, V.N.; Tcherkasskaya, O. Local flexibility in molecular function paradigm. Mol Cell Proteomics 2006, 5, 1212-1223. [CrossRef]
- Liu, J.; Perumal, N.B.; Oldfield, C.J.; Su, E.W.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder in transcription factors. Biochemistry 2006, 45, 6873-6888. [CrossRef]
- Minezaki, Y.; Homma, K.; Kinjo, A.R.; Nishikawa, K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol 2006, 359, 1137-1149. [CrossRef]
- Coelho Ribeiro Mde, L.; Espinosa, J.; Islam, S.; Martinez, O.; Thanki, J.J.; Mazariegos, S.; Nguyen, T.; Larina, M.; Xue, B.; Uversky, V.N. Malleable ribonucleoprotein machine: protein intrinsic disorder in the Saccharomyces cerevisiae spliceosome. PeerJ 2013, 1, e2. [CrossRef]
- Korneta, I.; Bujnicki, J.M. Intrinsic disorder in the human spliceosomal proteome. PLoS Comput Biol 2012, 8, e1002641. [CrossRef]
- Zhao, B.; Katuwawala, A.; Oldfield, C.J.; Hu, G.; Wu, Z.; Uversky, V.N.; Kurgan, L. Intrinsic Disorder in Human RNA-Binding Proteins. J Mol Biol 2021, 433, 167229. [CrossRef]
- Sambrook, J. Adenovirus amazes at Cold Spring Harbor. Nature 1977, 268, 101-104.
- Black, D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 2003, 72, 291-336. [CrossRef]
- Graveley, B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet 2001, 17, 100-107, doi:S0168-9525(00)02176-4 [pii].
- Stamm, S.; Ben-Ari, S.; Rafalska, I.; Tang, Y.; Zhang, Z.; Toiber, D.; Thanaraj, T.A.; Soreq, H. Function of alternative splicing. Gene 2005, 344, 1-20. [CrossRef]
- Brett, D.; Hanke, J.; Lehmann, G.; Haase, S.; Delbruck, S.; Krueger, S.; Reich, J.; Bork, P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett 2000, 474, 83-86, doi:S0014-5793(00)01581-7 [pii].
- Johnson, J.M.; Castle, J.; Garrett-Engele, P.; Kan, Z.; Loerch, P.M.; Armour, C.D.; Santos, R.; Schadt, E.E.; Stoughton, R.; Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 2003, 302, 2141-2144. [CrossRef]
- Minneman, K.P. Splice variants of G protein-coupled receptors. Molecular interventions 2001, 1, 108-116.
- Thai, T.H.; Kearney, J.F. Distinct and opposite activities of human terminal deoxynucleotidyltransferase splice variants. J Immunol 2004, 173, 4009-4019.
- Scheper, W.; Zwart, R.; Baas, F. Alternative splicing in the N-terminus of Alzheimer's presenilin 1. Neurogenetics 2004, 5, 223-227.
- Norris, J.D.; Fan, D.; Sherk, A.; McDonnell, D.P. A negative coregulator for the human ER. Mol Endocrinol 2002, 16, 459-468. [CrossRef]
- Ilik, I.A.; Malszycki, M.; Lubke, A.K.; Schade, C.; Meierhofer, D.; Aktas, T. SON and SRRM2 are essential for nuclear speckle formation. Elife 2020, 9. [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583-589. [CrossRef]
- Dayhoff, G.W., 2nd; Uversky, V.N. Rapid prediction and analysis of protein intrinsic disorder. Protein Sci 2022, 31, e4496. [CrossRef]
- Romero, P.R.; Zaidi, S.; Fang, Y.Y.; Uversky, V.N.; Radivojac, P.; Oldfield, C.J.; Cortese, M.S.; Sickmeier, M.; LeGall, T.; Obradovic, Z.; et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A 2006, 103, 8390-8395.
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem Int Ed Engl 2005, 44, 7342-7372. [CrossRef]
- Witze, E.S.; Old, W.M.; Resing, K.A.; Ahn, N.G. Mapping protein post-translational modifications with mass spectrometry. Nat Methods 2007, 4, 798-806. [CrossRef]
- Deribe, Y.L.; Pawson, T.; Dikic, I. Post-translational modifications in signal integration. Nat Struct Mol Biol 2010, 17, 666-672. [CrossRef]
- Mann, M.; Jensen, O.N. Proteomic analysis of post-translational modifications. Nat Biotechnol 2003, 21, 255-261. [CrossRef]
- Marks, F. Protein Phosphorylation; VCH Weinheim: New York, Basel, Cambridge, Tokyo, 1996.
- Yang, X.J. Multisite protein modification and intramolecular signaling. Oncogene 2005, 24, 1653-1662. [CrossRef]
- Erler, J.; Zhang, R.; Petridis, L.; Cheng, X.; Smith, J.C.; Langowski, J. The role of histone tails in the nucleosome: a computational study. Biophys J 2014, 107, 2911-2922. [CrossRef]
- Mersfelder, E.L.; Parthun, M.R. The tale beyond the tail: histone core domain modifications and the regulation of chromatin structure. Nucleic Acids Res 2006, 34, 2653-2662. [CrossRef]
- Dunker, A.K.; Brown, C.J.; Lawson, J.D.; Iakoucheva, L.M.; Obradovic, Z. Intrinsic disorder and protein function. Biochemistry 2002, 41, 6573-6582.
- Dunker, A.K.; Brown, C.J.; Obradovic, Z. Identification and functions of usefully disordered proteins. Adv Protein Chem 2002, 62, 25-49.
- Iakoucheva, L.M.; Radivojac, P.; Brown, C.J.; O'Connor, T.R.; Sikes, J.G.; Obradovic, Z.; Dunker, A.K. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004, 32, 1037-1049.
- Radivojac, P.; Vacic, V.; Haynes, C.; Cocklin, R.R.; Mohan, A.; Heyen, J.W.; Goebl, M.G.; Iakoucheva, L.M. Identification, analysis, and prediction of protein ubiquitination sites. Proteins 2010, 78, 365-380.
- Uversky, V.N. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci 2003, 60, 1852-1871.
- Turoverov, K.K.; Kuznetsova, I.M.; Uversky, V.N. The protein kingdom extended: ordered and intrinsically disordered proteins, their folding, supramolecular complex formation, and aggregation. Prog Biophys Mol Biol 2010, 102, 73-84. [CrossRef]
- Williams, R.M.; Obradovic, Z.; Mathura, V.; Braun, W.; Garner, E.C.; Young, J.; Takayama, S.; Brown, C.J.; Dunker, A.K. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput 2001, 89-100.
- Uversky, V.N. What does it mean to be natively unfolded? Eur J Biochem 2002, 269, 2-12, doi:2649 [pii].
- Dunker, A.K.; Obradovic, Z. The protein trinity--linking function and disorder. Nat Biotechnol 2001, 19, 805-806.
- Uversky, V.N. Paradoxes and wonders of intrinsic disorder: Complexity of simplicity. Intrinsically Disord Proteins 2016, 4, e1135015. [CrossRef]
- DeForte, S.; Uversky, V.N. Order, Disorder, and Everything in Between. Molecules 2016, 21. [CrossRef]
- Uversky, V.N. Dancing Protein Clouds: The Strange Biology and Chaotic Physics of Intrinsically Disordered Proteins. J Biol Chem 2016, 291, 6681-6688. [CrossRef]
- Uversky, V.N. Functional roles of transiently and intrinsically disordered regions within proteins. FEBS J 2015, 282, 1182-1189. [CrossRef]
- Oldfield, C.J.; Cheng, Y.; Cortese, M.S.; Romero, P.; Uversky, V.N.; Dunker, A.K. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry 2005, 44, 12454-12470. [CrossRef]
- Patil, A.; Nakamura, H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett 2006, 580, 2041-2045.
- Ekman, D.; Light, S.; Bjorklund, A.K.; Elofsson, A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol 2006, 7, R45.
- Haynes, C.; Oldfield, C.J.; Ji, F.; Klitgord, N.; Cusick, M.E.; Radivojac, P.; Uversky, V.N.; Vidal, M.; Iakoucheva, L.M. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2006, 2, e100.
- Dosztanyi, Z.; Chen, J.; Dunker, A.K.; Simon, I.; Tompa, P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res 2006, 5, 2985-2995.
- Singh, G.P.; Dash, D. Intrinsic disorder in yeast transcriptional regulatory network. Proteins 2007, 68, 602-605.
- Singh, G.P.; Ganapathi, M.; Dash, D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins 2007, 66, 761-765.
- Schulz, G.E. Nucleotide Binding Proteins. In Molecular Mechanism of Biological Recognition, Balaban, M., Ed.; Elsevier/North-Holland Biomedical Press: New York, 1979; pp. 79-94.
- Kriwacki, R.W.; Hengst, L.; Tennant, L.; Reed, S.I.; Wright, P.E. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A 1996, 93, 11504-11509.
- Mohan, A.; Oldfield, C.J.; Radivojac, P.; Vacic, V.; Cortese, M.S.; Dunker, A.K.; Uversky, V.N. Analysis of molecular recognition features (MoRFs). J Mol Biol 2006, 362, 1043-1059. [CrossRef]
- Cheng, Y.; Oldfield, C.J.; Meng, J.; Romero, P.; Uversky, V.N.; Dunker, A.K. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry 2007, 46, 13468-13477. [CrossRef]
- Disfani, F.M.; Hsu, W.L.; Mizianty, M.J.; Oldfield, C.J.; Xue, B.; Dunker, A.K.; Uversky, V.N.; Kurgan, L. MoRFpred, a computational tool for sequence-based prediction and characterization of short disorder-to-order transitioning binding regions in proteins. Bioinformatics 2012, 28, i75-83. [CrossRef]
- Landsteiner, K. The Specificity of Serological Reactions; Courier Dover Publications: Mineola, New York, 1936.
- Pauling, L. A theory of the structure and process of formation of antibodies. J Am Chem Soc 1940, 62, 2643-2657.
- Karush, F. Heterogeneity of the binding sites of bovine serum albumin. J Am Chem Soc 1950, 72, 2705-2713.
- Meador, W.E.; Means, A.R.; Quiocho, F.A. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science 1993, 262, 1718-1721.
- Uversky, V.N. A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn 2003, 21, 211-234, doi:d=3013&c=4118&p=11847&do=detail [pii].
- Fuxreiter, M.; Tompa, P. Fuzzy complexes: a more stochastic view of protein function. Adv Exp Med Biol 2012, 725, 1-14. [CrossRef]
- Tompa, P.; Fuxreiter, M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci 2008, 33, 2-8. [CrossRef]
- Uversky, V.N. Multitude of binding modes attainable by intrinsically disordered proteins: a portrait gallery of disorder-based complexes. Chem Soc Rev 2011, 40, 1623-1634. [CrossRef]
- Permyakov, S.E.; Millett, I.S.; Doniach, S.; Permyakov, E.A.; Uversky, V.N. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins 2003, 53, 855--862.
- Sigalov, A.; Aivazian, D.; Stern, L. Homooligomerization of the cytoplasmic domain of the T cell receptor zeta chain and of other proteins containing the immunoreceptor tyrosine-based activation motif. Biochemistry 2004, 43, 2049-2061. [CrossRef]
- Sigalov, A.B.; Zhuravleva, A.V.; Orekhov, V.Y. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie 2007, 89, 419-421. [CrossRef]
- Bjarnadottir, T.K.; Gloriam, D.E.; Hellstrand, S.H.; Kristiansson, H.; Fredriksson, R.; Schioth, H.B. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics 2006, 88, 263-273. [CrossRef]
- Anantharaman, V.; Abhiman, S.; de Souza, R.F.; Aravind, L. Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene 2011, 475, 63-78. [CrossRef]
- Southan, C.; Sharman, J.L.; Benson, H.E.; Faccenda, E.; Pawson, A.J.; Alexander, S.P.; Buneman, O.P.; Davenport, A.P.; McGrath, J.C.; Peters, J.A.; et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res 2016, 44, D1054-1068. [CrossRef]
- Fredriksson, R.; Lagerstrom, M.C.; Lundin, L.G.; Schioth, H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol 2003, 63, 1256-1272. [CrossRef]
- Flock, T.; Hauser, A.S.; Lund, N.; Gloriam, D.E.; Balaji, S.; Babu, M.M. Selectivity determinants of GPCR-G-protein binding. Nature 2017, 545, 317-322. [CrossRef]
- Isberg, V.; de Graaf, C.; Bortolato, A.; Cherezov, V.; Katritch, V.; Marshall, F.H.; Mordalski, S.; Pin, J.P.; Stevens, R.C.; Vriend, G.; et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol Sci 2015, 36, 22-31. [CrossRef]
- Fonin, A.V.; Darling, A.L.; Kuznetsova, I.M.; Turoverov, K.K.; Uversky, V.N. Multi-functionality of proteins involved in GPCR and G protein signaling: making sense of structure-function continuum with intrinsic disorder-based proteoforms. Cell Mol Life Sci 2019, 76, 4461-4492. [CrossRef]
- Neves, S.R.; Ram, P.T.; Iyengar, R. G protein pathways. Science 2002, 296, 1636-1639. [CrossRef]
- Marinissen, M.J.; Gutkind, J.S. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci 2001, 22, 368-376. [CrossRef]
- Latorraca, N.R.; Venkatakrishnan, A.J.; Dror, R.O. GPCR Dynamics: Structures in Motion. Chem Rev 2017, 117, 139-155. [CrossRef]
- Venkatakrishnan, A.J.; Flock, T.; Prado, D.E.; Oates, M.E.; Gough, J.; Madan Babu, M. Structured and disordered facets of the GPCR fold. Curr Opin Struct Biol 2014, 27, 129-137. [CrossRef]
- Bushdid, C.; Magnasco, M.O.; Vosshall, L.B.; Keller, A. Humans can discriminate more than 1 trillion olfactory stimuli. Science 2014, 343, 1370-1372. [CrossRef]
- Malnic, B.; Hirono, J.; Sato, T.; Buck, L.B. Combinatorial receptor codes for odors. Cell 1999, 96, 713-723. [CrossRef]
- Reddy, G.; Zak, J.D.; Vergassola, M.; Murthy, V.N. Antagonism in olfactory receptor neurons and its implications for the perception of odor mixtures. Elife 2018, 7. [CrossRef]
- Gánti, T. Chemoton theory: theory of living systems; Springer Science & Business Media: 2003.
- Kulkarni, P.; Bhattacharya, S.; Achuthan, S.; Behal, A.; Jolly, M.K.; Kotnala, S.; Mohanty, A.; Rangarajan, G.; Salgia, R.; Uversky, V. Intrinsically Disordered Proteins: Critical Components of the Wetware. Chem Rev 2022, 122, 6614-6633. [CrossRef]
- Katsnelson, A. Did Disordered Proteins Help Launch Life on Earth? ACS Cent Sci 2020, 6, 1854-1857. [CrossRef]
- Kulkarni, P.; Uversky, V.N. Intrinsically Disordered Proteins: The Dark Horse of the Dark Proteome. Proteomics 2018, 18, e1800061. [CrossRef]
- Penny, D.; Foulds, L.R.; Hendy, M.D. Testing the theory of evolution by comparing phylogenetic trees constructed from five different protein sequences. Nature 1982, 297, 197-200. [CrossRef]
- Futuyma, D. Evolutionary biology, 3rd edn Sinauer Associates. Sunderland.[Google Scholar] 1998.
- Zuckerkandl, E.; Pauling, L. Evolutionary divergence and convergence in proteins. In Evolving genes and proteins; Elsevier: 1965; pp. 97-166.
- Schluter, H.; Apweiler, R.; Holzhutter, H.G.; Jungblut, P.R. Finding one's way in proteomics: a protein species nomenclature. Chem Cent J 2009, 3, 11. [CrossRef]
- Uhlen, M.; Bjorling, E.; Agaton, C.; Szigyarto, C.A.; Amini, B.; Andersen, E.; Andersson, A.C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Molecular & Cellular Proteomics 2005, 4, 1920-1932. [CrossRef]
- Farrah, T.; Deutsch, E.W.; Omenn, G.S.; Sun, Z.; Watts, J.D.; Yamamoto, T.; Shteynberg, D.; Harris, M.M.; Moritz, R.L. State of the Human Proteome in 2013 as Viewed through PeptideAtlas: Comparing the Kidney, Urine, and Plasma Proteomes for the Biology- and Disease-Driven Human Proteome Project. Journal of Proteome Research 2014, 13, 60-75. [CrossRef]
- Farrah, T.; Deutsch, E.W.; Hoopmann, M.R.; Hallows, J.L.; Sun, Z.; Huang, C.Y.; Moritz, R.L. The State of the Human Proteome in 2012 as Viewed through PeptideAtlas. Journal of Proteome Research 2013, 12, 162-171. [CrossRef]
- Reddy, P.J.; Ray, S.; Srivastava, S. The Quest of the Human Proteome and the Missing Proteins: Digging Deeper. Omics-a Journal of Integrative Biology 2015, 19, 276-282, doi:DOI 10.1089/omi.2015.0035.
- Kim, M.S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575-+. [CrossRef]
- Ponomarenko, E.A.; Poverennaya, E.V.; Ilgisonis, E.V.; Pyatnitskiy, M.A.; Kopylov, A.T.; Zgoda, V.G.; Lisitsa, A.V.; Archakov, A.I. The Size of the Human Proteome: The Width and Depth. Int J Anal Chem 2016, 2016, 7436849. [CrossRef]
- Smith, L.M.; Kelleher, N.L.; Consortium for Top Down, P. Proteoform: a single term describing protein complexity. Nat Methods 2013, 10, 186-187. [CrossRef]
- Uversky, V.N. p53 Proteoforms and Intrinsic Disorder: An Illustration of the Protein Structure-Function Continuum Concept. Int J Mol Sci 2016, 17, 1874. [CrossRef]
- Pejaver, V.; Hsu, W.L.; Xin, F.; Dunker, A.K.; Uversky, V.N.; Radivojac, P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci 2014, 23, 1077-1093. [CrossRef]
- Dunker, A.K.; Lawson, J.D.; Brown, C.J.; Williams, R.M.; Romero, P.; Oh, J.S.; Oldfield, C.J.; Campen, A.M.; Ratliff, C.M.; Hipps, K.W.; et al. Intrinsically disordered protein. J. Mol. Graph. Model. 2001, 19, 26-59. [CrossRef]
- Dunker, A.K.; Silman, I.; Uversky, V.N.; Sussman, J.L. Function and structure of inherently disordered proteins. Curr Opin Struct Biol 2008, 18, 756-764. [CrossRef]
- Dunker, A.K.; Uversky, V.N. Signal transduction via unstructured protein conduits. Nat Chem Biol 2008, 4, 229-230. [CrossRef]
- Uversky, V.N. Disordered competitive recruiter: fast and foldable. J Mol Biol 2012, 418, 267-268. [CrossRef]
- Uversky, V.N.; Dunker, A.K. The case for intrinsically disordered proteins playing contributory roles in molecular recognition without a stable 3D structure. F1000 biology reports 2013, 5, 1. [CrossRef]
- Dyson, H.J.; Wright, P.E. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol 2002, 12, 54-60. [CrossRef]
- Dyson, H.J.; Wright, P.E. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol 2005, 6, 197-208. [CrossRef]
- Vacic, V.; Oldfield, C.J.; Mohan, A.; Radivojac, P.; Cortese, M.S.; Uversky, V.N.; Dunker, A.K. Characterization of molecular recognition features, MoRFs, and their binding partners. J Proteome Res 2007, 6, 2351-2366. [CrossRef]
- Comolatti, R.; Hoel, E. Causal emergence is widespread across measures of causation. arXiv preprint arXiv:2202.01854 2022.
- Baranger, M. Chaos, complexity, and entropy - A physics talk for non-physicists. 2001.
- Klein, B.; Hoel, E.; Swain, A.; Griebenow, R.; Levin, M. Evolution and emergence: higher order information structure in protein interactomes across the tree of life. Integr Biol (Camb) 2021, 13, 283-294. [CrossRef]
- Klein, B.; Hoel, E. The emergence of informative higher scales in complex networks. Complexity 2020, 2020, 8932526.
- Sebe-Pedros, A.; de Mendoza, A.; Lang, B.F.; Degnan, B.M.; Ruiz-Trillo, I. Unexpected repertoire of metazoan transcription factors in the unicellular holozoan Capsaspora owczarzaki. Mol Biol Evol 2011, 28, 1241-1254. [CrossRef]
- Lowe, C.B.; Kellis, M.; Siepel, A.; Raney, B.J.; Clamp, M.; Salama, S.R.; Kingsley, D.M.; Lindblad-Toh, K.; Haussler, D. Three periods of regulatory innovation during vertebrate evolution. Science 2011, 333, 1019-1024. [CrossRef]
- Hindorff, L.A.; Sethupathy, P.; Junkins, H.A.; Ramos, E.M.; Mehta, J.P.; Collins, F.S.; Manolio, T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 2009, 106, 9362-9367. [CrossRef]
- Huang, S.; Guo, Y.P.; May, G.; Enver, T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol 2007, 305, 695-713. [CrossRef]
- Wheat, J.C.; Sella, Y.; Willcockson, M.; Skoultchi, A.I.; Bergman, A.; Singer, R.H.; Steidl, U. Single-molecule imaging of transcription dynamics in somatic stem cells. Nature 2020, 583, 431-436. [CrossRef]

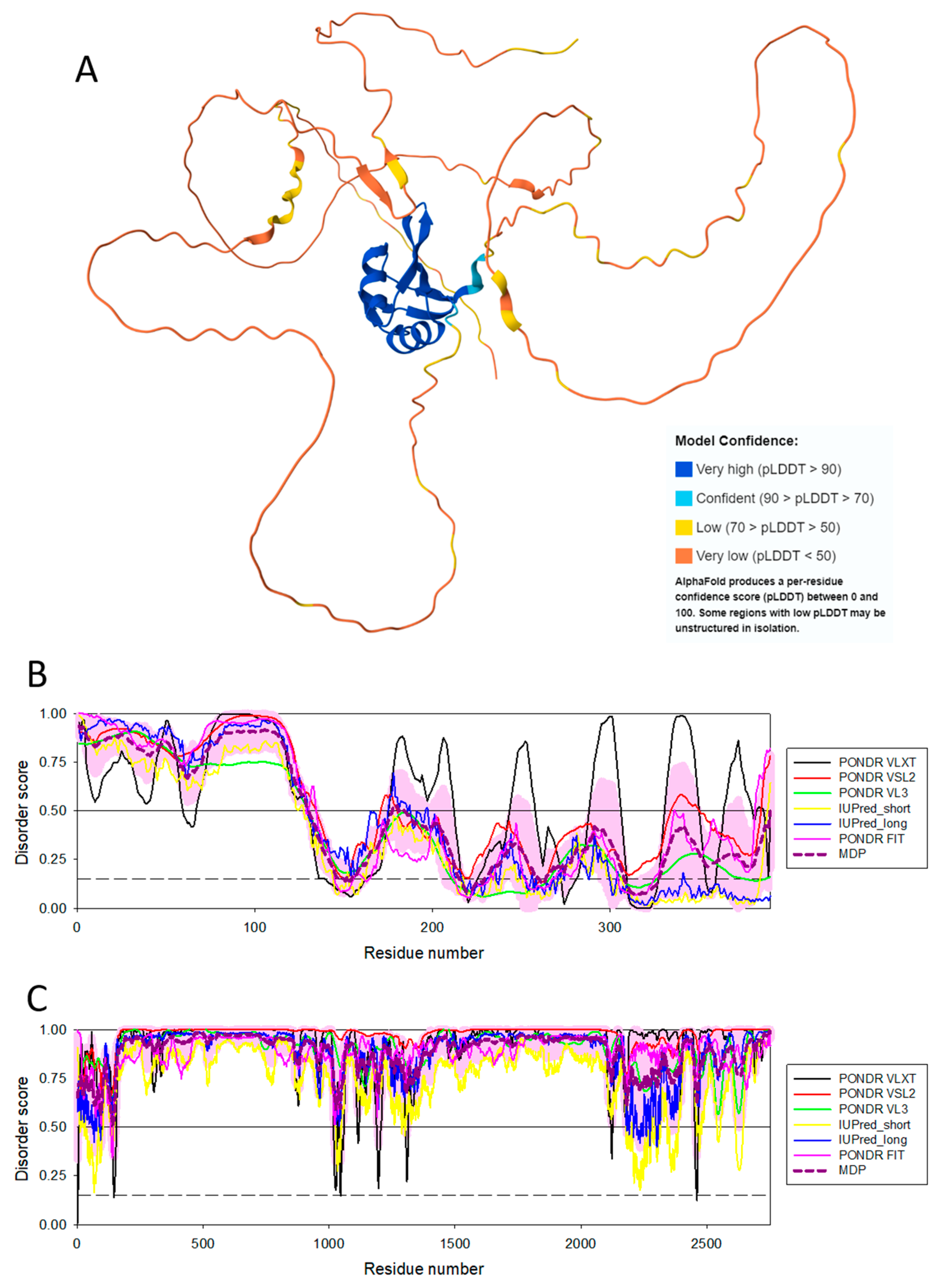
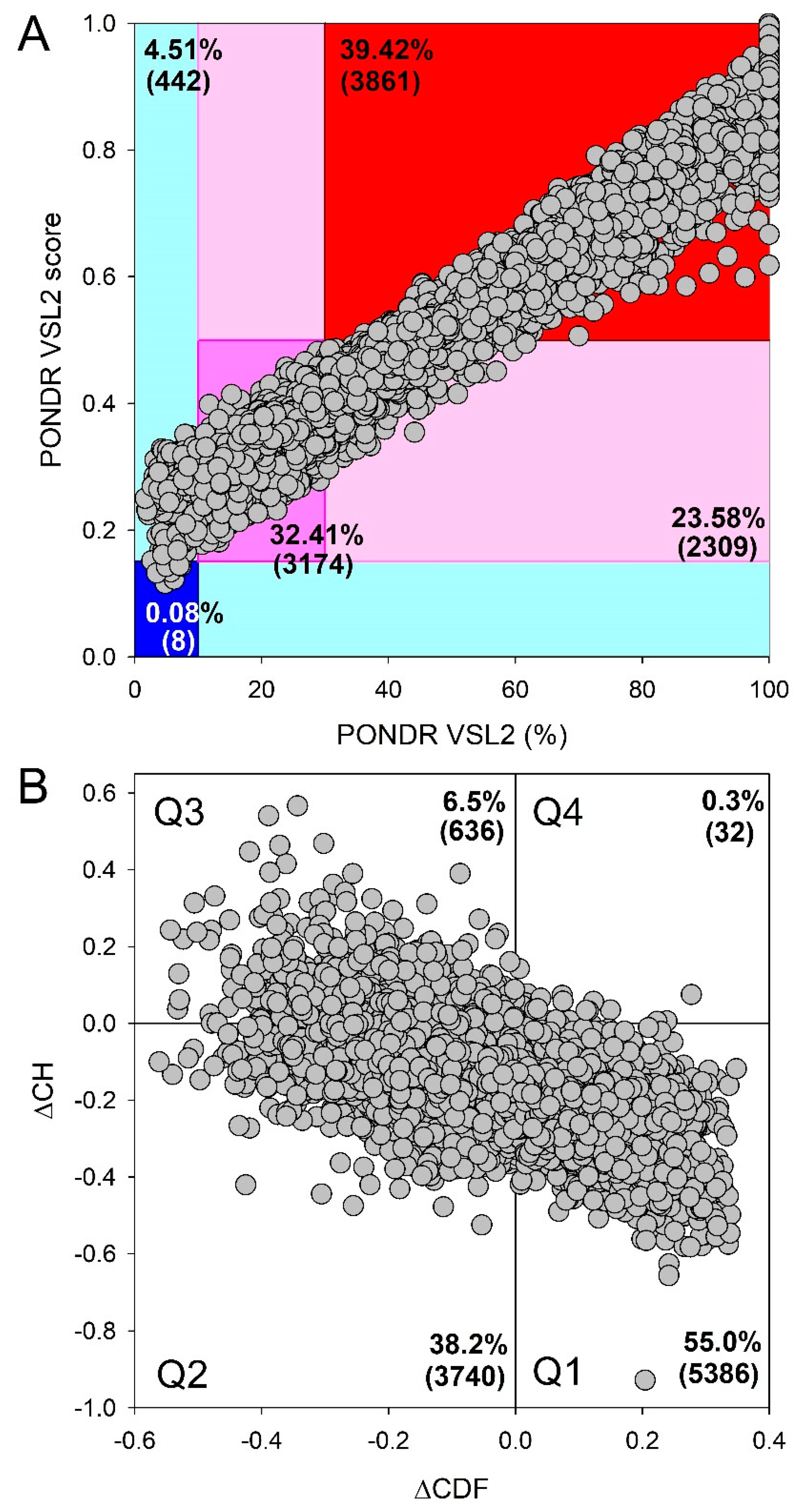
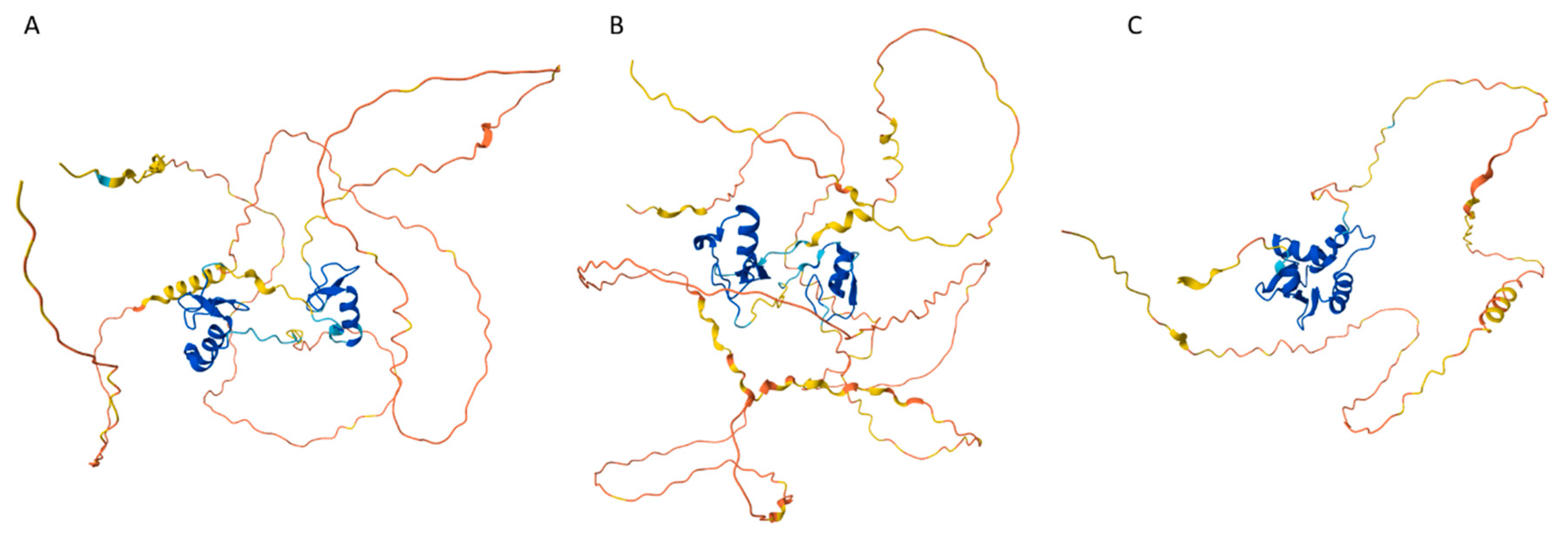
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




