1. Introduction
The bifurcating autoregressive (BAR) model is widely employed to analyze binary tree-structured data, as illustrated in
Figure 1, which frequently arise in various applications, most notably in cell lineage studies [
1,
2,
3,
4]. The BAR(1) model was initially proposed by [
5] to model cell lineage data. This model extends the first-order autoregressive AR(1) model by accounting for a unique aspect: observations on two sister cells, which share the same mother cell, are correlated. In practical settings, the BAR(1) model is useful for modeling the single-cell reproduction progress [c.f. [
3]. A number of studies have addressed the estimation and inference challenges associated with the BAR model [see, e.g., [
5,
6,
7,
8,
9,
10,
11,
12,
13], among others].
Maximum likelihood (ML) estimators for the model coefficients and the correlation between errors for the BAR(1) model were introduced by [
5] under the assumption of normally distributed errors. Building on this foundation, [
8] examined the asymptotic properties of the ML estimators for the BAR(
p) model, also assuming the normality of errors. In contrast, [
10] explored least squares (LS) estimation for BAR(
p) models without imposing distributional assumptions on the errors. In their study, they developed LS estimators for the model coefficients and introduced consistent estimators for the correlation between model errors. They further derived the asymptotic distribution of the LS estimators. However, the study did not address the finite-sample properties of these estimators. While LS estimators of BAR model coefficients are asymptotically unbiased, [
13] demonstrated that the finite sample bias of these estimators can be substantial, potentially leading to inaccurate inferences. This outcome is consistent with the well-known fact that LS estimators for the AR(1) model often exhibit significant bias, especially when the autoregressive parameter is near its limitations (
) (e.g., [
14]).
To address this issue, [
13] introduced two bias correction techniques for the LS estimator of the autoregressive parameter in the BAR(1) model: one based on asymptotic linear bias functions and another using a bootstrapping approach. Their results indicated that both methods effectively reduce the bias, with the bootstrap bias-correction method proving more efficient than the asymptotic linear bias function method, particularly when the parameter is close to the upper boundary (i.e., near
).
A related challenge in analyzing cell lineage data involves handling outliers. Several approaches have been proposed to develop robust estimators that mitigate the impact of outliers within the BAR model context, including quasi-maximum likelihood estimation methods (cf., [
15] and [
16]). In this direction, [
12] developed robust estimators for the first-order BAR model based on weighted absolute deviations. They demonstrated that, when the weights are fixed, the resulting estimator is an
-type estimator, offering robustness against outliers in the response space and showing greater efficiency than the least squares (LS) estimator in the presence of heavy-tailed error distributions. In contrast, when the weights are random and dependent on the points in the factor space, the weighted
(
) estimator shows robustness against outliers in the factor space. Additionally, they derived the asymptotic distribution of the
estimator and analyzed its large sample properties. However, they did not examine the bias of
estimators under finite sample conditions (i.e., small or moderate sample sizes). The present study seeks to address this gap in the literature.
In this paper, we concentrate on examining the bias of the weighted
(
) estimators for BAR(1) models. Our findings indicate that the
estimators of the BAR(1) autoregressive parameter (
, the parameter of interest) can show considerable bias, particularly when working with small to moderate sample sizes. To address this issue, we employ bootstrapping techniques to develop bias-corrected
estimators. We assess both the bias and mean squared error (MSE) of these estimators. It is important to note that bias is typically influenced by the true, unknown parameter values, and that bias correction may not always be advisable. This is because bias reduction can lead to increased variance, potentially resulting in a higher MSE than that of the uncorrected estimates [
17].
The layout of this paper is as follows. In the following section, we provide an overview of the BAR(1) model and the
estimation of its parameters.
Section 3 empirically examines the bias of the
estimators for the autoregressive parameter in the BAR(1) model. In
Section 4, we propose bias-corrected
estimators.
Section 5 presents a summary of the empirical findings, based on Monte Carlo simulations and a real data application, comparing the performance of the bias-corrected and uncorrected
estimators. Finally, the paper concludes in
Section 6 with a discussion of the key results and directions for future research.
2. The BAR(1) Model and Estimation
Consider the random variables , which represent observations on a perfect binary tree with g generations. The initial observation, , corresponds to generation 0, while the observations correspond to the observations in generation i, where . The total sample size is given by .
The first-order bifurcating autoregressive (BAR(1)) model is formulated as:
where
represents the observed value of a quantitative characteristic at time
t, and
, with
denoting the "mother" of
for all
, and
representing the greatest integer less than or equal to
u. The vector
contains the unknown model coefficients: the intercept,
, and the autoregressive parameter,
(also referred to as the inherited effect or maternal correlation), where
. This range for
ensures the stationary of the process.
The pairs
are supposed to be independently and identically distributed (i.i.d.) according to a joint distribution
F. Each pair
has a zero mean vector and a variance-covariance matrix given by:
where
represents the linear correlation between
and
, commonly referred to as the environmental effect or sister-sister correlation given their shared mother. Additionally,
represents the variance of the errors. It is further assumed that the pairs
and
are independent for
.
The underlying rationale for this correlation structure is rooted in the assumption that sister cells, particularly in the early stages of their development, are exposed to a common environment. Consequently, two distinct forms of correlation are expected: (i) environmental correlation between sisters, and (ii) maternal correlation deriving from inherited effects from the mother. In contrast, more distantly related cells, such as cousins, experience less environmental overlap, making it reasonable to assume that their environmental effects are independent.
[
12] introduced the weighted L
1 estimators of the intercept and the autoregressive coefficient,
and
, respectively, for the first-order BAR model by minimizing the following dispersion function
where
is a weight function chosen such that the product
remains bounded, thereby providing robustness against outliers in the design points (i.e., outlying values of
). When
, the estimator reduces to the standard
-norm estimator. The dispersion function
is non-negative, piecewise linear, and convex, provided that
. These properties are sufficient to guarantee that
attains a minimum, yielding the estimate
.
2.1. Weighted and Non-Weighted Estimates for BAR(1) Model
In this section, we introduce two types of estimators for the BAR(1) model based on the weight function,
, in (
2). These estimators were studied by [
12] in the context of bifurcating autoregressive (BAR) models.
2.1.1. Least Absolute Deviation (L1) Estimator
The first estimator is the Least Absolute Deviation (L
1) estimator, which is a non-weighted version. This estimator is derived by setting the weight function in (
2) to 1 for all
x (i.e.,
). The L
1 estimator is known to be sensitive to bad leverage points in a multivariate autoregressive context [
18]. [
12] demonstrated that this sensitivity persists in the BAR(1) model when confronted with bad leverage points caused by additive outliers.
2.1.2. Weighted Least Absolute Deviation (WL1) Estimators
The second type is the Weighted Least Absolute Deviation (
) estimator, which incorporates a weighting scheme defined by:
The weight is determined based on the Mahalanobis distance between
and a robust measure of location and dispersion,
, utilizing the minimum covariance determinant (MCD) estimates as outlined in [
19]. The constant
k serves as a cutoff point, set at the 95th percentile of the Chi-square distribution with one degree of freedom, while the parameter
is fixed at 2. The weighting scheme in (
3) is similar to that used by [
20].
This weighted estimator provides an advantage over the non-weighted L
1 estimator by being less sensitive to bad leverage points in both the response and factor spaces. For more detailed discussions, refer to [
21] in the context of linear regression, [
22] for autoregressive time series models, and [
12] for bifurcating autoregressive models.
[
12] derived the joint limiting distribution of the
estimators of the BAR(1) model coefficients. Under some regularity conditions, they showed that
where
,
,
,
, and
denotes the correlation coefficient between the signs of the pairs of errors
. The method of moments estimators for
and
are given by
and
where
with
and
. [
12] suggested to follow the estimation procedure proposed by [
23] and [
24] to estimate the parameter
:
3. Bias in the Estimators for the BAR(1) Model
In this section, we perform an empirical investigation using Monte Carlo simulations to assess the finite-sample bias of the estimator for the autoregressive coefficient , examining how it varies with the model parameters and . The simulations follow a specific setup. Perfect binary trees of sizes , and 255 (corresponding to different numbers of generations, , respectively) are generated for each combination of and , capturing a range of maternal and environmental effect correlation levels. The model intercept, , is fixed at 10 in all simulations. All generated trees are assumed to be stationary, with the initial observation randomly drawn from a larger simulated perfect binary tree of size 127.
Outliers are introduced by contaminating the errors through the use of two bivariate normal distributions. These distributions combine a standard bivariate normal distribution with contamination rates of 5% and 10%, drawn from an additional bivariate normal distribution with a mean of zero and a variance of 25. For each simulation scenario,
binary trees are generated, and the
estimator
is computed for each tree. Subsequently, we assess the empirical bias, absolute relative bias (ARB), variance, and mean squared error (MSE) of
as follows:
where
is the
estimator from iteration
.
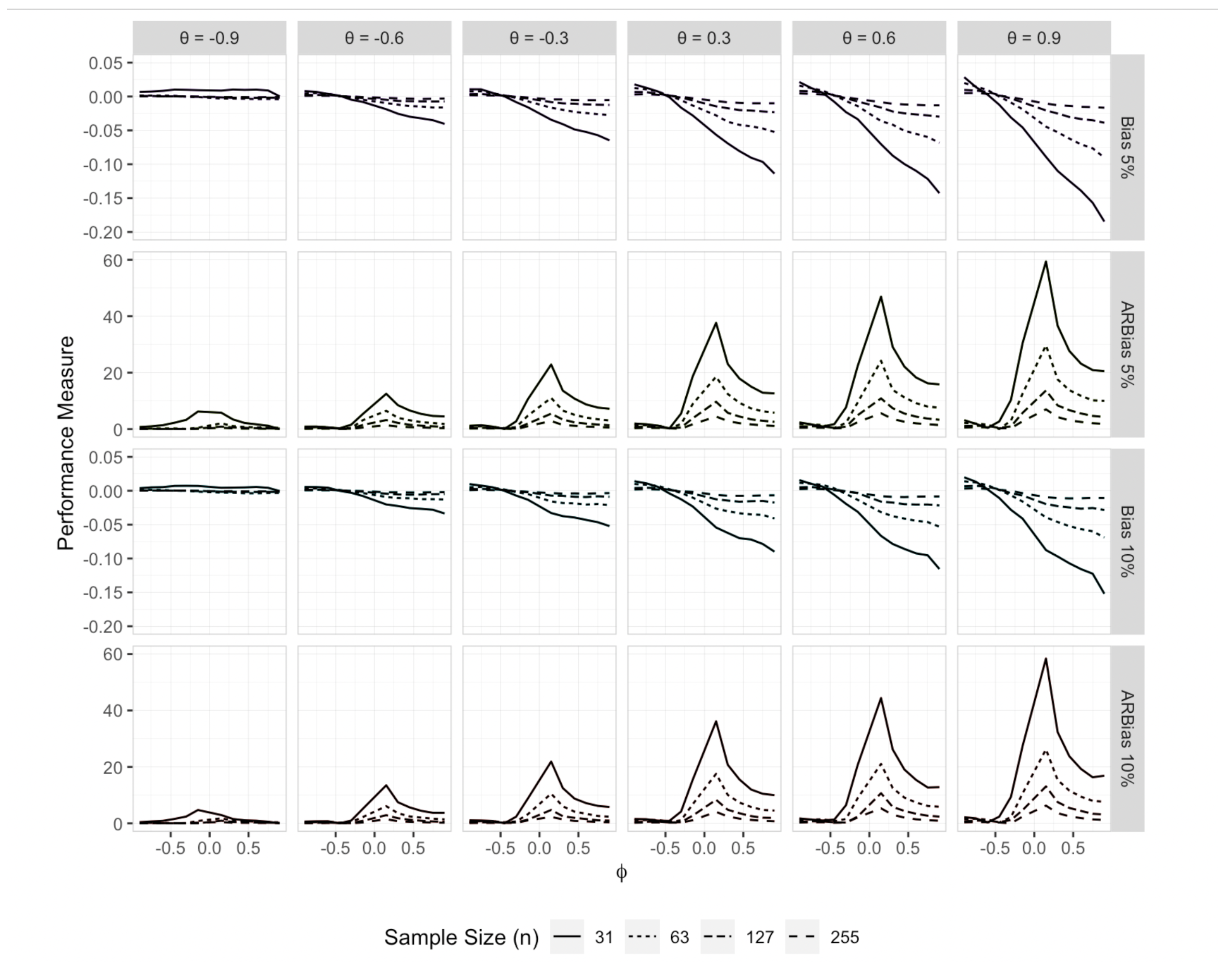
Figure 2 presents the empirical bias and ARB (
) of
. The results indicate that the bias of
can be significant across various combinations of
and
, particularly for smaller sample sizes. Specifically, it is evident that the volume of the bias is concerning when
is positive and
exceeds
. In general,
tends to underestimate
for values of
greater than
, with the degree of underestimation increasing as
approaches
. Furthermore, the bias appears to exhibit a linear relationship with
.
The extent of the bias is further emphasized by the ARB results, which show that for small sample sizes () and near zero (in the range to ), the bias of can range from 5% to 60% of the true value of as varies from to . For the smallest sample size (), the relative bias remains considerably large for values of greater than , provided does not approach . While the bias remains considerable in most cases for and , it becomes negligible for larger sample sizes (), except when is near zero (in the range to ) and is positive.
4. Bias-Corrected Estimators for the BAR(1) Model
In this section, we outline three bootstrap methods designed to correct the bias of the estimators, , for the autoregressive coefficient in the BAR(1) model. Specifically, we introduce the single bootstrap bias-corrected version of , as well as a fast double bootstrap bias-corrected version.
The model-based bootstrap technique has been widely used to correct bias in the least squares (LS) estimation of the AR(1) model, with its properties thoroughly explored in the literature (see [
25,
26,
27,
28], among others). This approach involves generating a large number of bootstrap samples based on the estimated model parameters and resampled residuals. These samples are then used to compute multiple replicates of the estimator, which allows for the estimation of the sampling distribution, variance, and bias of the estimator. Recently, [
13] extended this method to correct bias in the LS estimation of the BAR(1) model coefficients. In this section, we focus on describing two model-based bootstrap bias correction methods specifically designed for the BAR(1) model.
We begin by rewriting the BAR(1) model, originally presented in Eq. (
1), as follows:
for
. Here,
and
represent the observations for the two daughter nodes branching from
.
The single bootstrap bias correction procedure for the autoregressive coefficient in the BAR(1) model is described in Algorithm 1. It is important to note that the bias in the corrected estimate, , is of the order , whereas the original estimator, , shows bias of the order .
The double bootstrap technique involves performing the single bootstrap procedure twice. This method has been shown to enhance the accuracy of bootstrap bias correction in the AR(1) model [e.g., [
29,
30,
31,
32], and similarly for the BAR(1) model [see, [
13]. However, a notable drawback of the standard double bootstrap method is its high computational cost. To address this issue, a modified double bootstrap bias correction algorithm, referred to as the fast double bootstrap, was proposed by [
33]. This algorithm offers greater computational efficiency compared to the traditional double bootstrap, as it requires only one bootstrap resample in phase 2 (i.e.,
) for each bootstrap sample generated in phase 1, in contrast to the conventional double bootstrap, which generates
resamples within each phase 1 bootstrap sample. Consequently, while the computational complexity of the standard double bootstrap algorithm is of the order
, the fast double bootstrap reduces this to
. Despite this significant reduction in computational cost, [
13] observed that the bias correction performance of both algorithms remains comparable. The fast double bootstrap approach for correcting the bias in the
estimator of
under the BAR(1) model is outlined in Algorithm 2.
|
Algorithm 1: Single Bootstrap Bias-Corrected Estimation for (SBC). |
|
Input
Observed tree:
estimates of the BAR(1) model coefficients: and
Centered residuals: for
Number of bootstrap resamples: B
- 1:
for each to B do
- 2:
Set , the final observation from an initial binary tree § of size , where is the first observation in the original tree - 3:
for each to do
- 4:
Sample with replacement a pair from the set of pairs
- 5:
- 6:
end for
- 7:
Construct the bootstrap tree
- 8:
Compute the estimate from
- 9:
end for - 10:
Estimate the bias of as
-
Output:
The single bootstrap bias-corrected estimate of is given by:
|
§In each bootstrap iteration, the initial tree is generated from a BAR(1) model with coefficients set to the observed estimates, and , while the errors are sampled in pairs from the centered residuals. It is important to note that using from the observed tree as the initial observation in all bootstrap trees could introduce artificial correlation among the bootstrap samples. This issue may become more significant in higher-order BAR(p) models, where initial observations are required. In such cases, our method utilizes the final observations from the initial tree to avoid this potential problem.
§§Any appropriate initial tree size,
, can be chosen. The suggested size of
provides a reasonable compromise between computational efficiency and result stability.
|
Algorithm 2: Fast Double Bootstrap Bias-Corrected Estimation for (FDBC). |
|
Input:
Observed tree:
estimates of the BAR(1) model coefficients: and
Centered residuals: , for
Number of phase 1 bootstrap resamples:
Number of phase 2 bootstrap resamples:
- 1:
for each to do
- 2:
Set , where is the last observation in an initial binary tree of size , with as the first observation in
- 3:
for each to do
- 4:
Sample with replacement a pair from the set
- 5:
- 6:
end for
- 7:
Construct the bootstrap tree
- 8:
Compute the first-phase bootstrap estimates and from
- 9:
Obtain the first-phase bootstrap residuals:
- 10:
Compute centered bootstrap residuals:
- 11:
for each to do
- 12:
Apply steps 2-6 to using , , and the centered residuals
- 13:
Construct the second-phase bootstrap tree
- 14:
Compute the second-phase bootstrap estimate from
- 15:
end for
- 16:
end for - 17:
Estimate the single bootstrap bias of :
- 18:
Compute the double bootstrap bias adjustment factor:
-
Output:
The fast double bootstrap bias-corrected estimate for :
|
5. Empirical Results
In this section, we present the findings of an empirical investigation aimed at (i) evaluating the effectiveness of the proposed bias correction methods for the
estimator of the autoregressive coefficient in the BAR(1) model, and (ii) comparing the relative performance of these bias correction techniques. The study involved extensive simulations. All computations were performed using R version 4.1.3 [
34] on a machine with an Intel Xeon E5-2699Av4 processor, featuring a clock speed of 2.4 GHz, 44 cores, and 512 GB of memory. The simulations utilized 42 of the 44 available cores through the `parallel` package, while bifurcating trees were generated using the `bifurcatingr` package [
35].
The "core" process follows the model described in , adhering to the same variance-covariance matrix. The "observed" process is defined as:
In this definition, when
for all
t, the "observed" process aligns with the "core" process, and any deviations are attributed to the distribution of the errors. As noted by [
36], these deviations are referred to as Innovation Outliers (IO) model. In this, the distribution of errors is typically characterized by heavy tails. On the other hand, when
, the process corresponds to the Additive Outliers (AO) model [
36]. In this case,
follows a mixed distribution given by
, where
represents the contamination proportion,
denotes a point mass at zero, and
G is the contaminating distribution function. For our simulations,
G was selected as a
distribution. Therefore, the distribution of
can be expressed as:
where
denotes a Bivariate Contaminated Normal distribution with a specified mean vector and variance-covariance matrix.
The variable is generated as a sequence of independent and identically distributed random variables. In the AO model, outliers are introduced through the incorporation of external influences into the core process, resulting in outliers that arise from factors beyond the error distribution.
In the context of our study, an outlier introduced into the design is referred to as a leverage point. We differentiate between two types of leverage points: “good” and “bad.” As demonstrated by [
19], a leverage point is classified as “good” if it results in a small residual despite being an outlier in the design space. In contrast, it is considered “bad” if it results in a large residual. According to [
19], the IO model tends to produce “good” leverage points with relatively minimal impact on estimates. In contrast, the AO model generates “bad” leverage points that can significantly affect estimates, even those that are robust [
12].
5.1. Simulation Setup
The simulations contain a range of scenarios defined by varying sample sizes
n, maternal correlation levels
, and error correlations
, as outlined in
Section 3. For each scenario, we generate
trees and compute the
estimator for the coefficient
from each tree. Specifically, we evaluate the
estimator (
), the single bootstrap bias-corrected estimator (
), and the fast double bootstrap bias-corrected estimator (
).
We excluded the case of
from this simulation study, as the bias in
for this larger sample size was found to be negligible, as detailed in
Section 3. This exclusion was implemented to facilitate computation time.
For both the
and
estimators, the number of bootstrap replicates was set to
. The empirical bias and absolute relative bias (ARB) for each estimator were computed as outlined in Eqs. (
4) and (
5). Furthermore, the root mean squared error (RMSE) and mean absolute deviation (MAD) were calculated as follows:
where
is the
estimator from iteration
.
5.1.1. Innovation Outliers (IO) Model Setup
Under the Innovation Outlier (IO) model, where
, the “observed” process simplifies to the “core” process as specified in Eq. (
1). The error distributions examined in this study include the bivariate standard normal distribution with correlation
, as well as four variants of the bivariate contaminated normal distributions. The contaminated distributions are characterized by different levels of contamination, specifically (1) Contaminating variance of 25 with a contamination rate of 5%, (2) Contaminating variance of 25 with a contamination rate of 10%, (3) Contaminating variance of 49 with a contamination rate of 5%, (4) Contaminating variance of 49 with a contamination rate of 10%. These distributions are denoted as
,
,
,
, respectively. The chosen parameter settings for these bivariate contaminated normal distributions are intended to represent both “mild” and “extreme” contamination scenarios.
5.1.2. Additive Outliers (AO) Model Setup
Under the Additive Outlier (AO) model, the errors
follow an innovation process modeled by a bivariate normal distribution with zero mean, unit variance, and
. The distribution of
is given by:
where
represents the proportion of contamination,
is a point mass at zero, and
G is a contaminating distribution function. The “observed” process is defined as:
where
represents the model component,
is the error term, and
represents the additive outlier.
The distributions considered for include the bivariate standard normal distribution with correlation , as well as four bivariate contaminated normal distributions. These contaminated distributions have contaminating variances of 25 and 49, with contamination rates of 5% and 10%. Specifically, the distributions are (1) , (2) , (3) , (4) . In addition, It is assumed that is independent of .
5.2. Simulation Results
In this section, a results summary of the simulation study is presented in
Figure 3,
Figure 4,
Figure 5,
Figure 6,
Figure 7,
Figure 8,
Figure 9 and
Figure 10. Our analysis focuses on the absolute relative bias and root mean squared error (RMSE) for the
estimator of
, including both the single bootstrap and fast double bootstrap bias-corrected estimators. The key findings are outlined as follows:
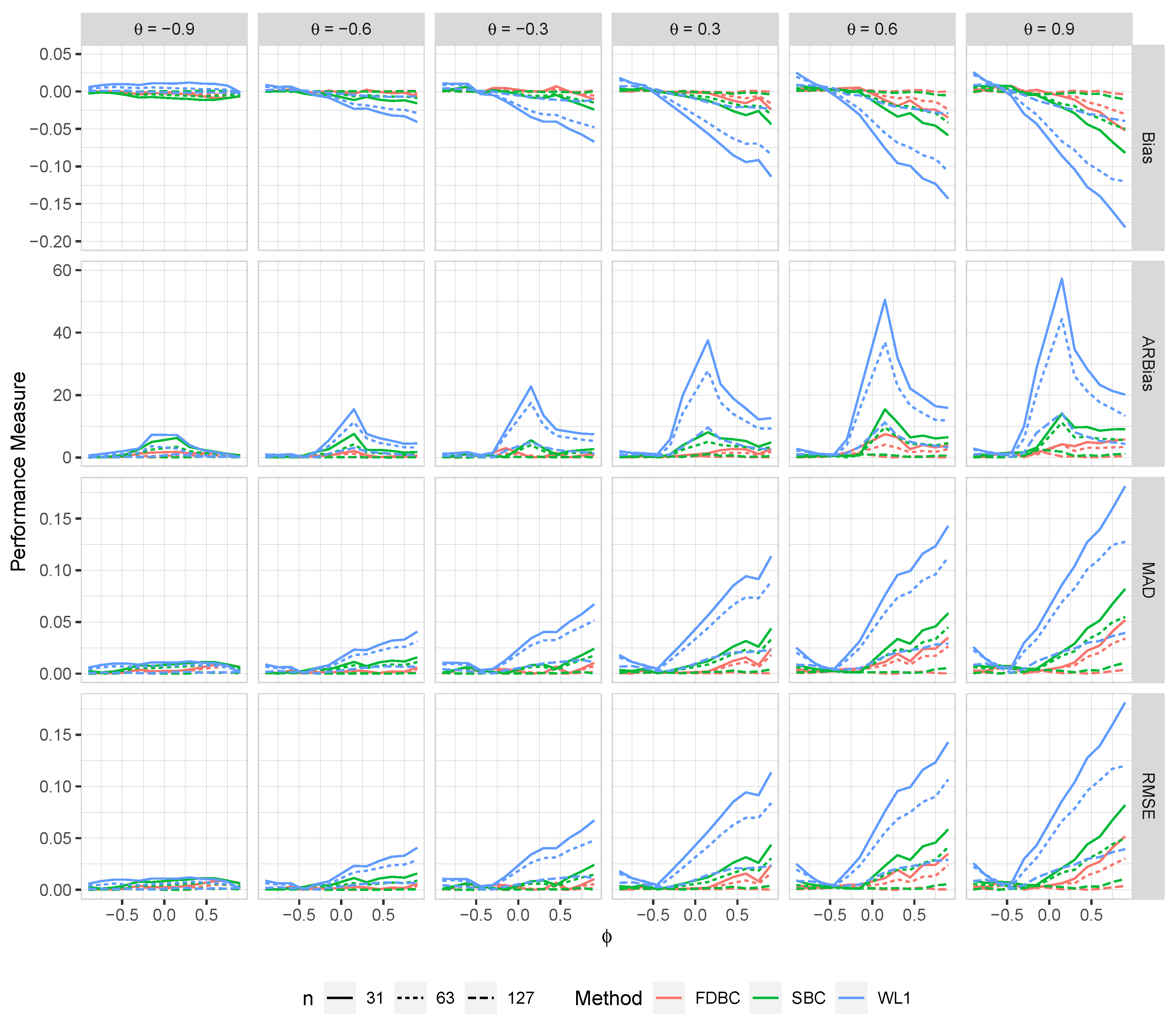
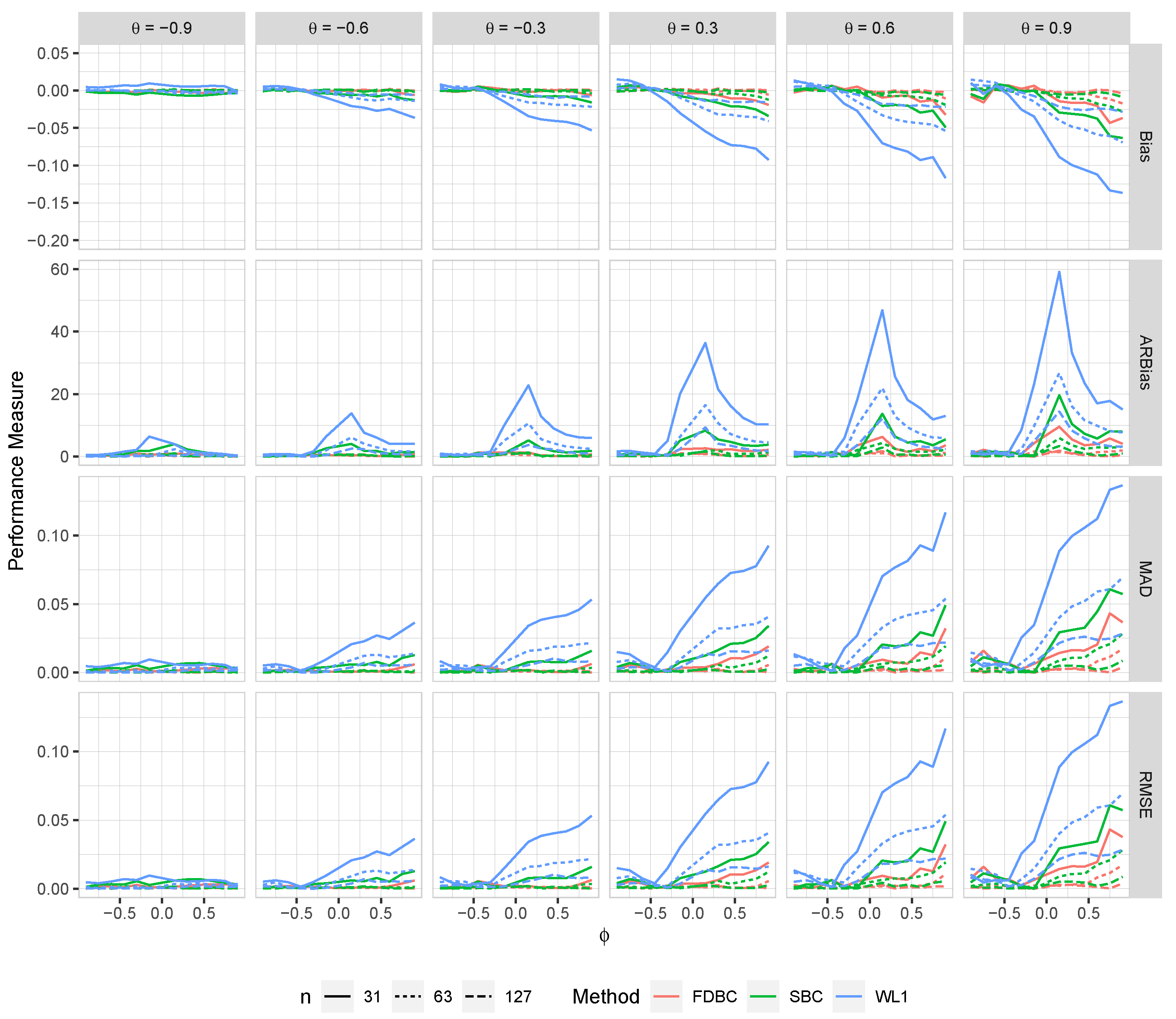
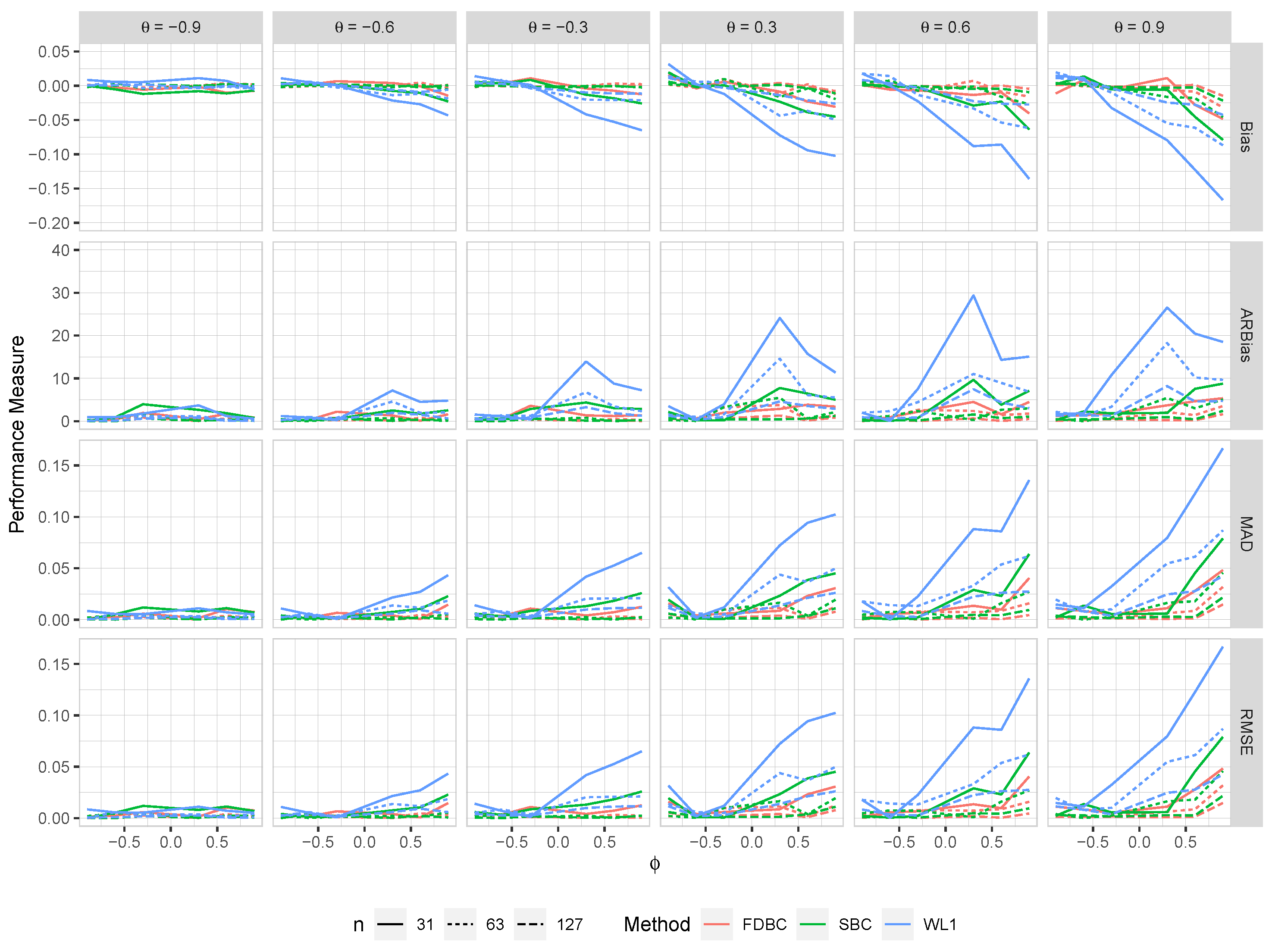
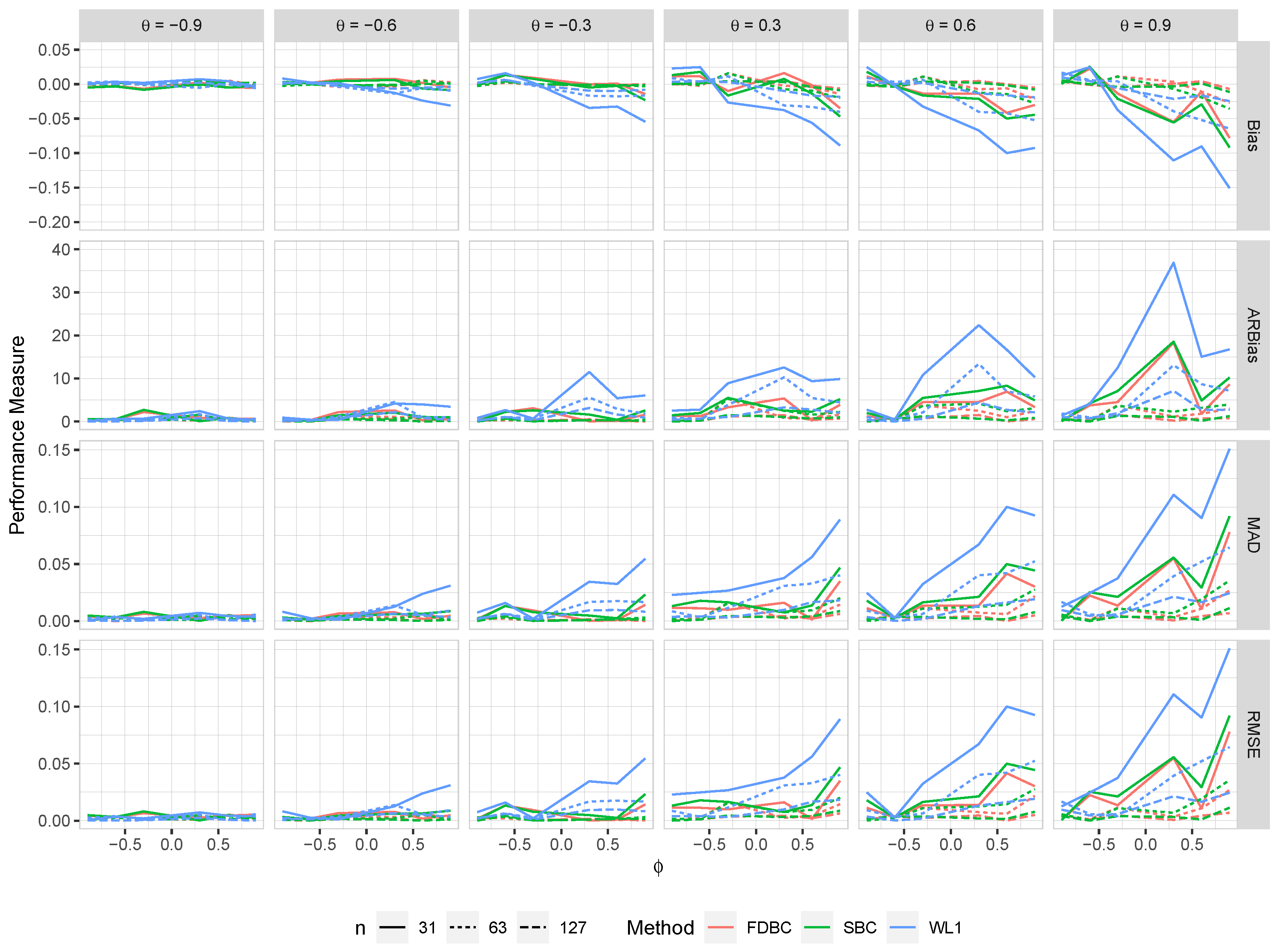
Generally, all proposed bias-correcting estimators substantially reduce the bias of the estimator across nearly all combinations of and , even with small sample sizes.
Both the single bootstrap () and fast double bootstrap () bias-corrected estimators effectively reduce bias. However, the fast double bootstrap estimator demonstrates superior performance compared to the single bootstrap estimator, particularly for small sample sizes (e.g., ). Despite this reduction, some residual bias persists when both and are positive, with the bias increasing as and approach the positive boundary.
For larger sample sizes, and , the fast double bootstrap estimator generally outperforms the single bootstrap estimator, though their performances are sometimes comparable. It is important to note that the fast double bootstrap method requires more computational time compared to the single bootstrap method.
Both bootstrap methods lead to significant reductions in RMSE and mean absolute deviation (MAD) compared to the estimator across all sample sizes and positive values of .
The bias reduction achieved by both bootstrap estimators is particularly notable for values near the boundaries (-1, +1), especially for small sample sizes (e.g., ). Additionally, the fast double bootstrap method outperforms the single bootstrap method in mitigating bias in the estimators.
Overall, these findings indicate that both the single and fast double bootstrap bias-corrected estimators are suitable for reducing bias in the estimation for the BAR(1) model under most combinations of and . Although the fast double bootstrap method is more effective in reducing bias, it requires more computational resources. Consequently, the single bootstrap approach may be preferred for its computational efficiency.
6. Conclusions
In this study, we revisited the weighted L1 (WL1) estimator within the framework of first-order bifurcating autoregressive BAR(1) models. While the WL1 estimator is known for its robustness to outliers, it can show substantial bias for small and moderate sample sizes, akin to the least squares estimator. The extent and direction of this bias are influenced by the autoregressive coefficient () and the correlation between model errors ().
To address this issue, we introduced bias-corrected versions of the WL1 estimator using the bootstrap method. Specifically, we developed the single bootstrap and fast double bootstrap bias-corrected WL1 estimators. Our extensive empirical analysis demonstrated that these bootstrap bias-corrected estimators effectively reduce the bias inherent in the WL1 estimator. The observed reduction in bias corresponded with decreased root mean squared error (RMSE), thereby enhancing the overall efficiency of the corrected estimators. Although the fast double bootstrap method is somewhat more computationally demanding than the single bootstrap, it consistently outperformed the latter in most of the scenarios considered in our simulations.
While our research primarily focused on BAR(1) models, future investigations will extend to higher-order BAR models (i.e., BAR()). Additionally, we plan to explore how bias and its correction impact related inferences, such as confidence intervals and hypothesis tests, to further assess the practical implications of our findings.
Author Contributions
Conceptualization, methodology, T.E. and S.M.; software, formal analysis and investigation, writing—original draft preparation, T.E.; writing—review and editing, T.E. and S.M. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The data and code supporting this study’s findings may be obtained from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Cowan, R. Statistical Concepts in the Analysis of Cell Lineage Data. 1983 Workshop Cell Growth Division. Melbourne: Latrobe University, 1984, pp. 18–22.
- Hawkins, E.D.; Markham, J.F.; McGuinness, L.P.; Hodgkin, P.D. A single-cell pedigree analysis of alternative stochastic lymphocyte fates. Proceedings of the National Academy of Sciences 2009, 106, 13457–13462. [Google Scholar] [CrossRef] [PubMed]
- Kimmel, M.; Axelrod, D. Branching Processes in Biology; Springer-Verlag: New York, 2005. [Google Scholar]
- Sandler, O.; Mizrahi, S.P.; Weiss, N.; Agam, O.; Simon, I.; Balaban, N.Q. Lineage correlations of single cell division time as a probe of cell-cycle dynamics. Nature 2015, 519, 468–471. [Google Scholar] [CrossRef] [PubMed]
- Cowan, R.; Staudte, R. The Bifurcating Autoregression Model in Cell Lineage Studies. Biometrics 1986, 42, 769–783. [Google Scholar] [CrossRef] [PubMed]
- Huggins, R.M. A law of large numbers for the bifurcating autoregressive process. 1995; 11, 273–278. [Google Scholar]
- Bui, Q.; Huggins, R. Inference for the random coefficients bifurcating autoregressive model for cell lineage studies. Journal of Statistical Planning and Inference 1999, 81, 253–262. [Google Scholar] [CrossRef]
- Huggins, R.M.; Basawa, I.V. Extensions of the Bifurcating Autoregressive Model for Cell Lineage Studies. Journal of Applied Probability 1999, 36, 1225–1233. [Google Scholar] [CrossRef]
- Huggins, R.; Basawa, I. Inference for the extended bifurcating autoregressive model for cell lineage studies. Australian & New Zealand Journal of Statistics 2000, 42, 423–432. [Google Scholar]
- Zhou, J.; Basawa, I. Least-squares estimation for bifurcating autoregressive processes. Statistics & Probability Letters 2005, 74, 77–88. [Google Scholar]
- Terpstra, J.T.; Elbayoumi, T. A law of large numbers result for a bifurcating process with an infinite moving average representation. Statistics & Probability Letters 2012, 82, 123–129. [Google Scholar]
- Elbayoumi, T.; Terpstra, J. Weighted L1-Estimates for the First-order Bifurcating Autoregressive Model. Communication in Statistics-Simulation and Computation 2016, 45, 2991–3013. [Google Scholar] [CrossRef]
- Elbayoumi, T.M.; Mostafa, S.A. On the estimation bias in first-order bifurcating autoregressive models. Stat, 10, e342. [CrossRef]
- Hurwicz, L. Least squares bias in time series. Statistical Inference in Dynamic Economic Models, 1950; 365–383. [Google Scholar]
- Huggins, R.M.; Marschner, I.C. Robust Analysis of the Bifurcating Autoregressive Model in Cell Lineage Studies. Australian & New Zealand Journal of Statistics 1991, 33, 209–220. [Google Scholar]
- Staudte, R.G. A bifurcating autoregression model for cell lineages with variable generation means. Journal of Theoretical Biology 1992, 156, 183–195. [Google Scholar] [CrossRef] [PubMed]
- MacKinnon, J.G.; Smith, A.A. Approximate bias correction in econometrics. Journal of Econometrics 1998, 85, 205–230. [Google Scholar] [CrossRef]
- Reber, J.C.; Terpstra, J.T.; Chen, X. Weighted L1-estimates for a VAR(p) time series model. Journal of Nonparametric Statistics 2008, 20, 395–411. [Google Scholar] [CrossRef]
- Rousseeuw, P.J.; Leroy, A.M. Robust Regression and Outlier Detection; John Wiley and Sons: New York, 1987. [Google Scholar]
- Naranjo, J.D.; Hettmansperger, T.P. Bounded Influence Rank Regression. 1994; 56, 209–220. [Google Scholar]
- Chang, W.H.; McKean, J.W.; Naranjo, J.D.; Sheather, S.J. High-Breakdown Rank Regression 1999. 94.
- Terpstra, J.T.; McKean, J.W.; Naranjo, J.D. Highly efficient weighted for autoregression Wilcoxon estimates for autoregression. 2000; 35, 45–80. [Google Scholar]
- McKean, J.M.; Schrader, R.M. A comparison of methods for studentizing the sample median. Communications in Statistics, Part B-Simulation and Computation 1984, 6, 751–773. [Google Scholar] [CrossRef]
- Sheather, S.J. Assessing the accuracy of the sample median: estimated standard errors versus interpolated confidence interval. In: Dodge, Y., ed. Statistical Data Analysis Based on the L1 Norm and Related Methods 1987, 6, 203–216. [Google Scholar]
- Berkowitz, J.; Kilian, L. Recent developments in bootstrapping time series. Econometric Reviews 2000, 19, 1–48. [Google Scholar] [CrossRef]
- Tanizaki, H.; Hamori, S.; Matsubayashi, Y. On least-squares bias in the AR(p) models: Bias correction using the bootstrap methods. Statistical Papers 2006, 47, 109–124. [Google Scholar] [CrossRef]
- Patterson, K. Bias Reduction through First-order Mean Correction, Bootstrapping and Recursive Mean Adjustment. Journal of Applied Statistics 2007, 34, 23–45. [Google Scholar] [CrossRef]
- Liu-Evans, G.D.; Phillips, G.D. Bootstrap, Jackknife and COLS: Bias and Mean Squared Error in Estimation of Autoregressive Models. Journal of Time Series Econometrics 2012, 4, 1–33. [Google Scholar] [CrossRef]
- Hall, P. The Bootstrap and Edgeworth Expansion; Springer-Verlag: New York, 1992. [Google Scholar]
- Lee, S.M.S.; Young, G.A. The effect of Monte Carlo approximation on coverage error of double-bootstrap confidence intervals. Journal of the Royal Statistical Society: Series B, 1999; 61, 353–1999. [Google Scholar]
- Shi, S.G. Accurate and Efficient Double-bootstrap Confidence Limit Method. Computational Statistics and Data Analysis 1992, 13, 21–32. [Google Scholar] [CrossRef]
- Chang, J.; Hall, P. Double-bootstrap methods that use a single double-bootstrap simulation. Biometrika 2015, 102, 203–214. [Google Scholar] [CrossRef]
- Ouysse, R. A Fast Iterated Bootstrap Procedure for Approximating the Small-Sample Bias. Communications in Statistics - Simulation and Computation 2013, 42, 1472–1494. [Google Scholar] [CrossRef]
- R Core Team. In R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2022.
- Elbayoumi, T.; Mostafa, S. bifurcatingr: Bifurcating Autoregressive Models, 2021. R package version 2.0.0.
- Fox, A.J. Outliers in Time Series. Journal of Royal Statistical Society B 1972, 34, 350–363. [Google Scholar] [CrossRef]
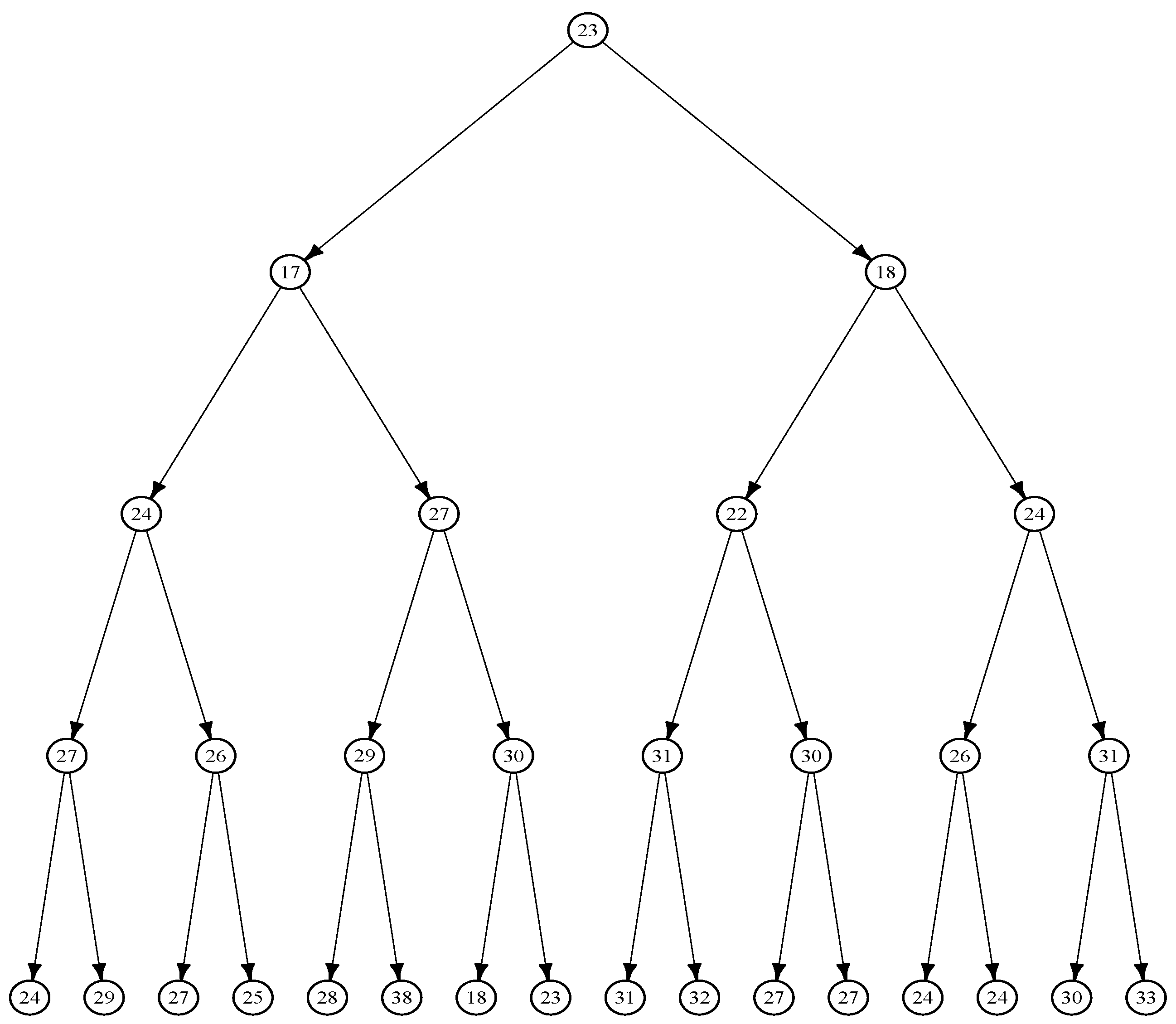
Figure 1.
The lifetimes (in minutes) of
E.Coli cells are represented in this tree, constructed using data from [
5].
Figure 1.
The lifetimes (in minutes) of
E.Coli cells are represented in this tree, constructed using data from [
5].
Figure 2.
Empirical bias and ARB (%) of the estimator of when the errors distribution is a 5% or 10% contaminated bivariate normal distribution.
Figure 2.
Empirical bias and ARB (%) of the estimator of when the errors distribution is a 5% or 10% contaminated bivariate normal distribution.
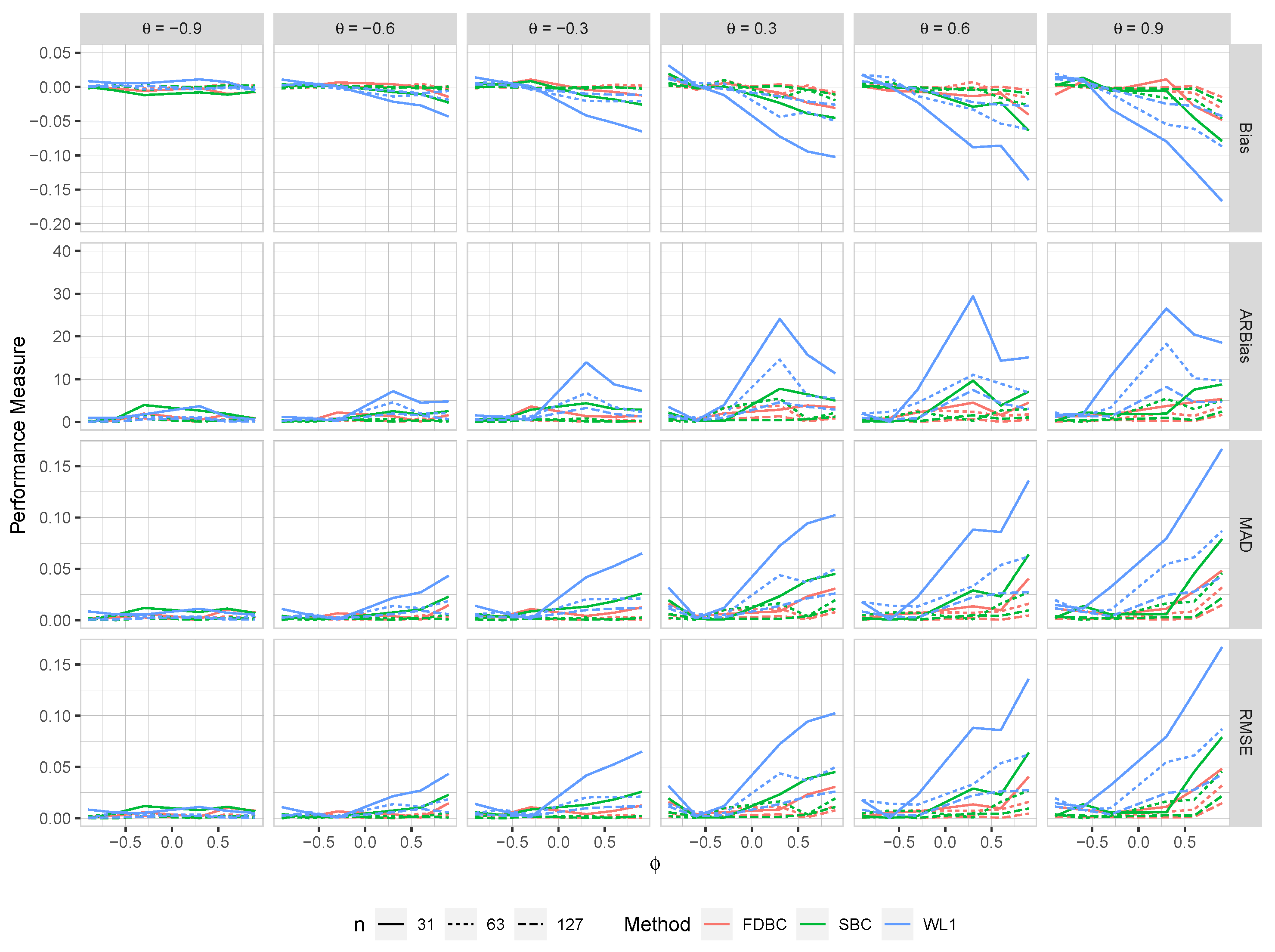
Figure 3.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
Figure 3.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
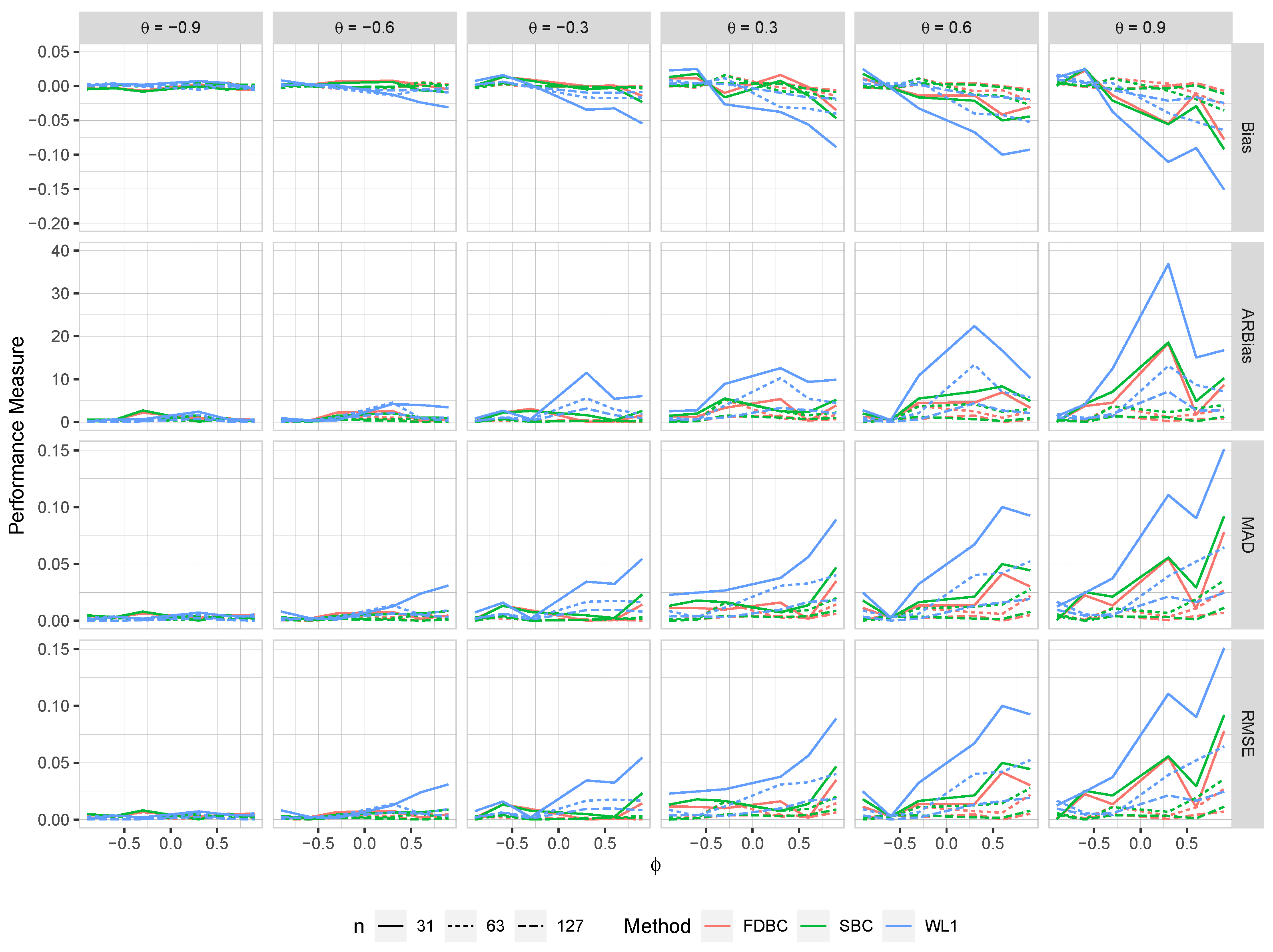
Figure 4.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
Figure 4.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
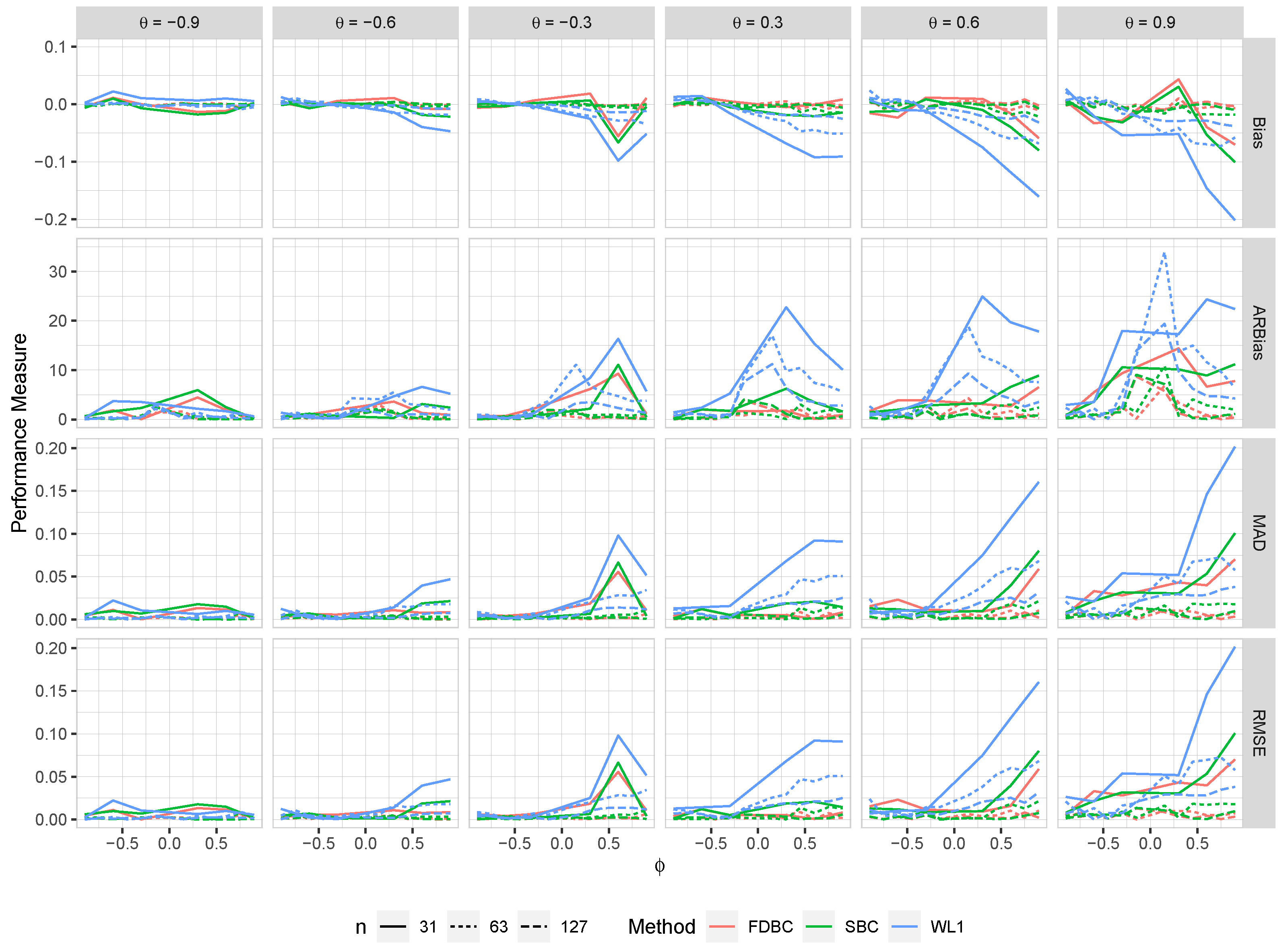
Figure 5.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
Figure 5.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
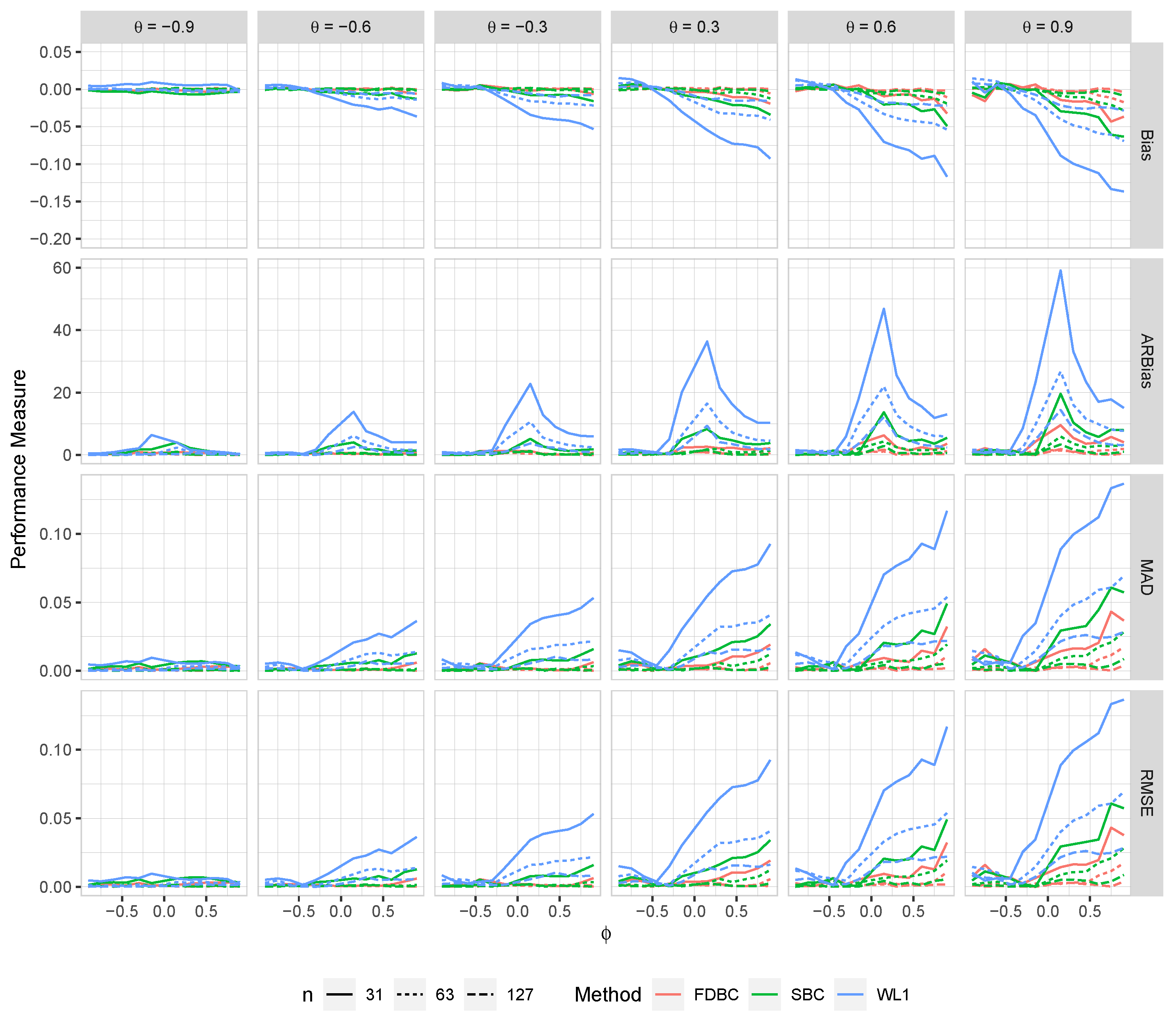
Figure 6.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
Figure 6.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an IO model with a bivariate contaminated normal distribution, .
Figure 7.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
Figure 7.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
Figure 8.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
Figure 8.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
Figure 9.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
Figure 9.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
Figure 10.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
Figure 10.
The bias, ARB, MAD, and RMSE of the , single bootstrap bias-corrected , and fast double bootstrap bias-corrected estimators as a function of at different levels of under an AO model with a bivariate contaminated normal distribution, .
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).