Submitted:
23 September 2024
Posted:
24 September 2024
You are already at the latest version
Abstract
Keywords:
1. Streptomyces Complex Metabolism—A Great Source of Valuable Targets for Engineering
1.1. Microbial Diversity as a Basis for Metabolic Diversity
1.2. Environment of Streptomyces, Biological Natural Product Producers
1.3. Primary Metabolism in Streptomyces as an Important Part of Secondary Metabolism
1.4. Streptomyces as Producers of Secondary Metabolites
1.5. Hosts for Heterologous Expression
2. Genetic Tools for Streptomyces
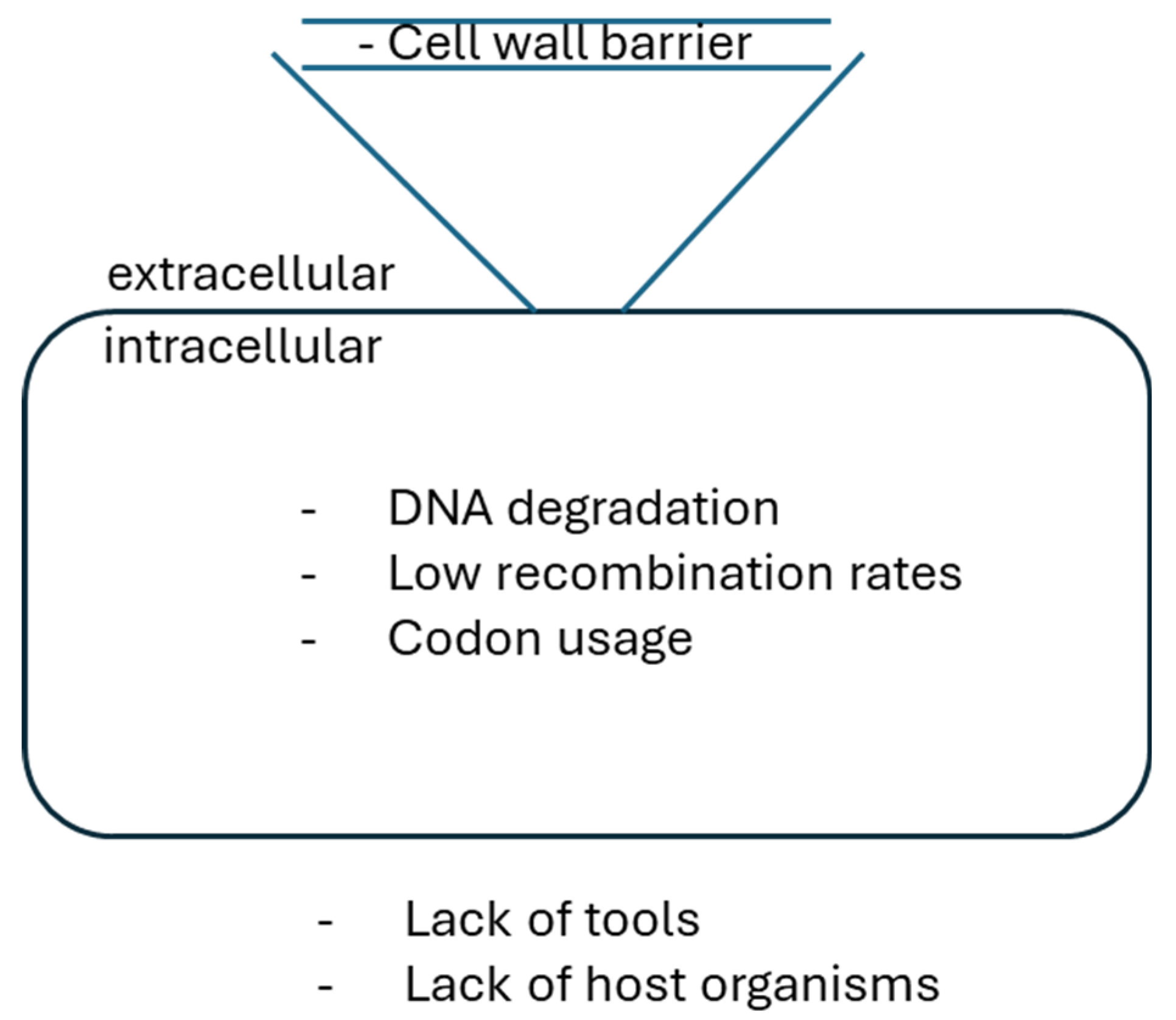
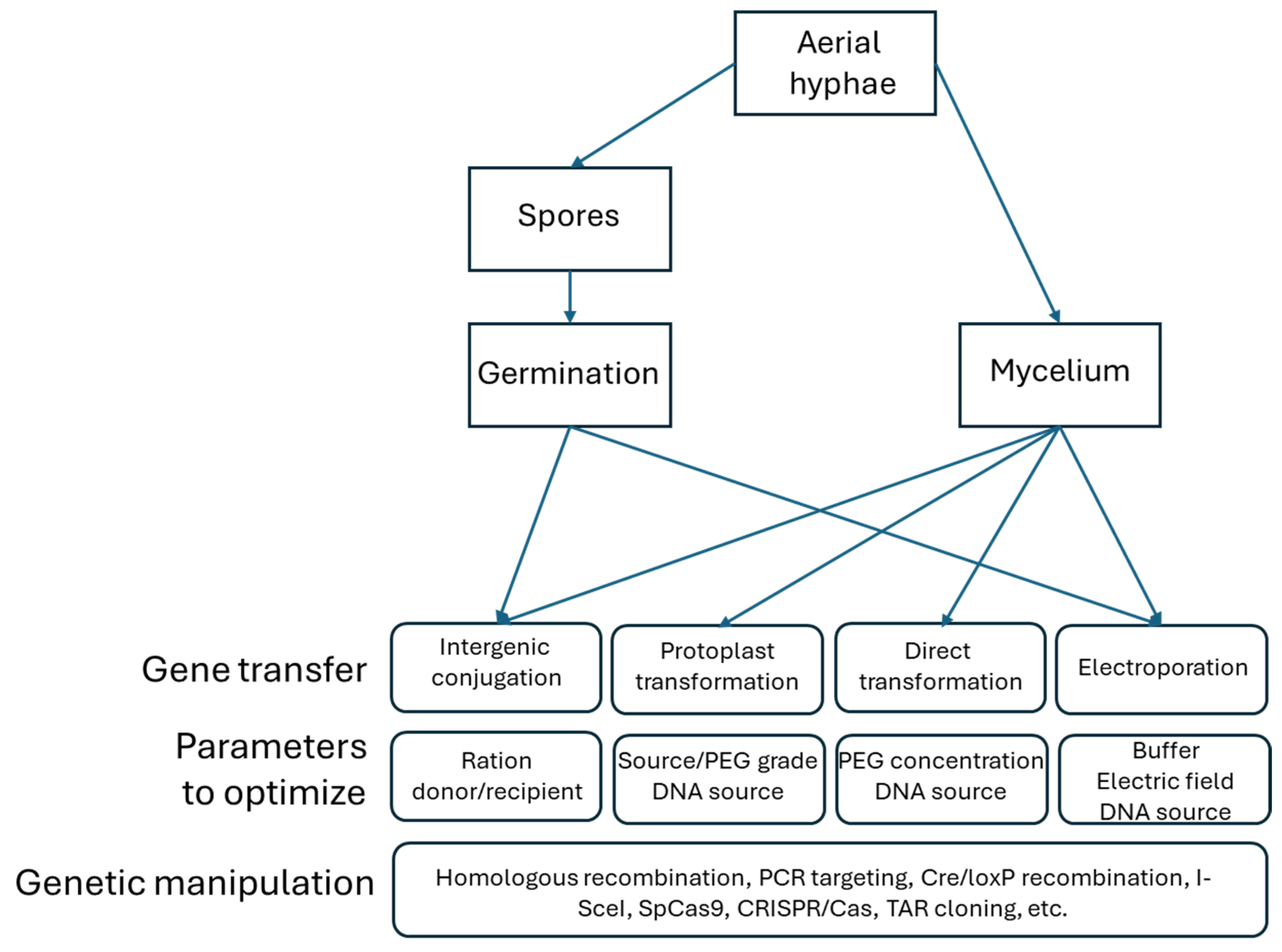
2.1. Methods for Heterologous DNA Transfer into Filamentous Actinobacteria
2.2. Synthetic Parts
2.2.1. Integrative and Replicative Plasmid Expression Systems for Actinomycetes-Cloning Vectors for the Genetic Manipulation of Filamentous Actinomycetes
2.2.2. Promoters
2.2.3. Reporter Genes
2.2.4. Ribosome-Binding Sites
2.2.5. Terminators
2.2.6. Riboswitches for Biosensors
2.3. Bioinformatics-Based Approaches for Natural Products Discovery
2.4. Assembly Strategies for Generation of Constructs for Genetic Engineering of Streptomyces
2.5. Genetic Approaches for Streptomyces Engineering
2.5.1. Transposon- and Homologous Recombination-Based Systems for Actinomycetes Engineering-I-SceI Meganuclease-Promoted Recombination System
2.5.2. PCR Targeting
2.5.3. Cre/loxP and Flp/FRT
2.5.4. CRISPR/Cas9-Based Editing Tools in Streptomyces
2.5.5. CRISPR/Cas9 TAR Cloning Approach

| Technology | Category | Feature | Reference |
|---|---|---|---|
| Target BGC acquisition | Genetic manipulation strategy | Transfer into a target host using a genomic library of cosmids, fosmids, BAC, PAC | [60,61,62] |
| Ligation and BGC assembly to the vector | Genetic manipulation strategy | Sticky/blunt end ligation, Gibson cloning, recombination in different hosts, etc. | [48,54,59] |
| Transfer of the BGC-encoded vector to theheterologous host for expression | Genetic manipulation strategy | Biparental conjugation, protoplast transformation | [54] |
| Target secondary metabolite production by expression of the BGC vector | Genetic manipulation strategy | Expression of integrative (e.g., pSET152, pIJ8600, pOJ436, pOJ444) or replicative (e.g., pUWL218, pUWL219, pUWL-SK, pUWL-KS) vectors | [60,61,62] |
| iCatch | Assembly strategy | Facilitate “catching” of large DNA regions like actinobacterial BGCs | [86] |
| DiPAC | Assembly strat-egy | Assembly of complete biosynthetic pathways by covering full BGCs with long-amplicon PCR | [87] |
| AGOS | Assembly strat-egy | Reconstruction and assembly of gene operons | [88] |
| PCR-targetingsystem | Genetic manipulation strategy | Nonpolar/in-frame deletion of genes/gene clusters in Streptomyces | [55] |
| Cre-loxP/Flp-FRP recombination systems | Genetic manipulation strategy | Knock out large DNA fragments in Streptomyces | [56,92] |
| I-SceI promoted recombination system | Genetic manipulation strategy | DNA double-strand breaks (DSBs), which promote double-crossover recombination events | [90] |
| SpCas9-based genome editing | Genetic manipulation strategy | Transcribed synthetic guide RNA to direct Cas proteins to any site on the genome. Editing plasmids: pCRISPomyces-1/2, pKCas9dO, pCRISPR-Cas9-ScaligD, and pWHU2653 | [94,95,96,97] |
| CRISPRi-mediated gene repression for single cells | Genetic manipulation strategy | Gene repression tool based on dCas9 or ddCpf1 and the base editors (BEs) for targeted base mutagenesis based on dCas9 or Cas9n | [95] |
| FnCpf1-based genome editing and CRISPRi | Genetic manipulation strategy | Editing plasmids: pKCCpf1, pKCCpf1-MsmE, and pSETddCpf1, etc. | [103] |
| CRISPR/Cas-based base editing tools | Genetic manipulation strategy | Editing plasmids: pCRISPR-cBEST/-aBEST, and pKC-dCas9-CDA-ULstr, etc. | [58,94,95,96] |
| Alternative CRISPR/Cas-based genome editing | Genetic manipulation strategy | Editing plasmids: pCRISPomyces-FnCpf1, pCRISPomyces-Sth1Cas9, and pCRISPomyces-SaCas9, etc. | [101,102,103,104] |
| TAR-cloning | Genetic manipulation strategy | Isolation of large chromosomal regions without the constructing a random clone library | [109,110] |
| Synthetic promoters | Genetics parts | Constitutive ermE, SF14P, kasOP, gapdh, rpsL promoters as well as inducible tipA nitA and xylA promoters | [59,60,72] |
| Ribosome-binding sites | Genetics parts | AAAGGAGG, diverse native or synthetic RBSs | [80,81] |
| Terminators | Genetics parts | Fdand TD1 | [59,82] |
| Reporter genes | Genetics parts | luxAB, amy, xylE, and gusA / eGFP, sfGFP, mRFP, mCherry | [59] |
| Riboswitches/Biosensors | Genetics parts | Non-protein coding RNAs that can regulate cellular processes including transcription and translation | [83,84] |
3. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Saiki, R. K., Gelfand, D. H., Stoffel, S., Scharf, S. J., Higuchi, R., Horn, G. T., Mullis, K. B., & Erlich, H. A. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science (New York, N.Y.), 239(4839), 487–491. [CrossRef]
- Liu, F., Xiong, J., Kumar, S., Yang, C., Ge, S., Li, S., Xia, N., & Swaminathan, K. (2011). Structural and biophysical characterization of Mycobacterium tuberculosis dodecin Rv1498A. Journal of structural biology, 175(1), 31–38. [CrossRef]
- Hibbing, M. E., Fuqua, C., Parsek, M. R., & Peterson, S. B. (2010). Bacterial competition: surviving and thriving in the microbial jungle. Nature reviews. Microbiology, 8(1), 15–25. [CrossRef]
- Stubbendieck, R. M., Vargas-Bautista, C., & Straight, P. D. (2016). Bacterial Communities: Interactions to Scale. Frontiers in microbiology, 7, 1234. [CrossRef]
- Crawford, J. M., & Clardy, J. (2011). Bacterial symbionts and natural products. Chemical communications (Cambridge, England), 47(27), 7559–7566. [CrossRef]
- Netzker, T., Fischer, J., Weber, J., Mattern, D. J., König, C. C., Valiante, V., Schroeckh, V., & Brakhage, A. A. (2015). Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Frontiers in microbiology, 6, 299. [CrossRef]
- Schrempf H, Cypionka H, Keller U (2016) Streptomyceten. Biologie in unserer Zeit 46:236-243. [CrossRef]
- Goodfellow, M.; Nouioui, I.; Sanderson, R.; Xie, F.; Bull, A.T. Rare Taxa and Dark Microbial Matter: Novel Bioactive Actinobacteria Abound in Atacama Desert Soils. Antonie Van Leeuwenhoek 2018, 111, 1315–1332.
- Seipke, R. F., Kaltenpoth, M., & Hutchings, M. I. (2012). Streptomyces as symbionts: an emerging and widespread theme?. FEMS microbiology reviews, 36(4), 862–876. [CrossRef]
- Haeder, S., Wirth, R., Herz, H., & Spiteller, D. (2009). Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proceedings of the National Academy of Sciences of the United States of America, 106(12), 4742–4746. [CrossRef]
- Kaltenpoth, M., Yildirim, E., Gürbüz, M. F., Herzner, G., & Strohm, E. (2012). Refining the roots of the beewolf-Streptomyces symbiosis: antennal symbionts in the rare genus Philanthinus (Hymenoptera, Crabronidae). Applied and environmental microbiology, 78(3), 822–827. [CrossRef]
- Pantigoso, H. A., Newberger, D., & Vivanco, J. M. (2022). The rhizosphere microbiome: Plant-microbial interactions for resource acquisition. Journal of applied microbiology, 133(5), 2864–2876. [CrossRef]
- Upadhyay, S. K., Srivastava, A. K., Rajput, V. D., Chauhan, P. K., Bhojiya, A. A., Jain, D., Chaubey, G., Dwivedi, P., Sharma, B., & Minkina, T. (2022). Root Exudates: Mechanistic Insight of Plant Growth Promoting Rhizobacteria for Sustainable Crop Production. Frontiers in microbiology, 13, 916488. [CrossRef]
- Duan, S., Feng, G., Limpens, E., Bonfante, P., Xie, X., & Zhang, L. (2024). Cross-kingdom nutrient exchange in the plant-arbuscular mycorrhizal fungus-bacterium continuum. Nature reviews. Microbiology, 10.1038/s41579-024-01073-7. Advance online publication. [CrossRef]
- Zhang, L., Zhou, J., George, T. S., Limpens, E., & Feng, G. (2022). Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends in plant science, 27(4), 402–411. [CrossRef]
- Jin, Z., Jiang, F., Wang, L., Declerck, S., Feng, G., & Zhang, L. (2024). Arbuscular mycorrhizal fungi and Streptomyces: brothers in arms to shape the structure and function of the hyphosphere microbiome in the early stage of interaction. Microbiome, 12(1), 83. [CrossRef]
- Le, K. D., Yu, N. H., Park, A. R., Park, D. J., Kim, C. J., & Kim, J. C. (2022). Streptomyces sp. AN090126 as a Biocontrol Agent against Bacterial and Fungal Plant Diseases. Microorganisms, 10(4), 791. [CrossRef]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. Practical Streptomyces Genetics, John Innes Foundation, Norwich, 2000.
- Ismail, S., Jiang, B., Nasimi, Z., Inam-Ul-Haq, M., Yamamoto, N., Danso Ofori, A., Khan, N., Arshad, M., Abbas, K., & Zheng, A. (2020). Investigation of Streptomycesscabies Causing Potato Scab by Various Detection Techniques, Its Pathogenicity and Determination of Host-Disease Resistance in Potato Germplasm. Pathogens (Basel, Switzerland), 9(9), 760. [CrossRef]
- Pérez, J., Muñoz-Dorado, J., Braña, A. F., Shimkets, L. J., Sevillano, L., & Santamaría, R. I. (2011). Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microbial biotechnology, 4(2), 175–183. [CrossRef]
- Yagüe, P., Lopez-Garcia, M. T., Rioseras, B., Sanchez, J., & Manteca, A. (2012). New insights on the development of Streptomyces and their relationships with secondary metabolite production. Current trends in microbiology, 8, 65–73.
- Bentley, S. D., Chater, K. F., Cerdeño-Tárraga, A. M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., Harper, D., Bateman, A., Brown, S., Chandra, G., Chen, C. W., Collins, M., Cronin, A., Fraser, A., Goble, A., Hidalgo, J., Hornsby, T., … Hopwood, D. A. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature, 417(6885), 141–147. [CrossRef]
- Krysenko, S.; Wohlleben, W. Role of Carbon, Nitrogen, Phosphate and Sulfur Metabolism in Secondary Metabolism Precursor Supply in Streptomyces spp. Microorganisms 2024, 12, 1571. [CrossRef]
- Hodgson D. A. (2000). Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Advances in microbial physiology, 42, 47–238. [CrossRef]
- Krysenko S, Wohlleben W. (2022) Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity. Medical Sciences; 10(3):40. [CrossRef]
- Görke, B., & Stülke, J. (2008). Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nature reviews. Microbiology, 6(8), 613–624. [CrossRef]
- Romero-Rodríguez, A., Rocha, D., Ruiz-Villafán, B., Guzmán-Trampe, S., Maldonado-Carmona, N., Vázquez-Hernández, M., Zelarayán, A., Rodríguez-Sanoja, R., & Sánchez, S. (2017). Carbon catabolite regulation in Streptomyces: new insights and lessons learned. World journal of microbiology & biotechnology, 33(9), 162. [CrossRef]
- Jahreis, K., Pimentel-Schmitt, E. F., Brückner, R., & Titgemeyer, F. (2008). Ins and outs of glucose transport systems in eubacteria. FEMS microbiology reviews, 32(6), 891–907. [CrossRef]
- Merrick, M. J., & Edwards, R. A. (1995). Nitrogen control in bacteria. Microbiological reviews, 59(4), 604–622. [CrossRef]
- Fink, D., Weissschuh, N., Reuther, J., Wohlleben, W., & Engels, A. (2002). Two transcriptional regulators GlnR and GlnRII are involved in regulation of nitrogen metabolism in Streptomyces coelicolor A3(2). Molecular microbiology, 46(2), 331–347. [CrossRef]
- Fink, D., Falke, D., Wohlleben, W., & Engels, A. (1999). Nitrogen metabolism in Streptomyces coelicolor A3(2): modification of glutamine synthetase I by an adenylyltransferase. Microbiology (Reading, England), 145 ( Pt 9), 2313–2322. [CrossRef]
- Ghoshroy, S., Binder, M., Tartar, A., & Robertson, D. L. (2010). Molecular evolution of glutamine synthetase II: Phylogenetic evidence of a non-endosymbiotic gene transfer event early in plant evolution. BMC evolutionary biology, 10, 198. [CrossRef]
- Reuther, J., & Wohlleben, W. (2007). Nitrogen metabolism in Streptomyces coelicolor: transcriptional and post-translational regulation. Journal of molecular microbiology and biotechnology, 12(1-2), 139–146. [CrossRef]
- Hillemann, D., Dammann, T., Hillemann, A., & Wohlleben, W. (1993). Genetic and biochemical characterization of the two glutamine synthetases GSI and GSII of the phosphinothricyl-alanyl-alanine producer, streptomyces viridochromogenes Tü494. Journal of general microbiology, 139(8), 1773–1783. [CrossRef]
- Krysenko S, Emani CS, Bäuerle M, Oswald M, Kulik A, Meyners C, Hillemann D, Merker M, Wohlers I, Hausch F, Brötz-Oesterhelt H, Mitulski A, Reiling N, Wohlleben W. n.d. Glna3Mt is able to Glutamylate Spermine but it is not essential for the detoxification of Spermine in Mycobacterium tuberculosis. bioRxiv. [CrossRef]
- Martín, J. F., & Liras, P. (2020). The Balance Metabolism Safety Net: Integration of Stress Signals by Interacting Transcriptional Factors in Streptomyces and Related Actinobacteria. Frontiers in microbiology, 10, 3120. [CrossRef]
- Romero-Rodríguez, A., Maldonado-Carmona, N., Ruiz-Villafán, B., Koirala, N., Rocha, D., & Sánchez, S. (2018). Interplay between carbon, nitrogen and phosphate utilization in the control of secondary metabolite production in Streptomyces. Antonie van Leeuwenhoek, 111(5), 761–781. [CrossRef]
- Ghorbel, S., Smirnov, A., Chouayekh, H., Sperandio, B., Esnault, C., Kormanec, J., & Virolle, M. J. (2006). Regulation of ppk expression and in vivo function of Ppk in Streptomyces lividans TK24. Journal of bacteriology, 188(17), 6269–6276. [CrossRef]
- Ferla, M. P., & Patrick, W. M. (2014). Bacterial methionine biosynthesis. Microbiology (Reading, England), 160(Pt 8), 1571–1584. [CrossRef]
- Kulkarni, A., Zeng, Y., Zhou, W., Van Lanen, S., Zhang, W., & Chen, S. (2015). A Branch Point of Streptomyces Sulfur Amino Acid Metabolism Controls the Production of Albomycin. Applied and environmental microbiology, 82(2), 467–477. [CrossRef]
- De Simeis, D., & Serra, S. (2021). Actinomycetes: A Never-Ending Source of Bioactive Compounds-An Overview on Antibiotics Production. Antibiotics (Basel, Switzerland), 10(5), 483. [CrossRef]
- Alam, K., Mazumder, A., Sikdar, S., Zhao, Y. M., Hao, J., Song, C., Wang, Y., Sarkar, R., Islam, S., Zhang, Y., & Li, A. (2022). Streptomyces: The biofactory of secondary metabolites. Frontiers in microbiology, 13, 968053. [CrossRef]
- Krysenko, S. Impact of Nitrogen-Containing Compounds on Secondary Metabolism in Streptomyces spp.—A Source of Metabolic Engineering Strategies. SynBio 2023, 1, 204-225. [CrossRef]
- Alam, K., Mazumder, A., Sikdar, S., Zhao, Y. M., Hao, J., Song, C., Wang, Y., Sarkar, R., Islam, S., Zhang, Y., & Li, A. (2022). Streptomyces: The biofactory of secondary metabolites. Frontiers in microbiology, 13, 968053. [CrossRef]
- Albarano, L., Esposito, R., Ruocco, N., & Costantini, M. (2020). Genome Mining as New Challenge in Natural Products Discovery. Marine drugs, 18(4), 199. [CrossRef]
- Miethke, M., Pieroni, M., Weber, T., Brönstrup, M., Hammann, P., Halby, L., Arimondo, P. B., Glaser, P., Aigle, B., Bode, H. B., Moreira, R., Li, Y., Luzhetskyy, A., Medema, M. H., Pernodet, J. L., Stadler, M., Tormo, J. R., Genilloud, O., Truman, A. W., Weissman, K. J., … Müller, R. (2021). Towards the sustainable discovery and development of new antibiotics. Nature reviews. Chemistry, 5(10), 726–749. [CrossRef]
- Hwang, S., Lee, Y., Kim, J. H., Kim, G., Kim, H., Kim, W., Cho, S., Palsson, B. O., & Cho, B. K. (2021). Streptomyces as Microbial Chassis for Heterologous Protein Expression. Frontiers in bioengineering and biotechnology, 9, 804295. [CrossRef]
- Mitousis, L.; Thoma, Y.; Musiol-Kroll, E.M. An Update on Molecular Tools for Genetic Engineering of Actinomycetes—The Source of Important Antibiotics and Other Valuable Compounds. Antibiotics 2020, 9, 494. [CrossRef]
- Myronovskyi, M., Welle, E., Fedorenko, V., & Luzhetskyy, A. (2011). Beta-glucuronidase as a sensitive and versatile reporter in actinomycetes. Applied and environmental microbiology, 77(15), 5370–5383. [CrossRef]
- Schulz, S., Schall, C., Stehle, T., Breitmeyer, C., Krysenko, S., Mitulski, A., & Wohlleben, W. (2022). Optimization of the precursor supply for an enhanced FK506 production in Streptomyces tsukubaensis. Frontiers in bioengineering and biotechnology, 10, 1067467. [CrossRef]
- Kim, W., Hwang, S., Lee, N., Lee, Y., Cho, S., Palsson, B., & Cho, B. K. (2020). Transcriptome and translatome profiles of Streptomyces species in different growth phases. Scientific data, 7(1), 138. [CrossRef]
- Wohlleben, W., Hartmann, V., Hillemann, D., Krey, K., Muth, G., Nussbaumer, B., & Pelzer, S. (1994). Transfer and establishment of DNA in Streptomyces (a brief review). Acta microbiologica et immunologica Hungarica, 41(4), 381–389.
- Kormanec, J., Rezuchova, B., Homerova, D., Csolleiova, D., Sevcikova, B., Novakova, R., & Feckova, L. (2019). Recent achievements in the generation of stable genome alterations/mutations in species of the genus Streptomyces. Applied microbiology and biotechnology, 103(14), 5463–5482. [CrossRef]
- Musiol-Kroll, E. M., Tocchetti, A., Sosio, M., & Stegmann, E. (2019). Challenges and advances in genetic manipulation of filamentous actinomycetes - the remarkable producers of specialized metabolites. Natural product reports, 36(9), 1351–1369. [CrossRef]
- Gust, B., Challis, G. L., Fowler, K., Kieser, T., & Chater, K. F. (2003). PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proceedings of the National Academy of Sciences of the United States of America, 100(4), 1541–1546. [CrossRef]
- Komatsu, M., Uchiyama, T., Omura, S., Cane, D. E., & Ikeda, H. (2010). Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proceedings of the National Academy of Sciences of the United States of America, 107(6), 2646–2651. [CrossRef]
- Lu, Z., Xie, P., & Qin, Z. (2010). Promotion of markerless deletion of the actinorhodin biosynthetic gene cluster in Streptomyces coelicolor. Acta biochimica et biophysica Sinica, 42(10), 717–721. [CrossRef]
- Lee, Y., Hwang, S., Kim, W., Kim, J. H., Palsson, B. O., & Cho, B. K. (2024). CRISPR-aided genome engineering for secondary metabolite biosynthesis in Streptomyces. Journal of industrial microbiology & biotechnology, 51, kuae009. [CrossRef]
- Lee, N., Hwang, S., Lee, Y., Cho, S., Palsson, B., & Cho, B. K. (2019). Synthetic Biology Tools for Novel Secondary Metabolite Discovery in Streptomyces. Journal of microbiology and biotechnology, 29(5), 667–686. [CrossRef]
- Aubry, C., Pernodet, J. L., & Lautru, S. (2019). Modular and Integrative Vectors for Synthetic Biology Applications in Streptomyces spp. Applied and environmental microbiology, 85(16), e00485-19. [CrossRef]
- Thoma, L., & Muth, G. (2016). Conjugative DNA-transfer in Streptomyces, a mycelial organism. Plasmid, 87-88, 1–9. [CrossRef]
- Wang, W., Zheng, G., & Lu, Y. (2021). Recent Advances in Strategies for the Cloning of Natural Product Biosynthetic Gene Clusters. Frontiers in bioengineering and biotechnology, 9, 692797. [CrossRef]
- Kieser, T., & Melton, R. E. (1988). Plasmid pIJ699, a multi-copy positive-selection vector for Streptomyces. Gene, 65(1), 83–91. [CrossRef]
- Torsvik, V., & Øvreås, L. (2002). Microbial diversity and function in soil: from genes to ecosystems. Current opinion in microbiology, 5(3), 240–245. [CrossRef]
- Omura, S., Ikeda, H., Ishikawa, J., Hanamoto, A., Takahashi, C., Shinose, M., Takahashi, Y., Horikawa, H., Nakazawa, H., Osonoe, T., Kikuchi, H., Shiba, T., Sakaki, Y., & Hattori, M. (2001). Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proceedings of the National Academy of Sciences of the United States of America, 98(21), 12215–12220. [CrossRef]
- Wnuk, E., Waśko, A., Walkiewicz, A., Bartmiński, P., Bejger, R., Mielnik, L., & Bieganowski, A. (2020). The effects of humic substances on DNA isolation from soils. PeerJ, 8, e9378. [CrossRef]
- Reyrat, J. M., Pelicic, V., Gicquel, B., & Rappuoli, R. (1998). Counterselectable markers: untapped tools for bacterial genetics and pathogenesis. Infection and immunity, 66(9), 4011–4017. [CrossRef]
- Jäger, W., Schäfer, A., Pühler, A., Labes, G., & Wohlleben, W. (1992). Expression of the Bacillus subtilis sacB gene leads to sucrose sensitivity in the gram-positive bacterium Corynebacterium glutamicum but not in Streptomyces lividans. Journal of bacteriology, 174(16), 5462–5465. [CrossRef]
- Hosted, T. J., & Baltz, R. H. (1997). Use of rpsL for dominance selection and gene replacement in Streptomyces roseosporus. Journal of bacteriology, 179(1), 180–186. [CrossRef]
- Martínez-García, E., Goñi-Moreno, A., Bartley, B., McLaughlin, J., Sánchez-Sampedro, L., Pascual Del Pozo, H., Prieto Hernández, C., Marletta, A. S., De Lucrezia, D., Sánchez-Fernández, G., Fraile, S., & de Lorenzo, V. (2020). SEVA 3.0: an update of the Standard European Vector Architecture for enabling portability of genetic constructs among diverse bacterial hosts. Nucleic acids research, 48(D1), D1164–D1170. [CrossRef]
- Fayed, B., Ashford, D. A., Hashem, A. M., Amin, M. A., El Gazayerly, O. N., Gregory, M. A., & Smith, M. C. (2015). Multiplexed integrating plasmids for engineering of the erythromycin gene cluster for expression in Streptomyces spp. and combinatorial biosynthesis. Applied and environmental microbiology, 81(24), 8402–8413. [CrossRef]
- Wang, W., Li, X., Wang, J., Xiang, S., Feng, X., & Yang, K. (2013). An engineered strong promoter for streptomycetes. Applied and environmental microbiology, 79(14), 4484–4492. [CrossRef]
- Seghezzi, N., Amar, P., Koebmann, B., Jensen, P. R., & Virolle, M. J. (2011). The construction of a library of synthetic promoters revealed some specific features of strong Streptomyces promoters. Applied microbiology and biotechnology, 90(2), 615–623. [CrossRef]
- Li, S., Wang, J., Li, X., Yin, S., Wang, W., & Yang, K. (2015). Genome-wide identification and evaluation of constitutive promoters in streptomycetes. Microbial cell factories, 14, 172. [CrossRef]
- Takano, E., White, J., Thompson, C. J., & Bibb, M. J. (1995). Construction of thiostrepton-inducible, high-copy-number expression vectors for use in Streptomyces spp. Gene, 166(1), 133–137. [CrossRef]
- Rodríguez-García, A., Combes, P., Pérez-Redondo, R., Smith, M. C., & Smith, M. C. (2005). Natural and synthetic tetracycline-inducible promoters for use in the antibiotic-producing bacteria Streptomyces. Nucleic acids research, 33(9), e87. [CrossRef]
- Horbal, L., Fedorenko, V., & Luzhetskyy, A. (2014). Novel and tightly regulated resorcinol and cumate-inducible expression systems for Streptomyces and other actinobacteria. Applied microbiology and biotechnology, 98(20), 8641–8655. [CrossRef]
- Phelan, R. M., Sachs, D., Petkiewicz, S. J., Barajas, J. F., Blake-Hedges, J. M., Thompson, M. G., Reider Apel, A., Rasor, B. J., Katz, L., & Keasling, J. D. (2017). Development of Next Generation Synthetic Biology Tools for Use in Streptomyces venezuelae. ACS synthetic biology, 6(1), 159–166. [CrossRef]
- Szafran, M. J., Gongerowska, M., Małecki, T., Elliot, M., & Jakimowicz, D. (2019). Transcriptional Response of Streptomyces coelicolor to Rapid Chromosome Relaxation or Long-Term Supercoiling Imbalance. Frontiers in microbiology, 10, 1605. [CrossRef]
- Na, D., & Lee, D. (2010). RBSDesigner: software for designing synthetic ribosome binding sites that yields a desired level of protein expression. Bioinformatics (Oxford, England), 26(20), 2633–2634. [CrossRef]
- Yi, J. S., Kim, M. W., Kim, M., Jeong, Y., Kim, E. J., Cho, B. K., & Kim, B. G. (2017). A Novel Approach for Gene Expression Optimization through Native Promoter and 5′ UTR Combinations Based on RNA-seq, Ribo-seq, and TSS-seq of Streptomyces coelicolor. ACS synthetic biology, 6(3), 555–565. [CrossRef]
- Hwang, S., Lee, N., Choe, D., Lee, Y., Kim, W., Kim, J. H., Kim, G., Kim, H., Ahn, N. H., Lee, B. H., Palsson, B. O., & Cho, B. K. (2022). System-Level Analysis of Transcriptional and Translational Regulatory Elements in Streptomyces griseus. Frontiers in bioengineering and biotechnology, 10, 844200. [CrossRef]
- Winkler W. C. (2005). Riboswitches and the role of noncoding RNAs in bacterial metabolic control. Current opinion in chemical biology, 9(6), 594–602. [CrossRef]
- Findeiß, S., Etzel, M., Will, S., Mörl, M., & Stadler, P. F. (2017). Design of Artificial Riboswitches as Biosensors. Sensors (Basel, Switzerland), 17(9), 1990. [CrossRef]
- Blin, K., Wolf, T., Chevrette, M. G., Lu, X., Schwalen, C. J., Kautsar, S. A., Suarez Duran, H. G., de Los Santos, E. L. C., Kim, H. U., Nave, M., Dickschat, J. S., Mitchell, D. A., Shelest, E., Breitling, R., Takano, E., Lee, S. Y., Weber, T., & Medema, M. H. (2017). antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic acids research, 45(W1), W36–W41. [CrossRef]
- Wang, J., Lu, A., Liu, J., Huang, W., Wang, J., Cai, Z., & Zhao, G. (2019). iCatch: a new strategy for capturing large DNA fragments using homing endonucleases. Acta biochimica et biophysica Sinica, 51(1), 97–103. [CrossRef]
- Greunke, C., Duell, E. R., D’Agostino, P. M., Glöckle, A., Lamm, K., & Gulder, T. A. M. (2018). Direct Pathway Cloning (DiPaC) to unlock natural product biosynthetic potential. Metabolic engineering, 47, 334–345. [CrossRef]
- Basitta, P., Westrich, L., Rösch, M., Kulik, A., Gust, B., & Apel, A. K. (2017). AGOS: A Plug-and-Play Method for the Assembly of Artificial Gene Operons into Functional Biosynthetic Gene Clusters. ACS synthetic biology, 6(5), 817–825. [CrossRef]
- Li, L., Zhao, Y., Ruan, L., Yang, S., Ge, M., Jiang, W., & Lu, Y. (2015). A stepwise increase in pristinamycin II biosynthesis by Streptomyces pristinaespiralis through combinatorial metabolic engineering. Metabolic engineering, 29, 12–25. [CrossRef]
- Goryshin, I. Y., Naumann, T. A., Apodaca, J., & Reznikoff, W. S. (2003). Chromosomal deletion formation system based on Tn5 double transposition: use for making minimal genomes and essential gene analysis. Genome research, 13(4), 644–653. [CrossRef]
- Xu, J., Zhang, J., Zhuo, J., Li, Y., Tian, Y., & Tan, H. (2017). Activation and mechanism of a cryptic oviedomycin gene cluster via the disruption of a global regulatory gene, adpA, in Streptomyces ansochromogenes. The Journal of biological chemistry, 292(48), 19708–19720. [CrossRef]
- Herrmann, S., Siegl, T., Luzhetska, M., Petzke, L., Jilg, C., Welle, E., Erb, A., Leadlay, P. F., Bechthold, A., & Luzhetskyy, A. (2012). Site-specific recombination strategies for engineering actinomycete genomes. Applied and environmental microbiology, 78(6), 1804–1812. [CrossRef]
- Bu, Q. T., Li, Y. P., Xie, H., Wang, J., Li, Z. Y., Chen, X. A., Mao, X. M., & Li, Y. Q. (2020). Comprehensive dissection of dispensable genomic regions in Streptomyces based on comparative analysis approach. Microbial cell factories, 19(1), 99. [CrossRef]
- Cobb, R. E., Wang, Y., & Zhao, H. (2015). High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS synthetic biology, 4(6), 723–728. [CrossRef]
- Tong, Y., Charusanti, P., Zhang, L., Weber, T., & Lee, S. Y. (2015). CRISPR-Cas9 Based Engineering of Actinomycetal Genomes. ACS synthetic biology, 4(9), 1020–1029. [CrossRef]
- Tong, Y., Whitford, C. M., Robertsen, H. L., Blin, K., Jørgensen, T. S., Klitgaard, A. K., Gren, T., Jiang, X., Weber, T., & Lee, S. Y. (2019). Highly efficient DSB-free base editing for streptomycetes with CRISPR-BEST. Proceedings of the National Academy of Sciences of the United States of America, 116(41), 20366–20375. [CrossRef]
- Huang, H., Zheng, G., Jiang, W., Hu, H., & Lu, Y. (2015). One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta biochimica et biophysica Sinica, 47(4), 231–243. [CrossRef]
- Zeng, H., Wen, S., Xu, W., He, Z., Zhai, G., Liu, Y., Deng, Z., & Sun, Y. (2015). Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA(sm) combined system. Applied microbiology and biotechnology, 99(24), 10575–10585. [CrossRef]
- Jia, H., Zhang, L., Wang, T., Han, J., Tang, H., & Zhang, L. (2017). Development of a CRISPR/Cas9-mediated gene-editing tool in Streptomyces rimosus. Microbiology (Reading, England), 163(8), 1148–1155. [CrossRef]
- Nidhi, S., Anand, U., Oleksak, P., Tripathi, P., Lal, J. A., Thomas, G., Kuca, K., & Tripathi, V. (2021). Novel CRISPR-Cas Systems: An Updated Review of the Current Achievements, Applications, and Future Research Perspectives. International journal of molecular sciences, 22(7), 3327. [CrossRef]
- Yeo, W. L., Heng, E., Tan, L. L., Lim, Y. W., Lim, Y. H., Hoon, S., Zhao, H., Zhang, M. M., & Wong, F. T. (2019). Characterization of Cas proteins for CRISPR-Cas editing in streptomycetes. Biotechnology and bioengineering, 116(9), 2330–2338. [CrossRef]
- Whitford, C. M., Gren, T., Palazzotto, E., Lee, S. Y., Tong, Y., & Weber, T. (2023). Systems Analysis of Highly Multiplexed CRISPR-Base Editing in Streptomycetes. ACS synthetic biology, 12(8), 2353–2366. [CrossRef]
- Li, K., Cai, D., Wang, Z., He, Z., & Chen, S. (2018). Development of an Efficient Genome Editing Tool in Bacillus licheniformis Using CRISPR-Cas9 Nickase. Applied and environmental microbiology, 84(6), e02608-17. [CrossRef]
- Zetsche, B., Heidenreich, M., Mohanraju, P., Fedorova, I., Kneppers, J., DeGennaro, E. M., Winblad, N., Choudhury, S. R., Abudayyeh, O. O., Gootenberg, J. S., Wu, W. Y., Scott, D. A., Severinov, K., van der Oost, J., & Zhang, F. (2017). Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nature biotechnology, 35(1), 31–34. [CrossRef]
- Pickar-Oliver, A., & Gersbach, C. A. (2019). The next generation of CRISPR-Cas technologies and applications. Nature reviews. Molecular cell biology, 20(8), 490–507. [CrossRef]
- Qiu, M., Li, Y., Ye, W., Zheng, X., & Wang, Y. (2021). A CRISPR/Cas9-mediated in situ complementation method for Phytophthora sojae mutants. Molecular plant pathology, 22(3), 373–381. [CrossRef]
- Beck, C., Gren, T., Ortiz-López, F. J., Jørgensen, T. S., Carretero-Molina, D., Martín Serrano, J., Tormo, J. R., Oves-Costales, D., Kontou, E. E., Mohite, O. S., Mingyar, E., Stegmann, E., Genilloud, O., & Weber, T. (2021). Activation and Identification of a Griseusin Cluster in Streptomyces sp. CA-256286 by Employing Transcriptional Regulators and Multi-Omics Methods. Molecules (Basel, Switzerland), 26(21), 6580. [CrossRef]
- Kim, D. R., & Kwak, Y. S. (2021). A Genome-Wide Analysis of Antibiotic Producing Genes in Streptomyces globisporus SP6C4. The plant pathology journal, 37(5), 503. [CrossRef]
- Lee, N. C., Larionov, V., & Kouprina, N. (2015). Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic acids research, 43(8), e55. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




