1. Introduction
When pathogens infect plants, the plants deploy two innate immune systems: pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI), to defend against the invasion of pathogens [
1,
2]. PTI serves as the primary line of defense, promptly recognizing and responding to a wide range of pathogen-associated molecular patterns (PAMPs) or microbe-associated molecular patterns (MAMPs). This triggers fundamental immune mechanisms such as ethylene production, oxidative bursts, callose deposition, and the expression of defense-related proteins [
3,
4], thereby impeding the invasion and spread of pathogens. However, pathogens evolve various strategies to counteract or evade PTI defenses. One common strategy is the release of effector molecules [
5], which can interfere with or inhibit key components of PTI, aiding pathogens in colonizing and proliferating within the plant host.
Fortunately, plants possess a second, more specialized immune system: ETI [
5]. When pathogens release effectors to suppress PTI, the plant’s resistance genes (
R genes) can directly or indirectly recognize these effectors, activating ETI immune responses. ETI typically leads to a hypersensitive response (HR) at the infection site, where cells rapidly undergo programmed cell death, limiting the further spread of pathogens [
6]. These two immune systems are interconnected, working together to maintain the health and survival of plants [
2,
7,
8].
R genes are critical components of the immune system’s ETI, playing a key role [
6]. Most
R genes encode disease resistance proteins with nucleotide-binding site and leucine-rich repeat (NBS-LRR) domains—these genes are thus often termed NLR genes [
9]. The
R genes discussed in this paper are NLR genes, excluding other types. Beyond the canonical NBS and LRR domains, the N-termini of R proteins frequently feature additional domains or motifs, such as the Toll/Interleukin-1 receptor (TIR) domains, the powdery mildew resistance protein 8 (RPW8) domains, and coiled-coil (CC) motifs [
1,
10]. Proteins harboring the TIR and RPW8 domains are classified as TNL and RNL, respectively [
10,
11]. Similarly, proteins with CC motifs are designated as CNL [
9]. A significant proportion of R proteins, possessing only the NBS and LRR domains, are collectively categorized as NL (NBS-LRR).
Comparative genomics research has shown that NLR genes are extensively distributed among land plants and charophytes, indicating a deep evolutionary origin [
1,
10,
12]. Additionally, NLR genes have been identified in a limited number of green algae species, further supporting their ancient lineage [
13]. Phylogenetic analyses suggest that the emergence of
R genes preceded the evolutionary split between charophytes and land plants [
1,
10]. Following their origin, NLR genes have undergone adaptive evolution, significantly contributing to the transition of plants from aquatic to land habitats [
10].
Citrus is one of the world’s most significant fruits, providing essential nutrients such as vitamin C and citric acid. Cultivation of citrus spans over 140 countries and regions globally, thriving in warm and moist climates below the northern latitudes [
14]. In 2020, the global citrus cultivation area reached 521,000 hectares, yielding a production of 96.81 million metric tons [
15]. However, citrus faces various threats from diseases, such as citrus HLB, a devastating ailment that severely impacts the yield and quality of citrus. Addressing this challenge necessitates a deep understanding of citrus’s mechanisms of disease resistance. The study of the diversity of disease-resistant genes is foundational for elucidating these defense mechanisms. Currently, there is no systematic research reported on citrus NLR genes.
This study integrates comparative genomics and phylogenetics to systematically explore the NLR genes in the citrus genome, unraveling the mechanisms underlying the diversity, origin, and evolutionary patterns of citrus NLR genes. We observed substantial diversity in NLR genes among different citrus species. Further in-depth investigation revealed that gene duplication and recombination are the primary mechanisms shaping the diversity of NLR genes in citrus. This study holds significant implications for a profound understanding of citrus NLR genes and for breeding disease-resistant citrus varieties.
2. Materials and Methods
2.1. Data Used in This Study
At the outset of this study (September 2020), genomic data for 10 citrus species were accessible, namely
C. medica,
C. ichangensis,
C. hindsii,
Atlantia buxifolia,
C. trifoliata,
C. grandis,
C. mangshanensis,
C. sinensis Osbeck cv. newhall,
C. sinensis Osbeck cv. Valencia, and
C. clementina. The genome of
C. sinensis Osbeck cv. newhall was assembled and annotated by our research team [
29], available for download from Zenodo database (
https://zenodo.org/record/8176803). Genomic data for the remaining 9 citrus species can be obtained from the CPBD (
http://citrus.hzau.edu.cn/download.php) [
22].
2.2. Identification of NLR Genes
The nine seed sequences of the NB-ARC domain (PF00931) were obtained from the European Bioinformatics Institute (EBI) database and stored in Stockholm format. A Hidden Markov Model (HMM) of NB-ARC was constructed using HMMER package 3.1b2 [
30]. Then, this model was used to search for proteins containing NB-ARC domain among the protein sequences of 10 citrus genomes (
Table S1). The Blastp algorithm was also employed to search for citrus proteins containing NB-ARC domain, using the NLR proteins of
Arabidopsis thaliana [
10] as queries, with a cutoff E-value of 10
-3. The results obtained from both methods were merged and redundant sequences were removed to obtain citrus proteins containing NB-ARC domain. Multiple sequence alignment of citrus proteins containing the NB-ARC domain and previously published NB-ARC or NAT domain proteins from both eukaryotes and prokaryotes [
31] was performed using MAFFT software [
32] with G-INS-i algorithm, followed by manual edited using MEGA11 [
33]. ModelFinder [
34] was utilized to search for the best model, and IQ-TREE software [
35] was employed with JTT+R10 model for phylogenetic analysis. Proteins with MalT and SWACOS domains were used as outgroups [
31]. The protein domain architecture was annotated using CD-Search [
36]. NLR genes were searched in the nucleotide sequences of 10 citrus genomes using NLR-Annotator software [
37] with default parameters, for testing analysis of the identification results.
2.3. Phylogenetic Analysis
To investigate the origin of NLR genes in citrus, 3168 NLR gene sequences from 16 species including citrus,
Sapindaceae, and
Anacardiaceae, were aligned using MAFFT [
32] with G-INS-i algorithm, manual edited using MEGA11 [
33]. The ModelFinder [
34] was used to research the best model, and then Q.mammal+R10 was employed for phylogenetic tree construction using IQ-TREE [
35].
The citrus NLR gene sequences were aligned using MAFFT [
32] for multiple sequence alignment, followed by manual edited using MEGA11 [
33]. The ModelFinde [
34] was employed to identify the best model, and subsequently, JTT+F+R10 was used for phylogenetic tree construction using IQ-TREE [
35]. The phylogenetic analysis of citrus species based on whole genomes was conducted using the STAG algorithm implemented in OrthoFinder [
19].
2.4. Positive Selection Analysis
Due to the significant divergence of NLR genes, alignment and positive selection analysis were not feasible for all sequences. Consequently, we divided them into 65 groups based on phylogenetic analysis (Figure 4). Nucleotide sequences were aligned using MAFFT [
32] and manually refined with MEGA11 [
33]. A likelihood ratio test for positive selection was conducted by comparing the M7 (beta) and M8 (beta&v) models using the PAML4.9 software package [
38,
39]. For gene groups demonstrating positive selection via the likelihood ratio test, positively selected sites were identified through the Bayes empirical Bayes (BEB) procedure.
2.5. Recombination Analysis
Recombination was detected using the RDP5 package [
40]. The aligned nucleotide sequences were imported into RDP5 and analyzed with the RDP, GENECONV, Chimaera, MaxChi, BootScan, SiScan, and 3Seq methods. Potential recombination events were deemed significant only if supported by at least four methods with a p-value < 10
−6.
3. Results
3.1. Identification and Diversity of Citrus NLR Genes
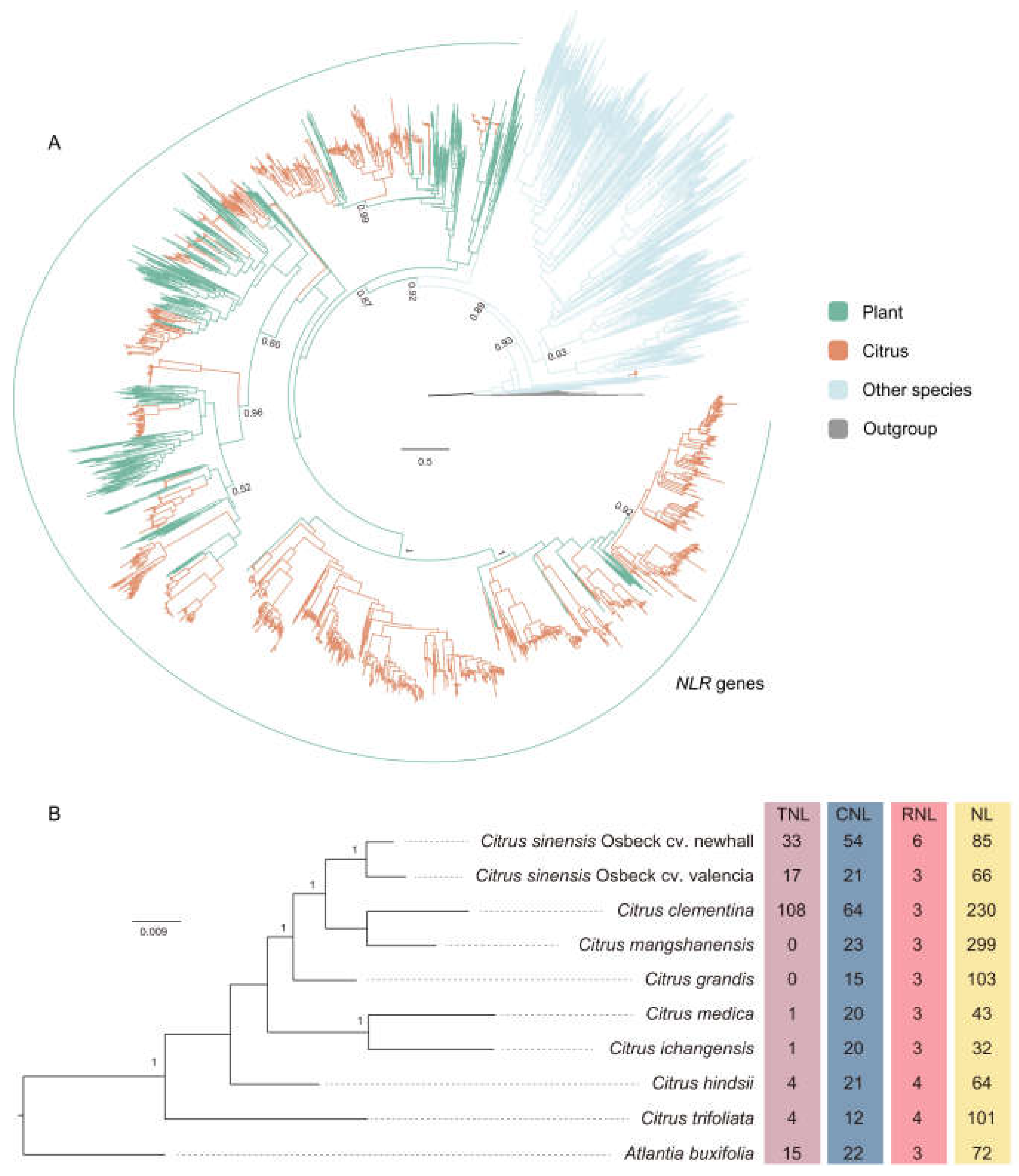
This study utilized comparative genomics and phylogenetics to identify NLR genes in the proteomes of 10 citrus species. Initially, protein sequences were screened for the NB-ARC domain using the HMM model, resulting in the identification of 1,875 sequences containing the NB-ARC domain (
Table S1). Subsequently, 1,845 sequences with the NB-ARC domain were identified via Blastp homology searches (
Table S1). After redundancy reduction, sequences containing the NB-ARC or NAT domains were used to construct a phylogenetic tree, along with reported homologous sequences from representative plants and other species (including animals, bacteria, fungi, and archaea). We identified citrus proteins with the NB-ARC domain that formed monophyletic groups with plant R genes, designating these as citrus NLR protein sequences (
Figure 1A). Some sequences with the NB-ARC domain formed monophyletic groups with non-plant homologs and were therefore excluded. Ultimately, we identified 1,585 NLR genes (
Table S1).
Utilizing the HMM method and the Pfam database, we annotated the domain structures of the 1,585 citrus NLR genes, identifying 183 as TNL and 35 as RNL (
Figure 1B). Notably, we also annotated the Rx_N (PF18052) domain, predicted to be a CC motif but suggested by research to be a four-helical bundle fold [
16]. The Rx_N domain is equivalent to the CC domain annotated in previous studies [
16]. Thus, in this context, RxN-NBS-LRR corresponds to CNL, as identified in prior research [
9,
17]. We collectively termed the NLR proteins containing the Rx_N domain as CNL, totaling 272 proteins (
Figure 1B). Additionally, a substantial number of NL proteins containing only NBS or NBS-LRR domains were annotated, totaling 1,095 (
Figure 1B).
Among the 10 citrus species, aside from the 108 TNL sequences in C.
clementina, the number of TNL sequences in the other nine species was relatively low. Only a few were found in C.
medica, C.
ichangensis, C.
hindsii, and C.
trifoliata, while they were absent in C.
mangshanensis and C.
grandis (
Figure 1B). RNL was the least abundant among the three types, evenly distributed across all species, ranging from 3 to 4 sequences each (
Figure 1B). There were six RNL sequences in C.
sinensis Osbeck cv. Newhall, possibly due to its diploid nature, resulting in a doubling of the quantity.
3.2. The Origin of Citrus NLR Genes
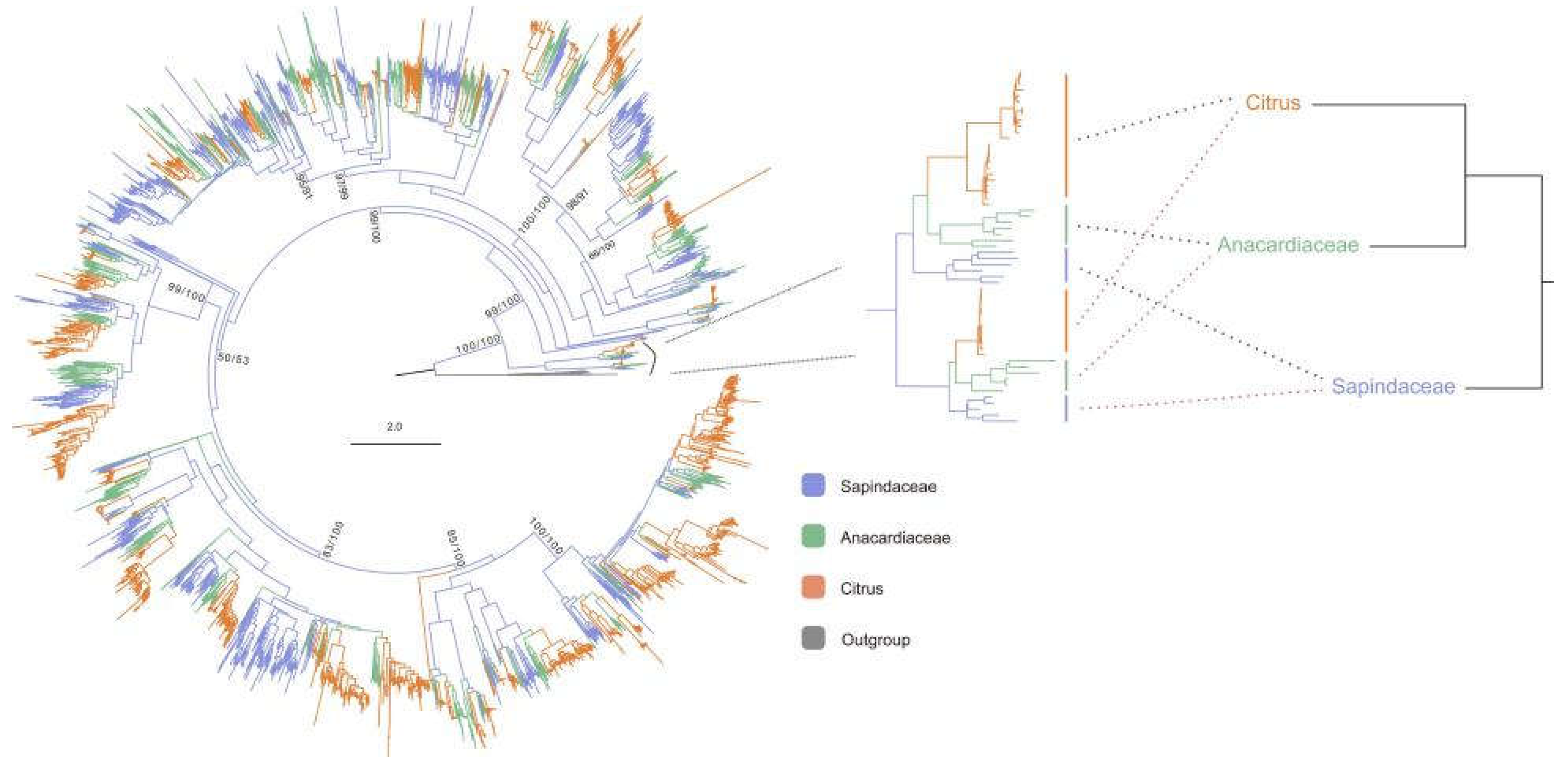
The study utilized 1,585 identified NLR proteins and 1,583 published NLR proteins from the Anacardiaceae and Sapindaceae families (
Table S2) to construct phylogenetic relationships (
Figure 2). The results indicate that citrus NLR proteins fall within the diversity of Sapindaceae, suggesting that citrus NLR genes originated comparatively later than those in Sapindaceae. Within the Sapindales order, Rutaceae is the oldest, followed by Sapindaceae, with Aurantioideae (Citrus genus) being the most recent [
18]. In most monophyletic groups, the phylogenetic relationships of NLR proteins align with the species relationships of Rutaceae, Sapindaceae, and the Citrus genus (
Figure 2). The coevolutionary relationships between NLR proteins and species suggest that NLR genes originated concurrently with the species’ origins.
3.3. The evolution of citrus NLR genes
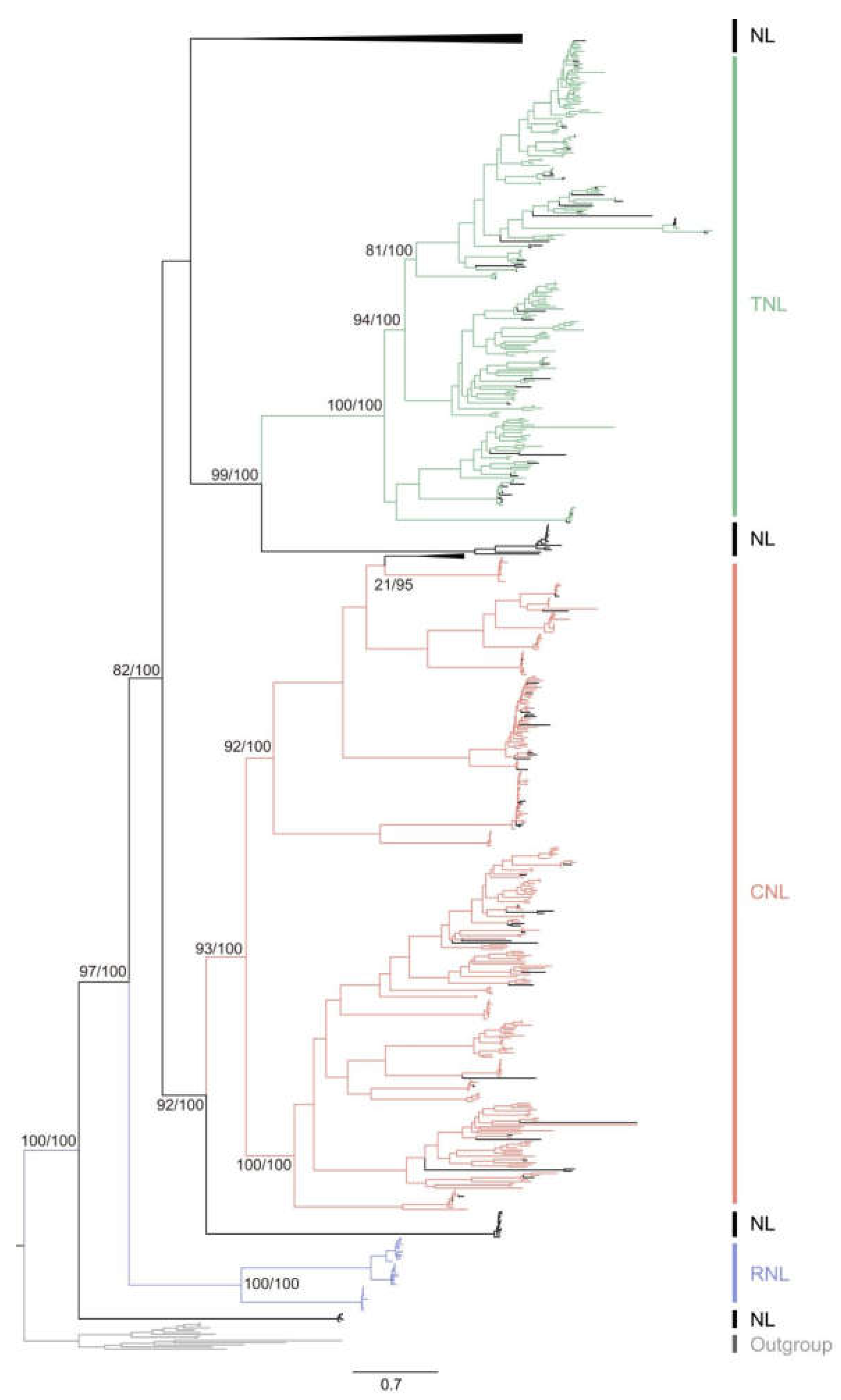
We employed maximum likelihood methods to construct phylogenetic relationships among citrus NLR proteins. Integrating domain architecture of NLRs, we identified five main clusters in the phylogenetic tree of citrus NLR proteins, comprising one cluster of TNLs, one cluster of CNLs, one cluster of RNLs, and two clusters of NLs (
Figure 3). NLs exhibited the highest diversity, while TNLs, RNLs, and CNLs fell within the diversity of NLs, indicating their origin from NLs with the acquisition of TIR, RPW8 domains, and CC motifs respectively. Additionally, NLs were found within both TNLs and CNLs clusters (
Figure 3), indicating TNLs and CNLs lost their TIR and Rx_N domains, respectively, transforming into NLs. Overall, citrus NLR genes underwent frequent events of domain acquisition and loss during evolution.
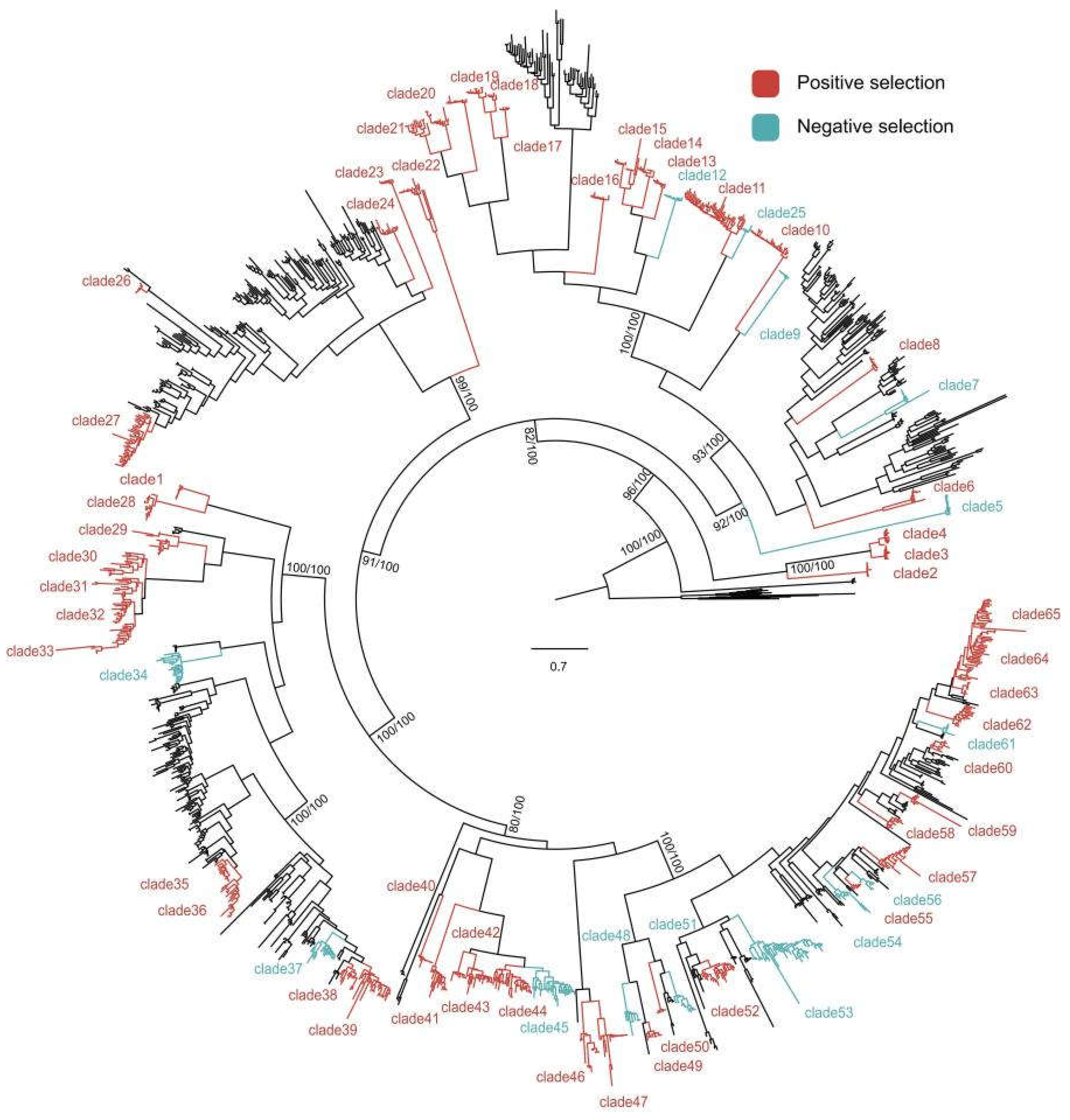
To test whether citrus NLR genes underwent adaptive evolution after their origin, positive selection analysis was conducted on 65 groups selected base on phylogenetic tree. amino acid residues under positive selection was detected in the majority of groups (52/65)(
Figure 4). Among the 52 gene groups where positive selection was detected, the number of positively selected sites varied(
Table S3). Our findings indicate that the vast majority of citrus NLR clusters underwent adaptive divergence.
3.4. Mechanism of NLR Genes Diversity Formation in Citrus
The phylogenetic tree of citrus was constructed using OrthoFinder software [
19] based on full genome of 10 citrus genomes (
Figure 1A).
Atlantia buxifolia is placed at the root of the tree, with the overall phylogenetic order largely consistent with previous reports [
20]. Comparing the number of NLR genes across different citrus species on the phylogenetic tree reveals considerable diversity in NLR gene counts even within the same subfamily. Among the 10 citrus species, substantial differences exist in the copy numbers of NLR genes. For instance,
C. clementina has 405 copies, whereas
C. ichangensis has only 56, making the former about eight times more abundant than the latter. In the case of the sweet orange subspecies
C. sinensis Osbeck cv. Newhall and
C. sinensis Osbeck cv. Valencia, 178 and 107 copies were identified, respectively. Notably, while
C. sinensis Osbeck cv. newhall is a diploid genome,
C. sinensis Osbeck cv. Valencia is a haploid genome. Although the genomic content of
C. sinensis Osbeck cv. Newhall is double that of
C. sinensis Osbeck cv. Valencia, the NLR gene copies are not proportionally higher. What evolutionary mechanisms have contributed to the genetic diversity of citrus NLR genes?
Based on the phylogenetic tree of citrus NLR proteins, we investigated horizontal gene transfer and gene duplication events within citrus NLR genes. Gene duplication events were identified when NLR genes from the same species formed a monophyletic group on the phylogenetic tree. Horizontal gene transfer events were identified when the genetic diversity of one species fell within that of another species.
Following statistical analysis, we generated a diagram depicting the levels of horizontal gene transfer (HGT) between various citrus species (
Figure 5A), revealing relatively frequent gene exchange among
C. sinensis Osbeck cv. Newhall,
C. sinensis Osbeck cv. Valencia,
C. mangshanensis, and
C. clementina. Notably, we observed gene exchange between
C. sinensis Osbeck cv. newhall and
C. clementina with
Atlantia buxifolia. Additionally, there was minimal gene exchange between
C. sinensis Osbeck cv. Newhall and
C. medica, while the remaining four citrus species (
C. ichangensis,
C. hindsii,
C. trifoliata, and
C. grandis) and other species showed no
NLR gene exchange. Correlation analysis between the number of
NLR genes and the frequency of horizontal gene transfer events revealed a non-significant positive relationship (
Figure 5B), suggesting that horizontal gene transfer contributes insignificantly to citrus NLR gene diversity.
Recombination has long been considered a significant driver for NLR gene diversification [
21]. We used seven algorithms within the RDP software (Martin et al., 2015) to search for recombination signals within citrus R genes. A single gene recombination event was considered when four or more algorithms detected positive signals. In this study, 173 recombination events were identified, with C.
clementina exhibiting the highest number at 81 occurrences (
Table S4). Correlation analysis between the number of NLR genes and the number of recombination events across citrus species showed a significant positive relationship (
Figure 5C), indicating that gene recombination is one of the mechanisms contributing to citrus NLR gene diversity.
A total of 293 gene duplication events were recorded in this study, with occurrences ranging from 6 to 116 across different citrus species.
C. clementina once again exhibited the highest number of gene duplication events at 116 occurrences (
Table S4). Correlation analysis between the number of NLR genes and the number of gene duplication events revealed a positive relationship, indicating that gene duplication also contributes to citrus NLR gene diversity. In summary, gene recombination and gene duplication are the main mechanisms shaping NLR gene diversity in citrus.
4. Discussion
In this study, we employed comparative genomics and phylogenetic methods to identify NLR genes across 10 citrus species. To validate the effectiveness of our mining approach, we used the NLR-Annotator software (Zhang, 2020) to search nucleotide genomes, resulting in the retrieval of 1847 citrus NLR gene sequences. The NLR counts extracted by NLR-Annotator across various citrus species closely matched the results obtained from Blastp and HMM of this study. Comparison with previously reported citrus NLR gene numbers in the ANNA database (Liu et al., 2021) revealed close correspondence between the quantities in A. buxifolia, C. grandis, C. medica, and C. ichangensis and the results obtained in this study through Blastp and HMM. Thus, the identification methods utilizing Blastp and HMM employed in this study are validated.
The ANNA database reports 529 NLR genes of C.
sinensis Osbeck cv. Valencia [
23], whereas our study’s Blastp and HMM analyses yielded 179 and 178, respectively, and the NLR-Annotator software result was 181. The substantial variance can be attributed to the utilization of the C.
sinensis Osbeck cv. Valencia v2.0 genome [
24] in prior studies, while we employed the v3.0 genome version [
25]. Factors such as genome assembly quality and annotation precision between the two genome versions may influence the NLR gene number. We conducted NLR gene mining on the nucleotide sequences of both sweet orange genome versions using the tblastn algorithm. We found comparable outcomes with no significant differences between the two genome versions. This underscores annotation precision as the primary factor influencing the quantity of NLR genes.
We integrated the results from Blastp and HMM, eliminating redundancies. Subsequently, we employed phylogenetic analysis to further screen the sequences, remove false positives, and discard NLR sequences generated by alternative splicing, thereby obtaining the NLRs for each citrus species in this study. We employed a more stringent identification method to ensure the accuracy of the results.
The distribution of TNLs varies significantly among citrus species. TNLs are sparsely present or absent in
C. medica,
C. ichangensis,
C. hindsii,
C. trifoliata,
C. grandis, and
C. mangshanensis, while in
C. clementina, their number can be as high as 108. To exclude the impact of gene annotation on the results, we used the tblastn algorithm to mine the genomes of
C. medica,
C. ichangensis,
C. hindsii,
C. trifoliata,
C. grandis, and
C. mangshanensis for TNLs. The identification results were consistent with our study, thus eliminating the influence of genome annotation on the identification results. The evolutionary fate of NLR genes is mainly determined by natural selection and genetic drift [
21]. In this study, positively selection amino acid residues were detected in all four clusters of TNLs selected, suggesting that the loss of TNLs in species such as
C. mangshanensis may be due to genetic drift.
In-depth analysis revealed that the expansion of TNL gene copies in
C. clementina is due to frequent gene recombination and duplication events. On one hand, previous studies have speculated multiple origins of TNLs [
26], while loss has been reported in Aquilegia coerulea and monocots [
27]. On the other hand, research has indicated that the evolutionary rate of TNLs is higher than that of non-TNLs [
26], suggesting that, compared to CNLs and RNLs, TNLs may be more prone to acquiring or losing the TIR domain, thus undergoing a more complex evolutionary process.
Differences in NLR gene copy numbers among citrus species are not uncommon, as seen in the Rosaceae family, where the wild strawberry (Fragaria vesca) has 144 copies, while the apple (Malus pumila) has 748 copies [
26]. This study further analyzes gene recombination and duplication as the primary mechanisms shaping NLR gene diversity in citrus. While horizontal gene transfer did not exhibit significant impact across all citrus species, it was the major driver of NLR gene copy number increase in
A. buxifolia. The number of gene duplications and recombinations in
C. medica and
A. buxifolia is the same, with 10 and 6 instances, respectively. Horizontal gene transfer occurred 9 times in
A. buxifolia and only once in
C. medica. The NLR gene copy number in
A. buxifolia is 1.67 times that in C.
medica. Therefore, horizontal gene transfer is the primary mechanism driving the increase in NLR gene copy number in
A. buxifolia.
Approximately 30 million years ago, the Rutaceae subfamily originated from the ancient Indian plate, which was then connected to Africa and Australia. As the plates collided and drifted, the Rutaceae subfamily gradually diversified, with the citrus genus originating over 90 million years ago in southern China [
20]. During the colonization of terrestrial environments, plants encountered new pathogen species and infection levels [
28]. Positively selected NLR genes play a crucial role in plant-pathogen interactions. This evolutionary process involved frequent gains and losses of TIR, RPW8, and LRR domains, enhancing R-protein diversity and enabling adaptation to various environmental challenges [
10]. The discovery of numerous adaptively evolved NLR genes in citrus suggests their significant role in global citrus colonization.
5. Conclusions
This study provides a comprehensive understanding of the diversity and evolution of NLR genes in citrus species, which were previously underexplored compared to other plants. By identifying 1,875 NLR genes across 10 citrus species and analyzing their variations, origins, and adaptive evolution. The findings are particularly significant for their potential application in breeding disease-resistant citrus varieties, making the study a foundational resource for future citrus genetic research and crop improvement efforts.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on
Preprints.org. Table S1: the identification of citrus NLR genes; Table S2: The study used NLR genes of Anacardiaceae and Sapindaceae; Table S3: Positive selection analysis of NLR gene groups in citrus; Table S4: Statistics of three events occurring in citrus NLR genes.
Author Contributions
ZWX participated in designing the study, obtained and analyzed the data. WSZ, YH and JXW participated in interpreting the results. ZJL contributed technical assistance. Corresponding author YXG designed the study, interpreted the results, wrote the manuscript and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (32060615), and Double Thousand Plan of Jiangxi Province (jxsq2020102134).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Acknowledgments
We thank Nian Wang (University of Florida), Han-Guan Zhu (Nanjing Normal University) and Gong Zhen (Nanjing Normal University) for guidance during the study research process.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Han, G. Origin and Evolution of the Plant Immune System. New Phytologist 2019, 222, 70–83. [Google Scholar] [CrossRef]
- Ngou, B.P.M.; Ahn, H.-K.; Ding, P.; Jones, J.D.G. Mutual Potentiation of Plant Immunity by Cell-Surface and Intracellular Receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, X.; Dai, L.; Wang, G. Recent Progress in Elucidating the Structure, Function and Evolution of Disease Resistance Genes in Plants. Journal of Genetics and Genomics 2007, 34, 765–776. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling Mechanisms in Pattern-Triggered Immunity (PTI). Molecular plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.; Chen, H.; Liu, F.; Fu, Z.Q. PTI and ETI: Convergent Pathways with Diverse Elicitors. Trends in Plant Science 2022, 27, 113–115. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-Recognition Receptors Are Required for NLR-Mediated Plant Immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Kim, C.-Y.; Song, H.; Lee, Y.-H. Ambivalent Response in Pathogen Defense: A Double-Edged Sword? Plant Communications 2022, 3, 100415–100415. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, Z.; Zhang, Y.; Li, Q.; Jiang, X.-M.; Jiang, Z.; Tang, J.; Chen, D.; Wang, Q.; Chen, J.-Q.; et al. An Angiosperm NLR Atlas Reveals That NLR Gene Reduction Is Associated with Ecological Specialization and Signal Transduction Component Deletion. Molecular Plant 2021, 14, 2015–2031. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, W.; Zhang, T.; Gong, Z.; Zhao, H.; Han, G.-Z. Out of Water: The Origin and Early Diversification of Plant R-Genes. Plant Physiology 2018, 177, 82–89. [Google Scholar] [CrossRef]
- Xiao, S. Broad-Spectrum Mildew Resistance in Arabidopsis Thaliana Mediated by RPW8. Science 2001, 291, 118–120. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Wang, Y.; Wu, P.; Wang, Q.; Yang, L.-T.; Pan, X.-H.; Wang, B.; Chen, J.-Q. A Primary Survey on Bryophyte Species Reveals Two Novel Classes of Nucleotide-Binding Site (NBS) Genes. PLOS ONE 2012, 7, e36700–e36700. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.-Q.; Xue, J.-Y.; Wang, Q.; Wang, B.; Chen, J.-Q. Revisiting the Origin of Plant NBS-LRR Genes. Trends in Plant Science 2019, 24, 9–12. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Zhang, P.; Trivedi, P.; Riera, N.; Wang, Y.; Liu, X.; Fan, G.; Tang, J.; Coletta-Filho, H.D.; et al. The Structure and Function of the Global Citrus Rhizosphere Microbiome. Nature Communications 2018, 9, 4894. [Google Scholar] [CrossRef]
- Faezeh Falaki Citrus Virus and Viroid Diseases. IntechOpen eBooks 2023, 1. [CrossRef]
- Hao, W.; Collier, S.A.; Moffett, P.; Chai, J. Structural Basis for the Interaction between the Potato Virus X Resistance Protein (Rx) and Its Cofactor Ran GTPase-Activating Protein 2 (RanGAP2). Journal of Biological Chemistry 2013, 288, 35868–35876. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.-H.; Wang, Y.; Chen, M.; Liu, J.; Lu, R.-S.; Zou, X.; Sun, X.-Q.; Zhang, Y.-M. Genome-Wide Identification and Evolutionary Analysis of NBS-LRR Genes from Secale Cereale. Frontiers in Genetics 2021, 12, 771814. [Google Scholar] [CrossRef]
- Joyce, E.M.; Appelhans, M.S.; Buerki, S.; Cheek, M.; de Vos, J.M.; Pirani, J.R.; Zuntini, A.R.; Bachelier, J.B.; Bayly, M.J.; Callmander, M.W.; et al. Phylogenomic Analyses of Sapindales Support New Family Relationships, Rapid Mid-Cretaceous Hothouse Diversification, and Heterogeneous Histories of Gene Duplication. Frontiers in Plant Science 2023, 14, 1063174. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biology 2019, 20, 238. [Google Scholar] [CrossRef]
- Huang, Y.; He, J.; Xu, Y.; Zheng, W.; Wang, S.; Chen, P.; Zeng, B.; Yang, S.; Jiang, X.; Liu, Z.X.; et al. Pangenome Analysis Provides Insight into the Evolution of the Orange Subfamily and a Key Gene for Citric Acid Accumulation in Citrus Fruits. Nature Genetics 2023, 55, 1964–1975. [Google Scholar] [CrossRef]
- Märkle, H.; Saur Isabel, M.L.; Stam, R. Evolution of Resistance (R) Gene Specificity. Essays in Biochemistry 2022, 66, 551–560. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus Evolution 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, X.; Liu, S.; Huang, Y.; Guo, Y.-X.; Xie, W.-Z.; Liu, H.; Muhammad, *!!! REPLACE !!!*; Xu, Q.; Chen, L.-L. Citrus Pan-Genome to Breeding Database (CPBD): A Comprehensive Genome Database for Citrus Breeding. Molecular Plant 2022, 15, 1503–1505. [Google Scholar] [CrossRef]
- Xu, Q.; Chen, L.-L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.-B.; Hao, B.-H.; Lyon, M.P.; et al. The Draft Genome of Sweet Orange (Citrus Sinensis). Nature Genetics 2012, 45, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Huang, Y.; Liu, Z.; He, J.; Jiang, X.; He, F.; Lu, Z.; Yang, S.; Chen, P.; Yu, H.; et al. Somatic Variations Led to the Selection of Acidic and Acidless Orange Cultivars. Nature Plants 2021, 7, 954–965. [Google Scholar] [CrossRef]
- Zhong, Y.; Yin, H.; Sargent, D.J.; Mickaël, M.; Cheng, Z. Species-Specific Duplications Driving the Recent Expansion of NBS-LRR Genes in Five Rosaceae Species. BMC Genomics 2015, 16. [Google Scholar] [CrossRef]
- Tarr, D.E.K.; Alexander, H.M. TIR-NBS-LRR Genes Are Rare in Monocots: Evidence from Diverse Monocot Orders. BMC Research Notes 2009, 2, 197. [Google Scholar] [CrossRef]
- Rensing, S.A.; Lang, D.; Zimmer, A.D.; Terry, A.; Salamov, A.; Shapiro, H.; Nishiyama, T.; Perroud, P.-F.; Lindquist, E.A.; Kamisugi, Y.; et al. The Physcomitrella Genome Reveals Evolutionary Insights into the Conquest of Land by Plants. Science 2007, 319, 64–69. [Google Scholar] [CrossRef]
- Gao, Y.; Xu, J.; Li, Z.; Zhang, Y.; Riera, N.; Xiong, Z.; Ouyang, Z.; Liu, X.; Lu, Z.; Seymour, D.; et al. Citrus Genomic Resources Unravel Putative Genetic Determinants of Huanglongbing Pathogenicity. iScience 2023, 26, 106024–106024. [Google Scholar] [CrossRef]
- Eddy, S.R. Accelerated Profile HMM Searches. PLoS Computational Biology 2011, 7, e1002195. [Google Scholar] [CrossRef]
- Urbach, J.M.; Ausubel, F.M. The NBS-LRR Architectures of Plant R-Proteins and Metazoan NLRs Evolved in Independent Events. Proceedings of the National Academy of Sciences 2017, 114, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Molecular Biology and Evolution 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Molecular Biology And Evolution 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nature Methods 2017, 14, 587–589. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional Classification of Proteins via Subfamily Domain Architectures. Nucleic acids research 2017, 45, D200–D203. [Google Scholar] [CrossRef]
- Zhang, W. NLR-Annotator: A Tool for de Novo Annotation of Intracellular Immune Receptor Repertoire. Plant Physiology 2020, 183, 418–420. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Molecular Biology and Evolution 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Yang, Z.; Nielsen, R.; Goldman, N.; Pedersen, A.-M.K. Codon-Substitution Models for Heterogeneous Selection Pressure at Amino Acid Sites. Genetics 2000, 155, 431–449. [Google Scholar] [CrossRef]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A Computer Program for Analyzing Recombination In, and Removing Signals of Recombination From, Nucleotide Sequence Datasets. Virus Evolution 2020, 7. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).