Submitted:
18 September 2024
Posted:
19 September 2024
You are already at the latest version
Abstract
Keywords:
Introduction
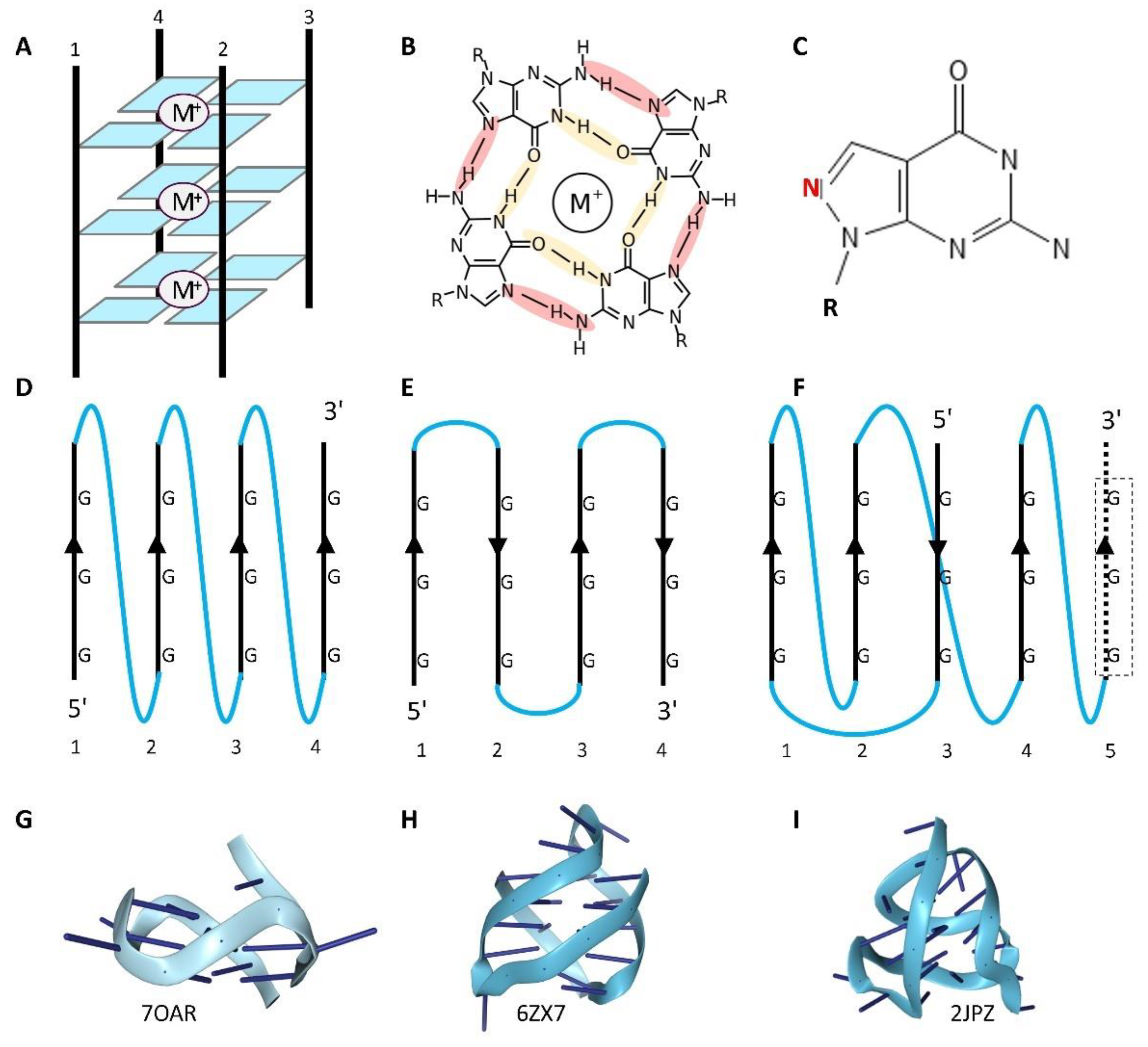
Biophysical and Computational Studies of the G-Quadruplex
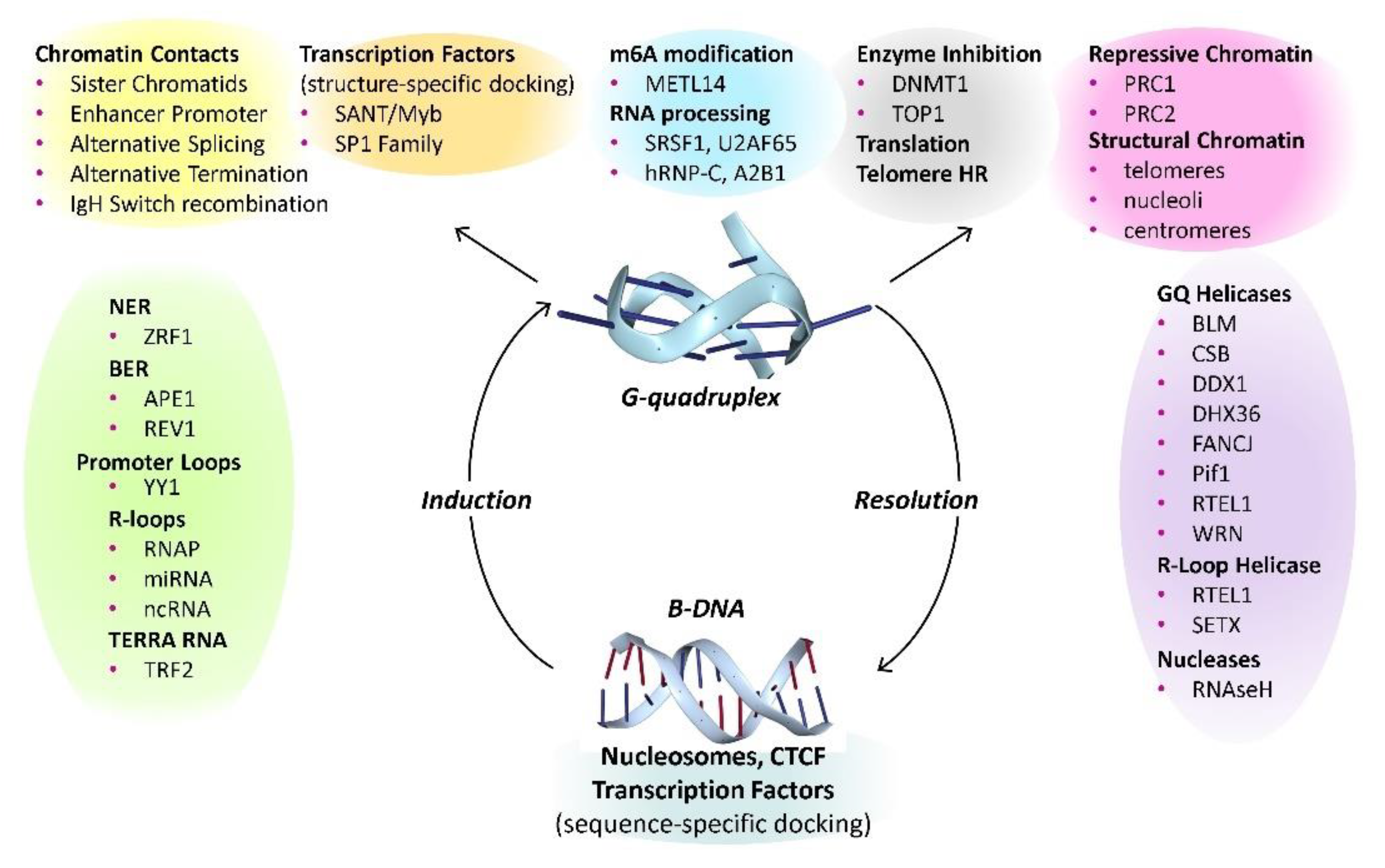
GQ Binding Proteins
The Accumulating Evidence for the Biological Importance of G-Quadruplexes
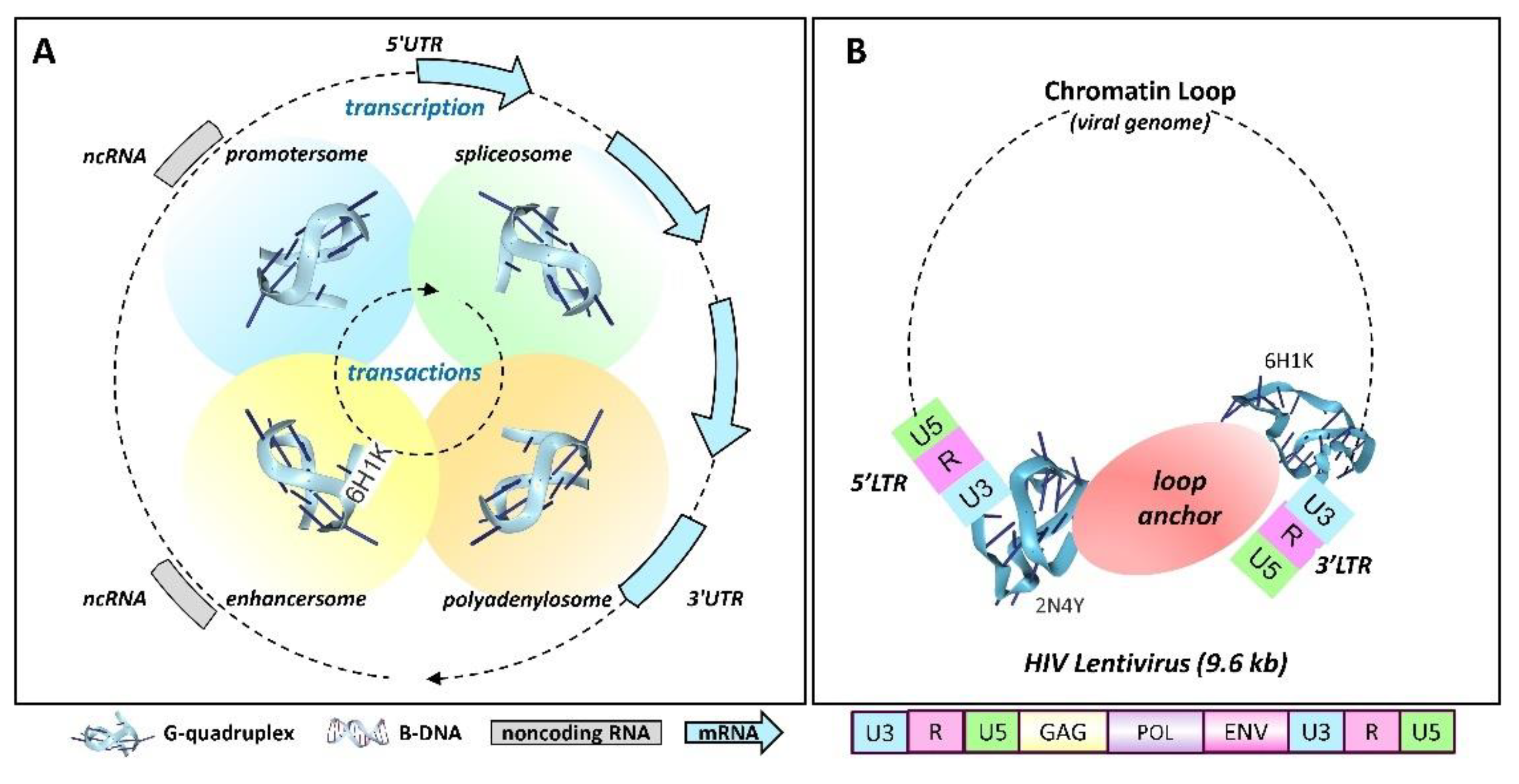
The GQ Architecture of Retroviruses
Cell Division
DNA Repair
GQ and Telomeres
Resolution of G-Quadruplexes
G-Quadruplexes and Gene Expression
G-Quadruplexes and Transcriptional Bursts
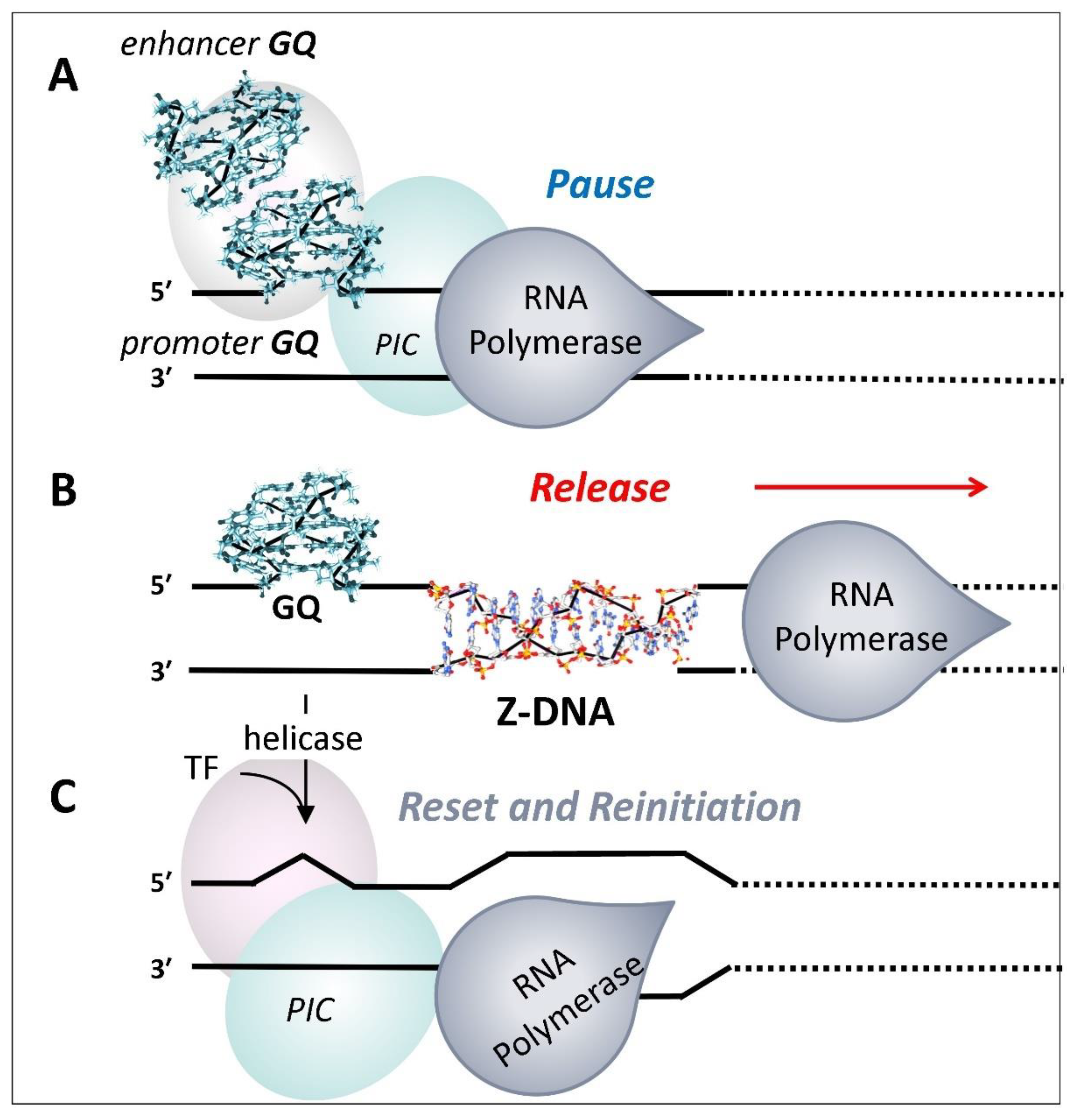
G-Quadruplexes and Promoter Pausing
G- and Z-flipons in Promoter Reset
Gene Repression
R Loop Resolution
Transactional Chromatin Looping and Transcript elongation
DNA G-Quadruplexes and Splicing
RNA G-Quadruplexes and Splicing
G-Quadruplexes and Translation
G-Quadruplexes and Development.
Summary and Outlook
References
- Herbert, A., A Genetic Instruction Code Based on DNA Conformation. Trends Genet 2019, 35, 887–890. [CrossRef] [PubMed]
- Herbert, A., Flipons and the logic of soft-wired genomes. 1st ed.; CRC Press: Boca Raton, 2024.
- Gellert, M.; Lipsett, M. N.; Davies, D. R., Helix Formation by Guanylic Acid. Proceedings of the National Academy of Sciences 1962, 48, (12), 2013-2018. [CrossRef]
- Arnott, S.; Chandrasekaran, R.; Marttila, C. M., Structures for polyinosinic acid and polyguanylic acid. Biochem J 1974, 141, (2), 537-43. [CrossRef] [PubMed]
- Sauer, M.; Paeschke, K., G-quadruplex unwinding helicases and their function in vivo. Biochem Soc Trans 2017, 45, (5), 1173-1182. [CrossRef]
- Blackburn, E. H.; Gall, J. G., A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol 1978, 120, (1), 33-53. [CrossRef]
- Sundquist, W. I.; Klug, A., Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature 1989, 342, (6251), 825-9. [CrossRef] [PubMed]
- Griffith, J. D.; Comeau, L.; Rosenfield, S.; Stansel, R. M.; Bianchi, A.; Moss, H.; de Lange, T., Mammalian telomeres end in a large duplex loop. Cell 1999, 97, (4), 503-14. [CrossRef]
- Huber, M. D.; Duquette, M. L.; Shiels, J. C.; Maizels, N., A conserved G4 DNA binding domain in RecQ family helicases. J Mol Biol 2006, 358, (4), 1071-80. [CrossRef]
- Maizels, N., G4-associated human diseases. EMBO Rep 2015, 16, (8), 910-22. [CrossRef]
- Duquette, M. L.; Handa, P.; Vincent, J. A.; Taylor, A. F.; Maizels, N., Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev 2004, 18, (13), 1618-29. [CrossRef]
- Okazaki, I. M.; Kinoshita, K.; Muramatsu, M.; Yoshikawa, K.; Honjo, T., The AID enzyme induces class switch recombination in fibroblasts. Nature 2002, 416, (6878), 340-5. [CrossRef] [PubMed]
- Qiao, Q.; Wang, L.; Meng, F. L.; Hwang, J. K.; Alt, F. W.; Wu, H., AID Recognizes Structured DNA for Class Switch Recombination. Mol Cell 2017, 67, (3), 361-373 e4.
- Ribeiro de Almeida, C.; Dhir, S.; Dhir, A.; Moghaddam, A. E.; Sattentau, Q.; Meinhart, A.; Proudfoot, N. J., RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol Cell 2018, 70, (4), 650-662 e8.
- Richard, P.; Manley, J. L., R Loops and Links to Human Disease. J Mol Biol 2017, 429, (21), 3168-3180. [CrossRef] [PubMed]
- Guo, J. U.; Bartel, D. P., RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science 2016, 353, (6306).
- Di Antonio, M.; Ponjavic, A.; Radzevicius, A.; Ranasinghe, R. T.; Catalano, M.; Zhang, X.; Shen, J.; Needham, L. M.; Lee, S. F.; Klenerman, D.; Balasubramanian, S., Single-molecule visualization of DNA G-quadruplex formation in live cells. Nat Chem 2020, 12, (9), 832-837. [CrossRef]
- Guo, J. K.; Blanco, M. R.; Walkup, W. G. t.; Bonesteele, G.; Urbinati, C. R.; Banerjee, A. K.; Chow, A.; Ettlin, O.; Strehle, M.; Peyda, P.; Amaya, E.; Trinh, V.; Guttman, M., Denaturing purifications demonstrate that PRC2 and other widely reported chromatin proteins do not appear to bind directly to RNA in vivo. Mol Cell 2024, 84, (7), 1271-1289 e12.
- Doolittle, W. F., Is junk DNA bunk? A critique of ENCODE. Proc Natl Acad Sci U S A 2013, 110, (14), 5294-300. [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S., The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol 2020, 21, 459–474. [CrossRef]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S., The Structure and Function of DNA G-Quadruplexes. Trends Chem 2020, 2, (2), 123-136. [CrossRef]
- Yadav, P.; Kim, N.; Kumari, M.; Verma, S.; Sharma, T. K.; Yadav, V.; Kumar, A., G-Quadruplex Structures in Bacteria: Biological Relevance and Potential as an Antimicrobial Target. J Bacteriol 2021, 203, (13), e0057720. [CrossRef]
- Wang, E.; Thombre, R.; Shah, Y.; Latanich, R.; Wang, J., G-Quadruplexes as pathogenic drivers in neurodegenerative disorders. Nucleic Acids Res 2021, 49, (9), 4816-4830. [CrossRef] [PubMed]
- Lejault, P.; Mitteaux, J.; Sperti, F. R.; Monchaud, D., How to untie G-quadruplex knots and why? Cell Chem Biol 2021, 28, (4), 436-455. [CrossRef]
- Sato, K.; Knipscheer, P., G-quadruplex resolution: From molecular mechanisms to physiological relevance. DNA Repair (Amst) 2023, 130, 103552. [CrossRef]
- Troisi, R.; Sica, F., Structural overview of DNA and RNA G-quadruplexes in their interaction with proteins. Curr Opin Struct Biol 2024, 87, 102846. [CrossRef] [PubMed]
- Sahayasheela, V. J.; Sugiyama, H., RNA G-quadruplex in functional regulation of noncoding RNA: Challenges and emerging opportunities. Cell Chem Biol 2024, 31, (1), 53-70. [CrossRef] [PubMed]
- Cammas, A.; Desprairies, A.; Dassi, E.; Millevoi, S., The shaping of mRNA translation plasticity by RNA G-quadruplexes in cancer progression and therapy resistance. NAR Cancer 2024, 6, (2), zcae025. [CrossRef]
- Sen, D.; Gilbert, W., A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 1990, 344, (6265), 410-4. [CrossRef] [PubMed]
- Fonseca Guerra, C.; Zijlstra, H.; Paragi, G.; Bickelhaupt, F. M., Telomere Structure and Stability: Covalency in Hydrogen Bonds, Not Resonance Assistance, Causes Cooperativity in Guanine Quartets. Chemistry – A European Journal 2011, 17, (45), 12612-12622.
- Sundaresan, S.; Uttamrao, P. P.; Kovuri, P.; Rathinavelan, T., The entangled world of DNA quadruplex folds. BioRxiv 2024, 2024. [CrossRef]
- Marusic, M.; Sket, P.; Bauer, L.; Viglasky, V.; Plavec, J., Solution-state structure of an intramolecular G-quadruplex with propeller, diagonal and edgewise loops. Nucleic Acids Res 2012, 40, (14), 6946-56. [CrossRef]
- Roschdi, S.; Yan, J.; Nomura, Y.; Escobar, C. A.; Petersen, R. J.; Bingman, C. A.; Tonelli, M.; Vivek, R.; Montemayor, E. J.; Wickens, M.; Kennedy, S. G.; Butcher, S. E., An atypical RNA quadruplex marks RNAs as vectors for gene silencing. Nat Struct Mol Biol 2022, 29, (11), 1113-1121. [CrossRef]
- Fay, M. M.; Lyons, S. M.; Ivanov, P., RNA G-Quadruplexes in Biology: Principles and Molecular Mechanisms. J Mol Biol 2017, 429, (14), 2127-2147. [CrossRef]
- Matsugami, A.; Okuizumi, T.; Uesugi, S.; Katahira, M., Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J Biol Chem 2003, 278, (30), 28147-53. [CrossRef]
- Palumbo, S. L.; Memmott, R. M.; Uribe, D. J.; Krotova-Khan, Y.; Hurley, L. H.; Ebbinghaus, S. W., A novel G-quadruplex-forming GGA repeat region in the c-myb promoter is a critical regulator of promoter activity. Nucleic Acids Res 2008, 36, (6), 1755-69. [CrossRef]
- Piazza, A.; Adrian, M.; Samazan, F.; Heddi, B.; Hamon, F.; Serero, A.; Lopes, J.; Teulade-Fichou, M. P.; Phan, A. T.; Nicolas, A., Short loop length and high thermal stability determine genomic instability induced by G-quadruplex-forming minisatellites. Embo J 2015, 34, (12), 1718-34. [CrossRef] [PubMed]
- Williams, J. D.; Houserova, D.; Johnson, B. R.; Dyniewski, B.; Berroyer, A.; French, H.; Barchie, A. A.; Bilbrey, D. D.; Demeis, J. D.; Ghee, K. R.; Hughes, A. G.; Kreitz, N. W.; McInnis, C. H.; Pudner, S. C.; Reeves, M. N.; Stahly, A. N.; Turcu, A.; Watters, B. C.; Daly, G. T.; Langley, R. J.; Gillespie, M. N.; Prakash, A.; Larson, E. D.; Kasukurthi, M. V.; Huang, J.; Jinks-Robertson, S.; Borchert, G. M., Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop 'G4 Kissing' interaction. Nucleic Acids Res 2020, 48, (11), 5907-5925. [CrossRef]
- Wu, F.; Niu, K.; Cui, Y.; Li, C.; Lyu, M.; Ren, Y.; Chen, Y.; Deng, H.; Huang, L.; Zheng, S.; Liu, L.; Wang, J.; Song, Q.; Xiang, H.; Feng, Q., Genome-wide analysis of DNA G-quadruplex motifs across 37 species provides insights into G4 evolution. Commun Biol 2021, 4, (1), 98. [CrossRef] [PubMed]
- Lee, C. Y.; McNerney, C.; Ma, K.; Zhao, W.; Wang, A.; Myong, S., R-loop induced G-quadruplex in non-template promotes transcription by successive R-loop formation. Nat Commun 2020, 11, (1), 3392. [CrossRef]
- Georgakopoulos-Soares, I.; Parada, G. E.; Wong, H. Y.; Medhi, R.; Furlan, G.; Munita, R.; Miska, E. A.; Kwok, C. K.; Hemberg, M., Alternative splicing modulation by G-quadruplexes. Nat Commun 2022, 13, (1), 2404. [CrossRef]
- Hegyi, H., Enhancer-promoter interaction facilitated by transiently forming G-quadruplexes. Sci Rep 2015, 5, 9165. [CrossRef]
- Zheng, K. W.; Xiao, S.; Liu, J. Q.; Zhang, J. Y.; Hao, Y. H.; Tan, Z., Co-transcriptional formation of DNA:RNA hybrid G-quadruplex and potential function as constitutional cis element for transcription control. Nucleic Acids Res 2013, 41, (10), 5533-41. [CrossRef]
- Varizhuk, A. M.; Protopopova, A. D.; Tsvetkov, V. B.; Barinov, N. A.; Podgorsky, V. V.; Tankevich, M. V.; Vlasenok, M. A.; Severov, V. V.; Smirnov, I. P.; Dubrovin, E. V.; Klinov, D. V.; Pozmogova, G. E., Polymorphism of G4 associates: from stacks to wires via interlocks. Nucleic Acids Res 2018, 46, (17), 8978-8992. [CrossRef] [PubMed]
- Kolesnikova, S.; Curtis, E. A., Structure and Function of Multimeric G-Quadruplexes. Molecules 2019, 24, (17).
- Sen, D.; Gilbert, W., Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature 1988, 334, (6180), 364-6. [CrossRef]
- Li, X.-m.; Zheng, K.-w.; Zhang, J.-y.; Liu, H.-h.; He, Y.-d.; Yuan, B.-f.; Hao, Y.-h.; Tan, Z., Guanine-vacancy–bearing G-quadruplexes responsive to guanine derivatives. Proceedings of the National Academy of Sciences 2015, 112, (47), 14581-14586. [CrossRef]
- Banco, M. T.; Ferre-D'Amare, A. R., The emerging structural complexity of G-quadruplex RNAs. Rna 2021, 27, (4), 390-402. [CrossRef]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palu, G.; Brazzale, A. R.; Richter, S. N.; Toppo, S., G-quadruplex forming sequences in the genome of all known human viruses: A comprehensive guide. PLoS Comput Biol 2018, 14, (12), e1006675. [CrossRef] [PubMed]
- Qian, S. H.; Shi, M. W.; Xiong, Y. L.; Zhang, Y.; Zhang, Z. H.; Song, X. M.; Deng, X. Y.; Chen, Z. X., EndoQuad: a comprehensive genome-wide experimentally validated endogenous G-quadruplex database. Nucleic Acids Res 2024, 52, (D1), D72-D80. [CrossRef] [PubMed]
- Spiegel, J.; Cuesta, S. M.; Adhikari, S.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S., G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol 2021, 22, (1), 117. [CrossRef] [PubMed]
- Brazda, V.; Cerven, J.; Bartas, M.; Mikyskova, N.; Coufal, J.; Pecinka, P., The Amino Acid Composition of Quadruplex Binding Proteins Reveals a Shared Motif and Predicts New Potential Quadruplex Interactors. Molecules 2018, 23, (9).
- Vasilyev, N.; Polonskaia, A.; Darnell, J. C.; Darnell, R. B.; Patel, D. J.; Serganov, A., Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proceedings of the National Academy of Sciences 2015, 112, (39), E5391-E5400. [CrossRef]
- Guedin, A.; Gros, J.; Alberti, P.; Mergny, J. L., How long is too long? Effects of loop size on G-quadruplex stability. Nucleic Acids Res 2010, 38, (21), 7858-68. [CrossRef]
- Zhang, A. Y.; Bugaut, A.; Balasubramanian, S., A sequence-independent analysis of the loop length dependence of intramolecular RNA G-quadruplex stability and topology. Biochemistry 2011, 50, (33), 7251-8. [CrossRef]
- Saha, A.; Duchambon, P.; Masson, V.; Loew, D.; Bombard, S.; Teulade-Fichou, M. P., Nucleolin Discriminates Drastically between Long-Loop and Short-Loop Quadruplexes. Biochemistry 2020, 59, (12), 1261-1272. [CrossRef]
- Ngo, K. H.; Liew, C. W.; Heddi, B.; Phan, A. T., Structural Basis for Parallel G-Quadruplex Recognition by an Ankyrin Protein. J Am Chem Soc 2024, 146, (20), 13709-13713. [CrossRef]
- Weaver, T. M.; Cortez, L. M.; Khoang, T. H.; Washington, M. T.; Agarwal, P. K.; Freudenthal, B. D., Visualizing Rev1 catalyze protein-template DNA synthesis. Proc Natl Acad Sci U S A 2020, 117, (41), 25494-25504. [CrossRef]
- Roychoudhury, S.; Pramanik, S.; Harris, H. L.; Tarpley, M.; Sarkar, A.; Spagnol, G.; Sorgen, P. L.; Chowdhury, D.; Band, V.; Klinkebiel, D.; Bhakat, K. K., Endogenous oxidized DNA bases and APE1 regulate the formation of G-quadruplex structures in the genome. Proc Natl Acad Sci U S A 2020, 117, (21), 11409-11420. [CrossRef] [PubMed]
- Pipier, A.; Devaux, A.; Lavergne, T.; Adrait, A.; Coute, Y.; Britton, S.; Calsou, P.; Riou, J. F.; Defrancq, E.; Gomez, D., Constrained G4 structures unveil topology specificity of known and new G4 binding proteins. Sci Rep 2021, 11, (1), 13469. [CrossRef]
- Mishra, S. K.; Tawani, A.; Mishra, A.; Kumar, A., G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci Rep 2016, 6, 38144. [CrossRef] [PubMed]
- Bourdon, S.; Herviou, P.; Dumas, L.; Destefanis, E.; Zen, A.; Cammas, A.; Millevoi, S.; Dassi, E., QUADRatlas: the RNA G-quadruplex and RG4-binding proteins database. Nucleic Acids Res 2023, 51, (D1), D240-D247. [CrossRef]
- Fleming, A. M.; Zhou, J.; Wallace, S. S.; Burrows, C. J., A Role for the Fifth G-Track in G-Quadruplex Forming Oncogene Promoter Sequences during Oxidative Stress: Do These "Spare Tires" Have an Evolved Function? ACS Cent Sci 2015, 1, (5), 226-233. [CrossRef] [PubMed]
- Handwerger, K. E.; Cordero, J. A.; Gall, J. G., Cajal bodies, nucleoli, and speckles in the Xenopus oocyte nucleus have a low-density, sponge-like structure. Mol Biol Cell 2005, 16, (1), 202-11. [CrossRef] [PubMed]
- Shin, Y.; Brangwynne, C. P., Liquid phase condensation in cell physiology and disease. Science 2017, 357, (6357).
- Iborra, F. J.; Pombo, A.; Jackson, D. A.; Cook, P. R., Active RNA polymerases are localized within discrete transcription "factories' in human nuclei. J Cell Sci 1996, 109 ( Pt 6), 1427-36. [CrossRef]
- Jackson, D. A., The amazing complexity of transcription factories. Brief Funct Genomic Proteomic 2005, 4, (2), 143-57. [CrossRef]
- Chubb, J. R.; Trcek, T.; Shenoy, S. M.; Singer, R. H., Transcriptional pulsing of a developmental gene. Curr Biol 2006, 16, (10), 1018-25. [CrossRef]
- Marshall, W. F.; Straight, A.; Marko, J. F.; Swedlow, J.; Dernburg, A.; Belmont, A.; Murray, A. W.; Agard, D. A.; Sedat, J. W., Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 1997, 7, (12), 930-9. [CrossRef]
- Ruggiero, E.; Tassinari, M.; Perrone, R.; Nadai, M.; Richter, S. N., Stable and Conserved G-Quadruplexes in the Long Terminal Repeat Promoter of Retroviruses. ACS Infect Dis 2019, 5, (7), 1150-1159. [CrossRef] [PubMed]
- Amrane, S.; Jaubert, C.; Bedrat, A.; Rundstadler, T.; Recordon-Pinson, P.; Aknin, C.; Guedin, A.; De Rache, A.; Bartolucci, L.; Diene, I.; Lemoine, F.; Gascuel, O.; Pratviel, G.; Mergny, J. L.; Andreola, M. L., Deciphering RNA G-quadruplex function during the early steps of HIV-1 infection. Nucleic Acids Res 2022, 50, (21), 12328-12343. [CrossRef]
- Sahakyan, A. B.; Murat, P.; Mayer, C.; Balasubramanian, S., G-quadruplex structures within the 3' UTR of LINE-1 elements stimulate retrotransposition. Nat Struct Mol Biol 2017, 24, (3), 243-247. [CrossRef] [PubMed]
- Sakamoto, M.; Ishiuchi, T., YY1-dependent transcriptional regulation manifests at the morula stage. MicroPubl Biol 2024, 2024.
- Kruisselbrink, E.; Guryev, V.; Brouwer, K.; Pontier, D. B.; Cuppen, E.; Tijsterman, M., Mutagenic capacity of endogenous G4 DNA underlies genome instability in FANCJ-defective C. elegans. Curr Biol 2008, 18, (12), 900-5. [CrossRef]
- Jones, M.; Rose, A., A DOG's View of Fanconi Anemia: Insights from C. elegans. Anemia 2012, 2012, 323721. [CrossRef]
- Tarailo-Graovac, M.; Wong, T.; Qin, Z.; Flibotte, S.; Taylor, J.; Moerman, D. G.; Rose, A. M.; Chen, N., Spectrum of variations in dog-1/FANCJ and mdf-1/MAD1 defective Caenorhabditis elegans strains after long-term propagation. BMC Genomics 2015, 16, (1), 210. [CrossRef]
- Sarkies, P.; Murat, P.; Phillips, L. G.; Patel, K. J.; Balasubramanian, S.; Sale, J. E., FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res 2012, 40, (4), 1485-98. [CrossRef]
- Liu, Y.; Zhu, X.; Wang, K.; Zhang, B.; Qiu, S., The Cellular Functions and Molecular Mechanisms of G-Quadruplex Unwinding Helicases in Humans. Front Mol Biosci 2021, 8, 783889. [CrossRef]
- Sarkies, P.; Reams, C.; Simpson, L. J.; Sale, J. E., Epigenetic instability due to defective replication of structured DNA. Mol Cell 2010, 40, (5), 703-13. [CrossRef]
- Kumagai, A.; Dunphy, W. G., MTBP, the partner of Treslin, contains a novel DNA-binding domain that is essential for proper initiation of DNA replication. Mol Biol Cell 2017, 28, (22), 2998-3012. [CrossRef]
- Poulet-Benedetti, J.; Tonnerre-Doncarli, C.; Valton, A. L.; Laurent, M.; Gerard, M.; Barinova, N.; Parisis, N.; Massip, F.; Picard, F.; Prioleau, M. N., Dimeric G-quadruplex motifs-induced NFRs determine strong replication origins in vertebrates. Nat Commun 2023, 14, (1), 4843. [CrossRef] [PubMed]
- Mitter, M.; Gasser, C.; Takacs, Z.; Langer, C. C. H.; Tang, W.; Jessberger, G.; Beales, C. T.; Neuner, E.; Ameres, S. L.; Peters, J. M.; Goloborodko, A.; Micura, R.; Gerlich, D. W., Conformation of sister chromatids in the replicated human genome. Nature 2020, 586, (7827), 139-144. [CrossRef]
- Hou, Y.; Li, F.; Zhang, R.; Li, S.; Liu, H.; Qin, Z. S.; Sun, X., Integrative characterization of G-Quadruplexes in the three-dimensional chromatin structure. Epigenetics 2019, 14, (9), 894-911. [CrossRef] [PubMed]
- De Magis, A.; Gotz, S.; Hajikazemi, M.; Fekete-Szucs, E.; Caterino, M.; Juranek, S.; Paeschke, K., Zuo1 supports G4 structure formation and directs repair toward nucleotide excision repair. Nat Commun 2020, 11, (1), 3907. [CrossRef] [PubMed]
- Ketkar, A.; Smith, L.; Johnson, C.; Richey, A.; Berry, M.; Hartman, J. H.; Maddukuri, L.; Reed, M. R.; Gunderson, J. E. C.; Leung, J. W. C.; Eoff, R. L., Human Rev1 relies on insert-2 to promote selective binding and accurate replication of stabilized G-quadruplex motifs. Nucleic Acids Res 2021, 49, (4), 2065-2084. [CrossRef]
- Sondka, Z.; Dhir, N. B.; Carvalho-Silva, D.; Jupe, S.; Madhumita; McLaren, K.; Starkey, M.; Ward, S.; Wilding, J.; Ahmed, M.; Argasinska, J.; Beare, D.; Chawla, M. S.; Duke, S.; Fasanella, I.; Neogi, A. G.; Haller, S.; Hetenyi, B.; Hodges, L.; Holmes, A.; Lyne, R.; Maurel, T.; Nair, S.; Pedro, H.; Sangrador-Vegas, A.; Schuilenburg, H.; Sheard, Z.; Yong, S. Y.; Teague, J., COSMIC: a curated database of somatic variants and clinical data for cancer. Nucleic Acids Res 2024, 52, (D1), D1210-D1217. [CrossRef]
- Liano, D.; Chowdhury, S.; Di Antonio, M., Cockayne Syndrome B Protein Selectively Resolves and Interact with Intermolecular DNA G-Quadruplex Structures. J Am Chem Soc 2021, 143, (49), 20988-21002.
- Kokic, G.; Wagner, F. R.; Chernev, A.; Urlaub, H.; Cramer, P., Structural basis of human transcription-DNA repair coupling. Nature 2021, 598, (7880), 368-372. [CrossRef] [PubMed]
- Fleming, A. M.; Zhu, J.; Ding, Y.; Esders, S.; Burrows, C. J., Oxidative Modification of Guanine in a Potential Z-DNA-Forming Sequence of a Gene Promoter Impacts Gene Expression. Chem Res Toxicol 2019, 32, (5), 899-909. [CrossRef]
- Ju, B. G.; Lunyak, V. V.; Perissi, V.; Garcia-Bassets, I.; Rose, D. W.; Glass, C. K.; Rosenfeld, M. G., A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 2006, 312, (5781), 1798-802. [CrossRef]
- Gray, L. T.; Puig Lombardi, E.; Verga, D.; Nicolas, A.; Teulade-Fichou, M. P.; Londono-Vallejo, A.; Maizels, N., G-quadruplexes Sequester Free Heme in Living Cells. Cell Chem Biol 2019, 26, (12), 1681-1691 e5.
- Li, Y.; Geyer, C. R.; Sen, D., Recognition of anionic porphyrins by DNA aptamers. Biochemistry 1996, 35, (21), 6911-22. [CrossRef] [PubMed]
- Rai, R.; Chen, Y.; Lei, M.; Chang, S., TRF2-RAP1 is required to protect telomeres from engaging in homologous recombination-mediated deletions and fusions. Nat Commun 2016, 7, 10881. [CrossRef] [PubMed]
- Mei, Y.; Deng, Z.; Vladimirova, O.; Gulve, N.; Johnson, F. B.; Drosopoulos, W. C.; Schildkraut, C. L.; Lieberman, P. M., TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci Rep 2021, 11, (1), 3509. [CrossRef]
- Lyonnais, S.; Hounsou, C.; Teulade-Fichou, M. P.; Jeusset, J.; Le Cam, E.; Mirambeau, G., G-quartets assembly within a G-rich DNA flap. A possible event at the center of the HIV-1 genome. Nucleic Acids Res 2002, 30, (23), 5276-83.
- Heddi, B.; Cheong, V. V.; Martadinata, H.; Phan, A. T., Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: Solution structure of a peptide-quadruplex complex. Proc Natl Acad Sci U S A 2015, 112, (31), 9608-13. [CrossRef]
- Chen, M. C.; Tippana, R.; Demeshkina, N. A.; Murat, P.; Balasubramanian, S.; Myong, S.; Ferre-D'Amare, A. R., Structural basis of G-quadruplex unfolding by the DEAH/RHA helicase DHX36. Nature 2018, 558, (7710), 465-469. [CrossRef] [PubMed]
- You, H.; Lattmann, S.; Rhodes, D.; Yan, J., RHAU helicase stabilizes G4 in its nucleotide-free state and destabilizes G4 upon ATP hydrolysis. Nucleic Acids Res 2017, 45, (1), 206-214. [CrossRef] [PubMed]
- Dai, Y. X.; Guo, H. L.; Liu, N. N.; Chen, W. F.; Ai, X.; Li, H. H.; Sun, B.; Hou, X. M.; Rety, S.; Xi, X. G., Structural mechanism underpinning Thermus oshimai Pif1-mediated G-quadruplex unfolding. EMBO Rep 2022, 23, (7), e53874. [CrossRef]
- Muellner, J.; Schmidt, K. H., Yeast Genome Maintenance by the Multifunctional PIF1 DNA Helicase Family. Genes (Basel) 2020, 11, (2).
- Varon, M.; Dovrat, D.; Heuze, J.; Tsirkas, I.; Singh, S. P.; Pasero, P.; Galletto, R.; Aharoni, A., Rrm3 and Pif1 division of labor during replication through leading and lagging strand G-quadruplex. Nucleic Acids Res 2024, 52, (4), 1753-1762. [CrossRef]
- Wu, W. Q.; Hou, X. M.; Li, M.; Dou, S. X.; Xi, X. G., BLM unfolds G-quadruplexes in different structural environments through different mechanisms. Nucleic Acids Res 2015, 43, (9), 4614-26. [CrossRef]
- Huet, J.; Cottrelle, P.; Cool, M.; Vignais, M. L.; Thiele, D.; Marck, C.; Buhler, J. M.; Sentenac, A.; Fromageot, P., A general upstream binding factor for genes of the yeast translational apparatus. Embo J 1985, 4, (13A), 3539-3547. [CrossRef]
- König, P.; Giraldo, R.; Chapman, L.; Rhodes, D., The Crystal Structure of the DNA-Binding Domain of Yeast RAP1 in Complex with Telomeric DNA. Cell 1996, 85, (1), 125-136. [CrossRef]
- Traczyk, A.; Liew, C. W.; Gill, D. J.; Rhodes, D., Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1. Nucleic Acids Res 2020, 48, (8), 4562-4571. [CrossRef] [PubMed]
- Herbert, A., ALU non-B-DNA conformations, flipons, binary codes and evolution. Royal Society Open Science 2020, 7, (6), 200222. [CrossRef] [PubMed]
- Esain-Garcia, I.; Kirchner, A.; Melidis, L.; Tavares, R. C. A.; Dhir, S.; Simeone, A.; Yu, Z.; Madden, S. K.; Hermann, R.; Tannahill, D.; Balasubramanian, S., G-quadruplex DNA structure is a positive regulator of MYC transcription. Proc Natl Acad Sci U S A 2024, 121, (7), e2320240121. [CrossRef]
- Shrestha, O. K.; Sharma, R.; Tomiczek, B.; Lee, W.; Tonelli, M.; Cornilescu, G.; Stolarska, M.; Nierzwicki, L.; Czub, J.; Markley, J. L.; Marszalek, J.; Ciesielski, S. J.; Craig, E. A., Structure and evolution of the 4-helix bundle domain of Zuotin, a J-domain protein co-chaperone of Hsp70. PLoS One 2019, 14, (5), e0217098. [CrossRef]
- Biffi, G.; Tannahill, D.; Balasubramanian, S., An intramolecular G-quadruplex structure is required for binding of telomeric repeat-containing RNA to the telomeric protein TRF2. J Am Chem Soc 2012, 134, (29), 11974-6. [CrossRef]
- Sharma, S.; Mukherjee, A. K.; Roy, S. S.; Bagri, S.; Lier, S.; Verma, M.; Sengupta, A.; Kumar, M.; Nesse, G.; Pandey, D. P.; Chowdhury, S., Human telomerase is directly regulated by non-telomeric TRF2-G-quadruplex interaction. Cell Rep 2021, 35, (7), 109154. [CrossRef]
- Boyer, L. A.; Latek, R. R.; Peterson, C. L., The SANT domain: a unique histone-tail-binding module? Nat Rev Mol Cell Biol 2004, 5, (2), 158-63. [CrossRef]
- Weintraub, A. S.; Li, C. H.; Zamudio, A. V.; Sigova, A. A.; Hannett, N. M.; Day, D. S.; Abraham, B. J.; Cohen, M. A.; Nabet, B.; Buckley, D. L.; Guo, Y. E.; Hnisz, D.; Jaenisch, R.; Bradner, J. E.; Gray, N. S.; Young, R. A., YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, (7), 1573-1588 e28.
- Li, L.; Williams, P.; Ren, W.; Wang, M. Y.; Gao, Z.; Miao, W.; Huang, M.; Song, J.; Wang, Y., YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat Chem Biol 2021, 17, (2), 161-168. [CrossRef]
- Wreczycka, K.; Franke, V.; Uyar, B.; Wurmus, R.; Bulut, S.; Tursun, B.; Akalin, A., HOT or not: examining the basis of high-occupancy target regions. Nucleic Acids Res 2019, 47, (11), 5735-5745. [CrossRef]
- Ramaker, R. C.; Hardigan, A. A.; Goh, S. T.; Partridge, E. C.; Wold, B.; Cooper, S. J.; Myers, R. M., Dissecting the regulatory activity and sequence content of loci with exceptional numbers of transcription factor associations. Genome Res 2020, 30, (7), 939-950. [CrossRef] [PubMed]
- Partridge, E. C.; Chhetri, S. B.; Prokop, J. W.; Ramaker, R. C.; Jansen, C. S.; Goh, S. T.; Mackiewicz, M.; Newberry, K. M.; Brandsmeier, L. A.; Meadows, S. K.; Messer, C. L.; Hardigan, A. A.; Coppola, C. J.; Dean, E. C.; Jiang, S.; Savic, D.; Mortazavi, A.; Wold, B. J.; Myers, R. M.; Mendenhall, E. M., Occupancy maps of 208 chromatin-associated proteins in one human cell type. Nature 2020, 583, (7818), 720-728. [CrossRef] [PubMed]
- Lago, S.; Nadai, M.; Cernilogar, F. M.; Kazerani, M.; Dominiguez Moreno, H.; Schotta, G.; Richter, S. N., Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat Commun 2021, 12, (1), 3885. [CrossRef] [PubMed]
- Bartman, C. R.; Hsu, S. C.; Hsiung, C. C.; Raj, A.; Blobel, G. A., Enhancer Regulation of Transcriptional Bursting Parameters Revealed by Forced Chromatin Looping. Mol Cell 2016, 62, (2), 237-247. [CrossRef]
- Hasegawa, Y.; Struhl, K., Promoter-specific dynamics of TATA-binding protein association with the human genome. Genome Res 2019, 29, (12), 1939-1950. [CrossRef]
- Henninger, J. E.; Oksuz, O.; Shrinivas, K.; Sagi, I.; LeRoy, G.; Zheng, M. M.; Andrews, J. O.; Zamudio, A. V.; Lazaris, C.; Hannett, N. M.; Lee, T. I.; Sharp, P. A.; Cisse, II; Chakraborty, A. K.; Young, R. A., RNA-Mediated Feedback Control of Transcriptional Condensates. Cell 2021, 184, (1), 207-225 e24.
- De Nicola, B.; Lech, C. J.; Heddi, B.; Regmi, S.; Frasson, I.; Perrone, R.; Richter, S. N.; Phan, A. T., Structure and possible function of a G-quadruplex in the long terminal repeat of the proviral HIV-1 genome. Nucleic Acids Res 2016, 44, (13), 6442-51. [CrossRef]
- Butovskaya, E.; Heddi, B.; Bakalar, B.; Richter, S. N.; Phan, A. T., Major G-Quadruplex Form of HIV-1 LTR Reveals a (3 + 1) Folding Topology Containing a Stem-Loop. J Am Chem Soc 2018, 140, (42), 13654-13662. [CrossRef]
- Krafcikova, P.; Demkovicova, E.; Halaganova, A.; Viglasky, V., Putative HIV and SIV G-Quadruplex Sequences in Coding and Noncoding Regions Can Form G-Quadruplexes. J Nucleic Acids 2017, 2017, 6513720. [CrossRef]
- Pathak, R., G-Quadruplexes in the Viral Genome: Unlocking Targets for Therapeutic Interventions and Antiviral Strategies. Viruses 2023, 15, (11).
- Ramskold, D.; Hendriks, G. J.; Larsson, A. J. M.; Mayr, J. V.; Ziegenhain, C.; Hagemann-Jensen, M.; Hartmanis, L.; Sandberg, R., Single-cell new RNA sequencing reveals principles of transcription at the resolution of individual bursts. Nat Cell Biol 2024. [CrossRef]
- Shen, J.; Varshney, D.; Simeone, A.; Zhang, X.; Adhikari, S.; Tannahill, D.; Balasubramanian, S., Promoter G-quadruplex folding precedes transcription and is controlled by chromatin. Genome Biol 2021, 22, (1), 143. [CrossRef]
- Baranello, L.; Wojtowicz, D.; Cui, K.; Devaiah, B. N.; Chung, H. J.; Chan-Salis, K. Y.; Guha, R.; Wilson, K.; Zhang, X.; Zhang, H.; Piotrowski, J.; Thomas, C. J.; Singer, D. S.; Pugh, B. F.; Pommier, Y.; Przytycka, T. M.; Kouzine, F.; Lewis, B. A.; Zhao, K.; Levens, D., RNA Polymerase II Regulates Topoisomerase 1 Activity to Favor Efficient Transcription. Cell 2016, 165, (2), 357-71. [CrossRef] [PubMed]
- Marchand, C.; Pourquier, P.; Laco, G. S.; Jing, N.; Pommier, Y., Interaction of Human Nuclear Topoisomerase I with Guanosine Quartet-forming and Guanosine-rich Single-stranded DNA and RNA Oligonucleotides. Journal of Biological Chemistry 2002, 277, (11), 8906-8911. [CrossRef] [PubMed]
- Schwalb, B.; Michel, M.; Zacher, B.; Fruhauf, K.; Demel, C.; Tresch, A.; Gagneur, J.; Cramer, P., TT-seq maps the human transient transcriptome. Science 2016, 352, (6290), 1225-8. [CrossRef]
- Herbert, A.; Pavlov, F.; Konovalov, D.; Poptsova, M., Conserved microRNAs and Flipons Shape Gene Expression during Development by Altering Promoter Conformations. Int J Mol Sci 2023, 24, (5).
- Herbert, A., Flipons and small RNAs accentuate the asymmetries of pervasive transcription by the reset and sequence-specific microcoding of promoter conformation. J Biol Chem 2023, 299, (9), 105140. [CrossRef]
- Herbert, A., The ancient Z-DNA and Z-RNA specific Zα fold has evolved modern roles in immunity and transcription through the natural selection of flipons. Royal Society Open Science 2024, 11, (6).
- Beknazarov, N.; Konovalov, D.; Herbert, A.; Poptsova, M., Z-DNA formation in promoters conserved between human and mouse are associated with increased transcription reinitiation rates. Sci Rep 2024, 14, (1), 17786. [CrossRef] [PubMed]
- Le, S. N.; Brown, C. R.; Harvey, S.; Boeger, H.; Elmlund, H.; Elmlund, D., The TAFs of TFIID Bind and Rearrange the Topology of the TATA-Less RPS5 Promoter. Int J Mol Sci 2019, 20, (13).
- Kouzine, F.; Wojtowicz, D.; Baranello, L.; Yamane, A.; Nelson, S.; Resch, W.; Kieffer-Kwon, K. R.; Benham, C. J.; Casellas, R.; Przytycka, T. M.; Levens, D., Permanganate/S1 Nuclease Footprinting Reveals Non-B DNA Structures with Regulatory Potential across a Mammalian Genome. Cell Syst 2017, 4, (3), 344-356. [CrossRef]
- Song, J.; Gooding, A. R.; Hemphill, W. O.; Love, B. D.; Robertson, A.; Yao, L.; Zon, L. I.; North, T. E.; Kasinath, V.; Cech, T. R., Structural basis for inactivation of PRC2 by G-quadruplex RNA. Science 2023, 381, (6664), 1331-1337. [CrossRef]
- Watanabe, T.; Totoki, Y.; Toyoda, A.; Kaneda, M.; Kuramochi-Miyagawa, S.; Obata, Y.; Chiba, H.; Kohara, Y.; Kono, T.; Nakano, T.; Surani, M. A.; Sakaki, Y.; Sasaki, H., Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 2008, 453, (7194), 539-43. [CrossRef]
- Ha, H.; Song, J.; Wang, S.; Kapusta, A.; Feschotte, C.; Chen, K. C.; Xing, J., A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics 2014, 15, (1), 545. [CrossRef]
- Ozata, D. M.; Yu, T.; Mou, H.; Gainetdinov, I.; Colpan, C.; Cecchini, K.; Kaymaz, Y.; Wu, P. H.; Fan, K.; Kucukural, A.; Weng, Z.; Zamore, P. D., Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat Ecol Evol 2020, 4, (1), 156-168.
- Li, L. C.; Okino, S. T.; Zhao, H.; Pookot, D.; Place, R. F.; Urakami, S.; Enokida, H.; Dahiya, R., Small dsRNAs induce transcriptional activation in human cells. Proc Natl Acad Sci U S A 2006, 103, (46), 17337-42. [CrossRef] [PubMed]
- Matsui, M.; Chu, Y.; Zhang, H.; Gagnon, K. T.; Shaikh, S.; Kuchimanchi, S.; Manoharan, M.; Corey, D. R.; Janowski, B. A., Promoter RNA links transcriptional regulation of inflammatory pathway genes. Nucleic Acids Res 2013, 41, (22), 10086-109. [CrossRef] [PubMed]
- Leonaite, B.; Han, Z.; Basquin, J.; Bonneau, F.; Libri, D.; Porrua, O.; Conti, E., Sen1 has unique structural features grafted on the architecture of the Upf1-like helicase family. Embo J 2017, 36, (11), 1590-1604. [CrossRef] [PubMed]
- Lansdorp, P.; van Wietmarschen, N., Helicases FANCJ, RTEL1 and BLM Act on Guanine Quadruplex DNA in Vivo. Genes (Basel) 2019, 10, (11).
- Nguyen, H. D.; Yadav, T.; Giri, S.; Saez, B.; Graubert, T. A.; Zou, L., Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol Cell 2017, 65, (5), 832-847 e4.
- Yan, Q.; Wulfridge, P.; Doherty, J.; Fernandez-Luna, J. L.; Real, P. J.; Tang, H. Y.; Sarma, K., Proximity labeling identifies a repertoire of site-specific R-loop modulators. Nat Commun 2022, 13, (1), 53. [CrossRef]
- Chernukhin, I.; Shamsuddin, S.; Kang, S. Y.; Bergstrom, R.; Kwon, Y. W.; Yu, W.; Whitehead, J.; Mukhopadhyay, R.; Docquier, F.; Farrar, D.; Morrison, I.; Vigneron, M.; Wu, S. Y.; Chiang, C. M.; Loukinov, D.; Lobanenkov, V.; Ohlsson, R.; Klenova, E., CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol Cell Biol 2007, 27, (5), 1631-48. [CrossRef]
- Gomes, N. P.; Espinosa, J. M., Gene-specific repression of the p53 target gene PUMA via intragenic CTCF-Cohesin binding. Genes Dev 2010, 24, (10), 1022-34. [CrossRef] [PubMed]
- Nanavaty, V.; Abrash, E. W.; Hong, C.; Park, S.; Fink, E. E.; Li, Z.; Sweet, T. J.; Bhasin, J. M.; Singuri, S.; Lee, B. H.; Hwang, T. H.; Ting, A. H., DNA Methylation Regulates Alternative Polyadenylation via CTCF and the Cohesin Complex. Mol Cell 2020, 78, (4), 752-764 e6.
- Mao, S. Q.; Ghanbarian, A. T.; Spiegel, J.; Martinez Cuesta, S.; Beraldi, D.; Di Antonio, M.; Marsico, G.; Hansel-Hertsch, R.; Tannahill, D.; Balasubramanian, S., DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol 2018, 25, (10), 951-957. [CrossRef]
- Alharbi, A. B.; Schmitz, U.; Bailey, C. G.; Rasko, J. E. J., CTCF as a regulator of alternative splicing: new tricks for an old player. Nucleic Acids Res 2021, 49, (14), 7825-7838. [CrossRef]
- Gajos, M.; Jasnovidova, O.; van Bommel, A.; Freier, S.; Vingron, M.; Mayer, A., Conserved DNA sequence features underlie pervasive RNA polymerase pausing. Nucleic Acids Res 2021, 49, (8), 4402-4420. [CrossRef]
- Ehara, H.; Kujirai, T.; Shirouzu, M.; Kurumizaka, H.; Sekine, S. I., Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT. Science 2022, 377, (6611), eabp9466. [CrossRef]
- Cramer, P.; Pesce, C. G.; Baralle, F. E.; Kornblihtt, A. R., Functional association between promoter structure and transcript alternative splicing. Proc Natl Acad Sci U S A 1997, 94, (21), 11456-60. [CrossRef] [PubMed]
- Cramer, P.; Caceres, J. F.; Cazalla, D.; Kadener, S.; Muro, A. F.; Baralle, F. E.; Kornblihtt, A. R., Coupling of transcription with alternative splicing: RNA pol II promoters modulate SF2/ASF and 9G8 effects on an exonic splicing enhancer. Mol Cell 1999, 4, (2), 251-8. [CrossRef]
- He, X.; Yuan, J.; Gao, Z.; Wang, Y., Promoter R-Loops Recruit U2AF1 to Modulate Its Phase Separation and RNA Splicing. J Am Chem Soc 2023, 145, (39), 21646-21660. [CrossRef] [PubMed]
- Shukla, S.; Kavak, E.; Gregory, M.; Imashimizu, M.; Shutinoski, B.; Kashlev, M.; Oberdoerffer, P.; Sandberg, R.; Oberdoerffer, S., CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 2011, 479, (7371), 74-9. [CrossRef]
- Marina, R. J.; Sturgill, D.; Bailly, M. A.; Thenoz, M.; Varma, G.; Prigge, M. F.; Nanan, K. K.; Shukla, S.; Haque, N.; Oberdoerffer, S., TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. Embo J 2016, 35, (3), 335-55. [CrossRef]
- Guo, Y.; Monahan, K.; Wu, H.; Gertz, J.; Varley, K. E.; Li, W.; Myers, R. M.; Maniatis, T.; Wu, Q., CTCF/cohesin-mediated DNA looping is required for protocadherin alpha promoter choice. Proc Natl Acad Sci U S A 2012, 109, (51), 21081-6. [CrossRef]
- Monahan, K.; Rudnick, N. D.; Kehayova, P. D.; Pauli, F.; Newberry, K. M.; Myers, R. M.; Maniatis, T., Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc Natl Acad Sci U S A 2012, 109, (23), 9125-30. [CrossRef]
- Lamas-Maceiras, M.; Singh, B. N.; Hampsey, M.; Freire-Picos, M. A., Promoter-Terminator Gene Loops Affect Alternative 3'-End Processing in Yeast. J Biol Chem 2016, 291, (17), 8960-8. [CrossRef]
- Tan-Wong, S. M.; Zaugg, J. B.; Camblong, J.; Xu, Z.; Zhang, D. W.; Mischo, H. E.; Ansari, A. Z.; Luscombe, N. M.; Steinmetz, L. M.; Proudfoot, N. J., Gene loops enhance transcriptional directionality. Science 2012, 338, (6107), 671-5. [CrossRef]
- von Hacht, A.; Seifert, O.; Menger, M.; Schutze, T.; Arora, A.; Konthur, Z.; Neubauer, P.; Wagner, A.; Weise, C.; Kurreck, J., Identification and characterization of RNA guanine-quadruplex binding proteins. Nucleic Acids Res 2014, 42, (10), 6630-44. [CrossRef]
- Zhang, J.; Harvey, S. E.; Cheng, C., A high-throughput screen identifies small molecule modulators of alternative splicing by targeting RNA G-quadruplexes. Nucleic Acids Res 2019, 47, (7), 3667-3679. [CrossRef] [PubMed]
- Jara-Espejo, M.; Fleming, A. M.; Burrows, C. J., Potential G-Quadruplex Forming Sequences and N(6)-Methyladenosine Colocalize at Human Pre-mRNA Intron Splice Sites. ACS Chem Biol 2020, 15, (6), 1292-1300. [CrossRef]
- Darnell, R. B.; Ke, S.; Darnell, J. E., Jr., Pre-mRNA processing includes N(6) methylation of adenosine residues that are retained in mRNA exons and the fallacy of "RNA epigenetics". Rna 2018, 24, (3), 262-267. [CrossRef] [PubMed]
- Wei, G.; Almeida, M.; Pintacuda, G.; Coker, H.; Bowness, J. S.; Ule, J.; Brockdorff, N., Acute depletion of METTL3 implicates N (6)-methyladenosine in alternative intron/exon inclusion in the nascent transcriptome. Genome Res 2021, 31, (8), 1395-1408. [CrossRef] [PubMed]
- Fleming, A. M.; Nguyen, N. L. B.; Burrows, C. J., Colocalization of m(6)A and G-Quadruplex-Forming Sequences in Viral RNA (HIV, Zika, Hepatitis B, and SV40) Suggests Topological Control of Adenosine N (6)-Methylation. ACS Cent Sci 2019, 5, (2), 218-228. [CrossRef]
- Yoshida, A.; Oyoshi, T.; Suda, A.; Futaki, S.; Imanishi, M., Recognition of G-quadruplex RNA by a crucial RNA methyltransferase component, METTL14. Nucleic Acids Res 2022, 50, (1), 449-457. [CrossRef]
- Patil, D. P.; Chen, C. K.; Pickering, B. F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S. R., m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, (7620), 369-373. [CrossRef] [PubMed]
- Ye, H.; Li, T.; Rigden, D. J.; Wei, Z., m6ACali: machine learning-powered calibration for accurate m6A detection in MeRIP-Seq. Nucleic Acids Res 2024, 52, (9), 4830-4842. [CrossRef]
- Iwasaki, Y.; Ookuro, Y.; Iida, K.; Nagasawa, K.; Yoshida, W., Destabilization of DNA and RNA G-quadruplex structures formed by GGA repeat due to N(6)-methyladenine modification. Biochem Biophys Res Commun 2022, 597, 134-139. [CrossRef]
- Shi, H.; Wei, J.; He, C., Where, When, and How: Context-Dependent Functions of RNA Methylation Writers, Readers, and Erasers. Mol Cell 2019, 74, (4), 640-650. [CrossRef]
- Ke, S.; Pandya-Jones, A.; Saito, Y.; Fak, J. J.; Vagbo, C. B.; Geula, S.; Hanna, J. H.; Black, D. L.; Darnell, J. E., Jr.; Darnell, R. B., m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 2017, 31, (10), 990-1006. [CrossRef] [PubMed]
- Mestre-Fos, S.; Penev, P. I.; Suttapitugsakul, S.; Hu, M.; Ito, C.; Petrov, A. S.; Wartell, R. M.; Wu, R.; Williams, L. D., G-Quadruplexes in Human Ribosomal RNA. J Mol Biol 2019, 431, (10), 1940-1955. [CrossRef]
- Scognamiglio, P. L.; Di Natale, C.; Leone, M.; Poletto, M.; Vitagliano, L.; Tell, G.; Marasco, D., G-quadruplex DNA recognition by nucleophosmin: new insights from protein dissection. Biochim Biophys Acta 2014, 1840, (6), 2050-9. [CrossRef] [PubMed]
- Okuwaki, M.; Saotome-Nakamura, A.; Yoshimura, M.; Saito, S.; Hirawake-Mogi, H.; Sekiya, T.; Nagata, K., RNA-recognition motifs and glycine and arginine-rich region cooperatively regulate the nucleolar localization of nucleolin. J Biochem 2021, 169, (1), 87-100. [CrossRef] [PubMed]
- Santos, T.; Salgado, G. F.; Cabrita, E. J.; Cruz, C., Nucleolin: a binding partner of G-quadruplex structures. Trends Cell Biol 2022, 32, (7), 561-564. [CrossRef]
- Tian, B.; Manley, J. L., Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 2017, 18, (1), 18-30. [CrossRef]
- Leppek, K.; Das, R.; Barna, M., Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 2018, 19, (3), 158-174. [CrossRef]
- Schuster, S. L.; Hsieh, A. C., The Untranslated Regions of mRNAs in Cancer. Trends Cancer 2019, 5, (4), 245-262. [CrossRef]
- Mayr, C., What Are 3' UTRs Doing? Cold Spring Harb Perspect Biol 2019, 11, (10).
- Lee, D. S. M.; Ghanem, L. R.; Barash, Y., Integrative analysis reveals RNA G-quadruplexes in UTRs are selectively constrained and enriched for functional associations. Nat Commun 2020, 11, (1), 527. [CrossRef]
- Sauer, M.; Juranek, S. A.; Marks, J.; De Magis, A.; Kazemier, H. G.; Hilbig, D.; Benhalevy, D.; Wang, X.; Hafner, M.; Paeschke, K., DHX36 prevents the accumulation of translationally inactive mRNAs with G4-structures in untranslated regions. Nat Commun 2019, 10, (1), 2421. [CrossRef]
- Benhalevy, D.; Gupta, S. K.; Danan, C. H.; Ghosal, S.; Sun, H. W.; Kazemier, H. G.; Paeschke, K.; Hafner, M.; Juranek, S. A., The Human CCHC-type Zinc Finger Nucleic Acid-Binding Protein Binds G-Rich Elements in Target mRNA Coding Sequences and Promotes Translation. Cell Rep 2017, 18, (12), 2979-2990. [CrossRef] [PubMed]
- Dong, L.; Mao, Y.; Zhou, A.; Liu, X. M.; Zhou, J.; Wan, J.; Qian, S. B., Relaxed initiation pausing of ribosomes drives oncogenic translation. Sci Adv 2021, 7, (8).
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S. R.; Qian, S. B., Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, (7574), 591-4. [CrossRef] [PubMed]
- Zaccara, S.; Jaffrey, S. R., A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 2020, 181, (7), 1582-1595 e18.
- Cirillo, L. A.; Lin, F. R.; Cuesta, I.; Friedman, D.; Jarnik, M.; Zaret, K. S., Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 2002, 9, (2), 279-89. [CrossRef]
- Zaret, K. S., Pioneer Transcription Factors Initiating Gene Network Changes. Annu Rev Genet 2020, 54, 367-385. [CrossRef] [PubMed]
- Herbert, A., Nucleosomes and flipons exchange energy to alter chromatin conformation, the readout of genomic information, and cell fate. Bioessays 2022, 44, (12), e2200166. [CrossRef]
- Czech, B.; Munafo, M.; Ciabrelli, F.; Eastwood, E. L.; Fabry, M. H.; Kneuss, E.; Hannon, G. J., piRNA-Guided Genome Defense: From Biogenesis to Silencing. Annu Rev Genet 2018, 52, 131-157. [CrossRef]
- Zyner, K. G.; Simeone, A.; Flynn, S. M.; Doyle, C.; Marsico, G.; Adhikari, S.; Portella, G.; Tannahill, D.; Balasubramanian, S., G-quadruplex DNA structures in human stem cells and differentiation. Nat Commun 2022, 13, (1), 142. [CrossRef]
- Skourti-Stathaki, K.; Torlai Triglia, E.; Warburton, M.; Voigt, P.; Bird, A.; Pombo, A., R-Loops Enhance Polycomb Repression at a Subset of Developmental Regulator Genes. Mol Cell 2019, 73, (5), 930-945 e4.
- Yang, Q.; Lin, J.; Liu, M.; Li, R.; Tian, B.; Zhang, X.; Xu, B.; Liu, M.; Zhang, X.; Li, Y.; Shi, H.; Wu, L., Highly sensitive sequencing reveals dynamic modifications and activities of small RNAs in mouse oocytes and early embryos. Sci Adv 2016, 2, (6), e1501482. [CrossRef]
- Zhang, Y.; Zhang, X.; Shi, J.; Tuorto, F.; Li, X.; Liu, Y.; Liebers, R.; Zhang, L.; Qu, Y.; Qian, J.; Pahima, M.; Liu, Y.; Yan, M.; Cao, Z.; Lei, X.; Cao, Y.; Peng, H.; Liu, S.; Wang, Y.; Zheng, H.; Woolsey, R.; Quilici, D.; Zhai, Q.; Li, L.; Zhou, T.; Yan, W.; Lyko, F.; Zhang, Y.; Zhou, Q.; Duan, E.; Chen, Q., Dnmt2 mediates intergenerational transmission of paternally acquired metabolic disorders through sperm small non-coding RNAs. Nat Cell Biol 2018, 20, (5), 535-540. [CrossRef]
- Paloviita, P.; Hyden-Granskog, C.; Yohannes, D. A.; Paluoja, P.; Kere, J.; Tapanainen, J. S.; Krjutskov, K.; Tuuri, T.; Vosa, U.; Vuoristo, S., Small RNA expression and miRNA modification dynamics in human oocytes and early embryos. Genome Res 2021, 31, (8), 1474-1485. [CrossRef]
- Tomar, A.; Gomez-Velazquez, M.; Gerlini, R.; Comas-Armangue, G.; Makharadze, L.; Kolbe, T.; Boersma, A.; Dahlhoff, M.; Burgstaller, J. P.; Lassi, M.; Darr, J.; Toppari, J.; Virtanen, H.; Kuhnapfel, A.; Scholz, M.; Landgraf, K.; Kiess, W.; Vogel, M.; Gailus-Durner, V.; Fuchs, H.; Marschall, S.; Hrabe de Angelis, M.; Kotaja, N.; Korner, A.; Teperino, R., Epigenetic inheritance of diet-induced and sperm-borne mitochondrial RNAs. Nature 2024, 630, (8017), 720-727. [CrossRef]
- Maldonado, R.; Langst, G., The chromatin - triple helix connection. Biol Chem 2023, 404, (11-12), 1037-1049.
- Leisegang, M. S.; Warwick, T.; Stotzel, J.; Brandes, R. P., RNA-DNA triplexes: molecular mechanisms and functional relevance. Trends Biochem Sci 2024, 49, (6), 532-544. [CrossRef] [PubMed]
- Zhou, Z.; Giles, K. E.; Felsenfeld, G., DNA.RNA triple helix formation can function as a cis-acting regulatory mechanism at the human beta-globin locus. Proc Natl Acad Sci U S A 2019, 116, (13), 6130-6139. [CrossRef] [PubMed]
- Maldonado, R.; Schwartz, U.; Silberhorn, E.; Langst, G., Nucleosomes Stabilize ssRNA-dsDNA Triple Helices in Human Cells. Mol Cell 2019, 73, (6), 1243-1254 e6.
- Kohestani, H.; Wereszczynski, J., The effects of RNA.DNA-DNA triple helices on nucleosome structures and dynamics. Biophys J 2023, 122, (7), 1229-1239.
- Jimenez-Garcia, E.; Vaquero, A.; Espinas, M. L.; Soliva, R.; Orozco, M.; Bernues, J.; Azorin, F., The GAGA factor of Drosophila binds triple-stranded DNA. J Biol Chem 1998, 273, (38), 24640-8. [CrossRef] [PubMed]
- Leisegang, M. S.; Bains, J. K.; Seredinski, S.; Oo, J. A.; Krause, N. M.; Kuo, C. C.; Gunther, S.; Senturk Cetin, N.; Warwick, T.; Cao, C.; Boos, F.; Izquierdo Ponce, J.; Haydar, S.; Bednarz, R.; Valasarajan, C.; Fuhrmann, D. C.; Preussner, J.; Looso, M.; Pullamsetti, S. S.; Schulz, M. H.; Jonker, H. R. A.; Richter, C.; Rezende, F.; Gilsbach, R.; Pfluger-Muller, B.; Wittig, I.; Grummt, I.; Ribarska, T.; Costa, I. G.; Schwalbe, H.; Brandes, R. P., HIF1alpha-AS1 is a DNA:DNA:RNA triplex-forming lncRNA interacting with the HUSH complex. Nat Commun 2022, 13, (1), 6563. [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




