Submitted:
20 August 2024
Posted:
21 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Reagents and Culture Media
2.2. Strains, Media and Culture Conditions
2.3. Fermentation of Isolates
2.4. Preparation of extracts from pigmented strains
2.5. Biological Activity Tests
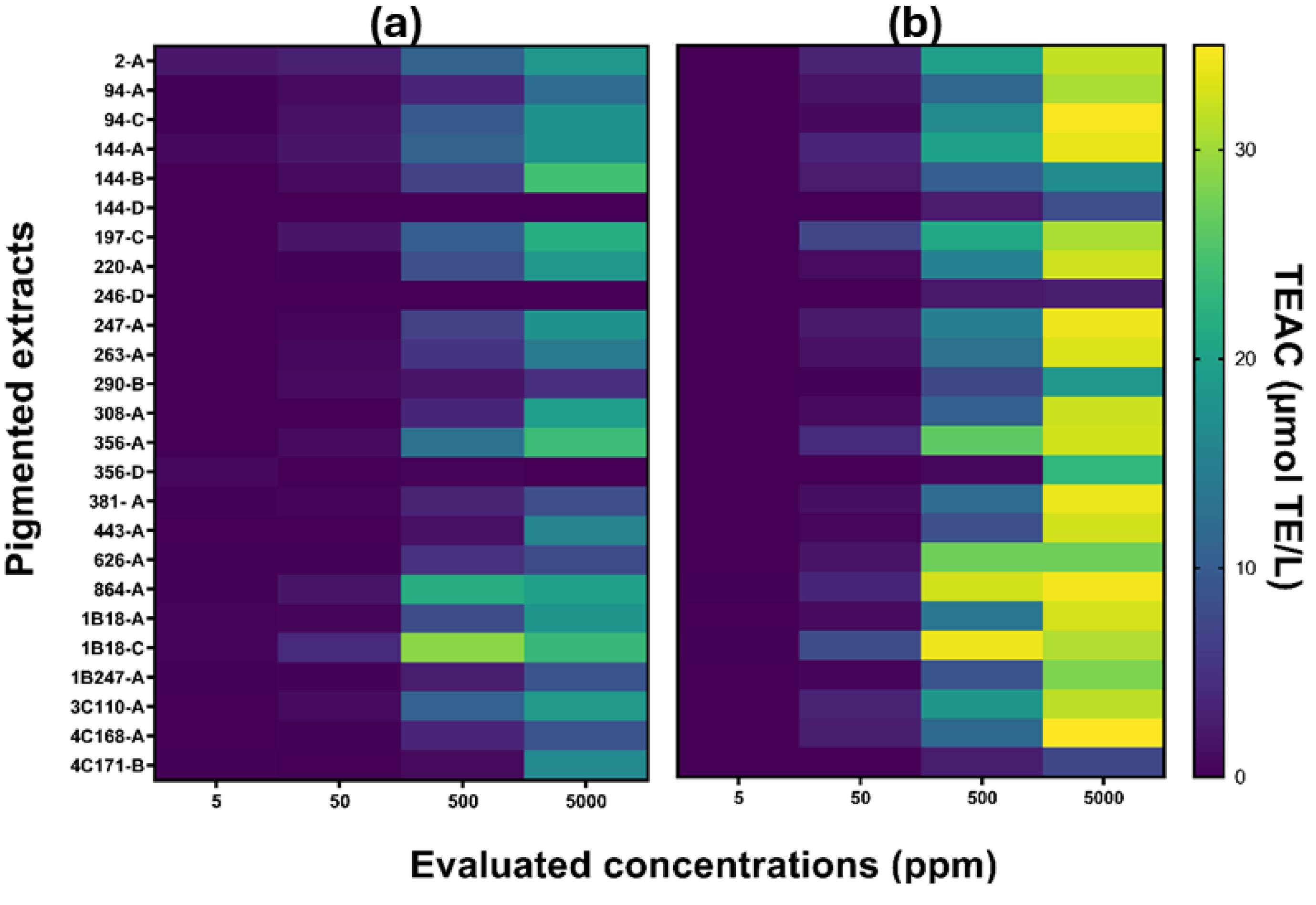
2.5.1. Antioxidant Capacity
2.5.2. Antibacterial Activity
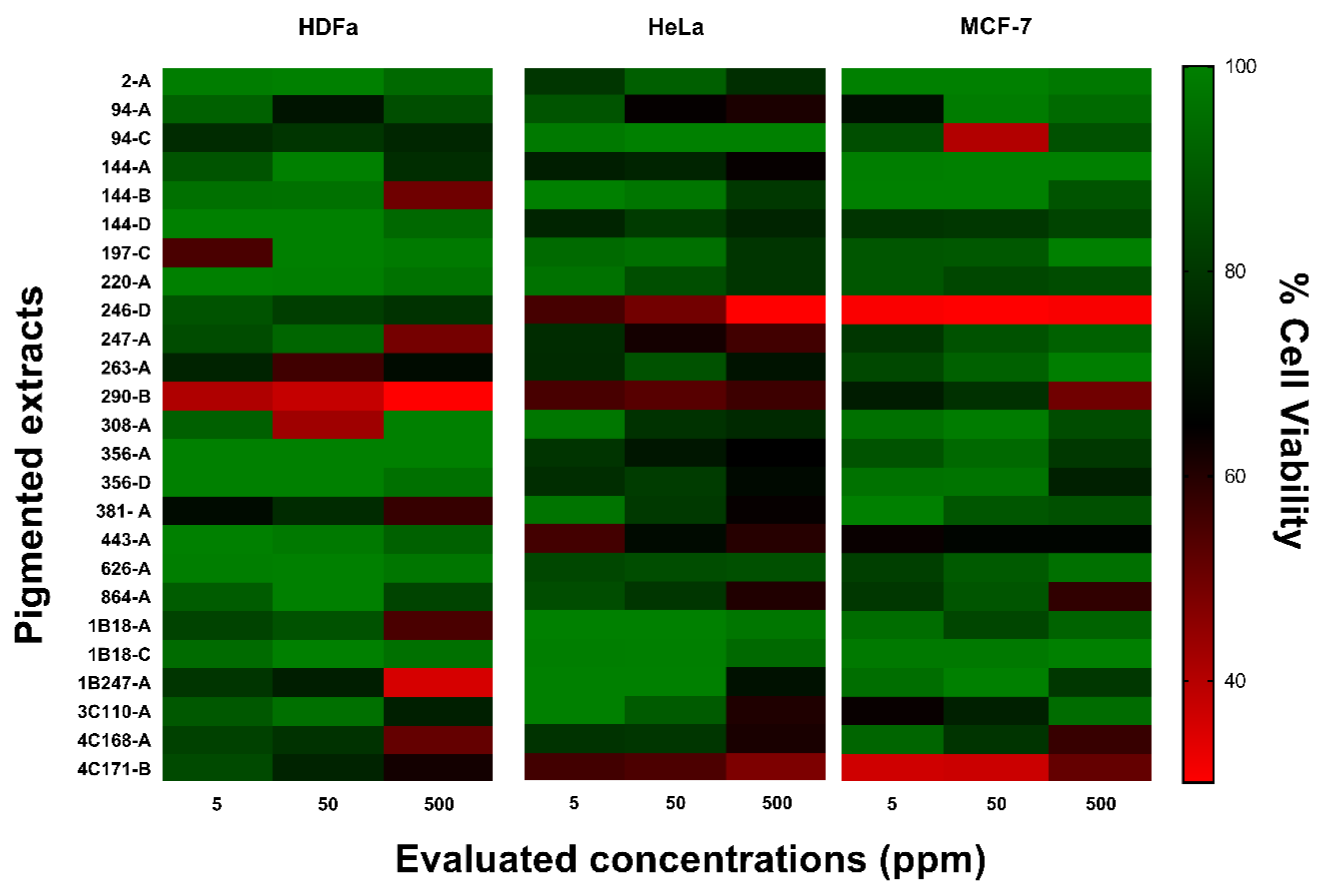
2.5.3. Cytotoxic Activity
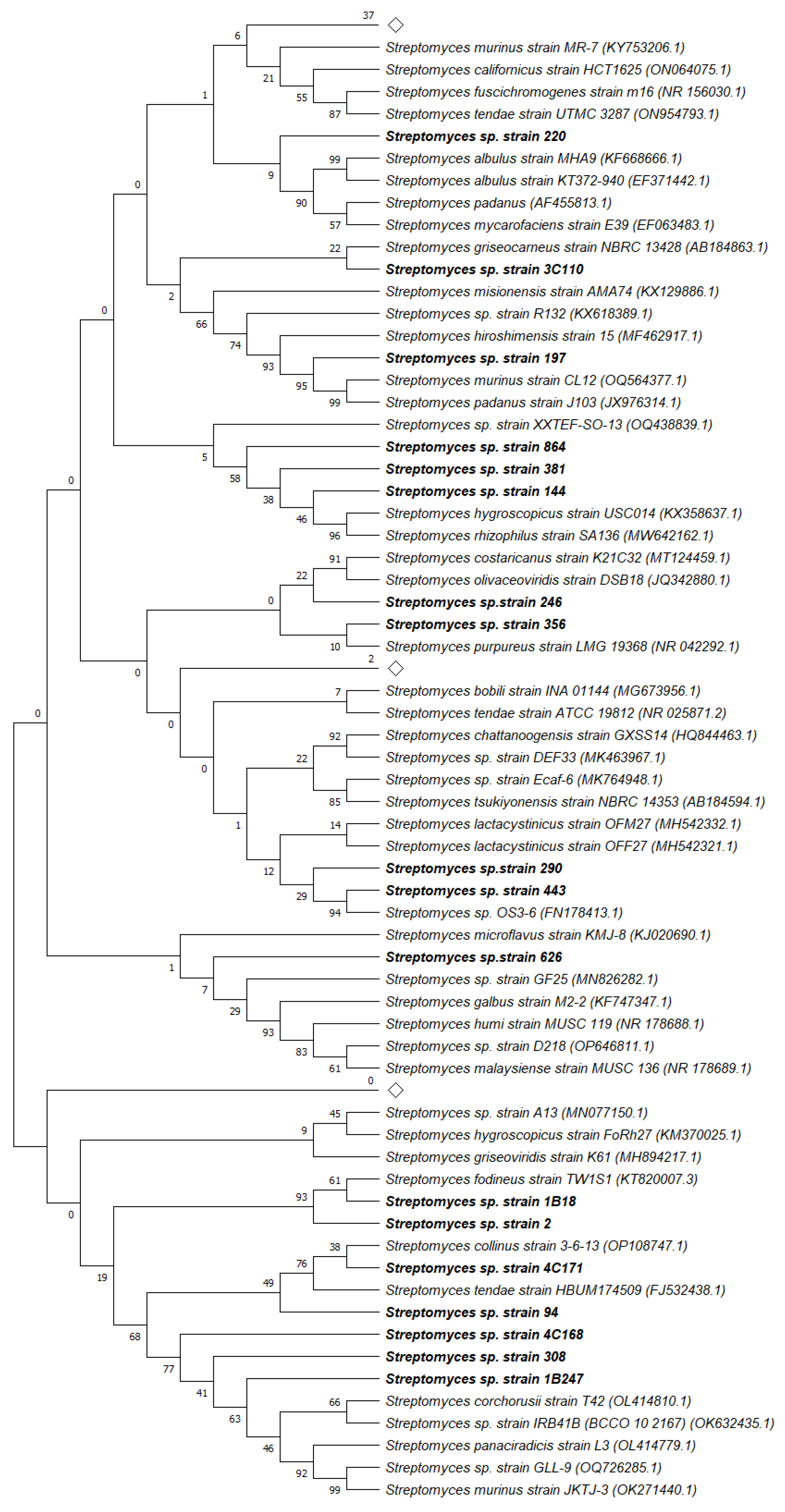
2.6. Molecular Identification
2.7. High-Performance Liquid Chromatography Coupled with Mass Spectrometry
3. Results and Discussion
3.1. Pigmentation of the Strains in the Different Culture Media
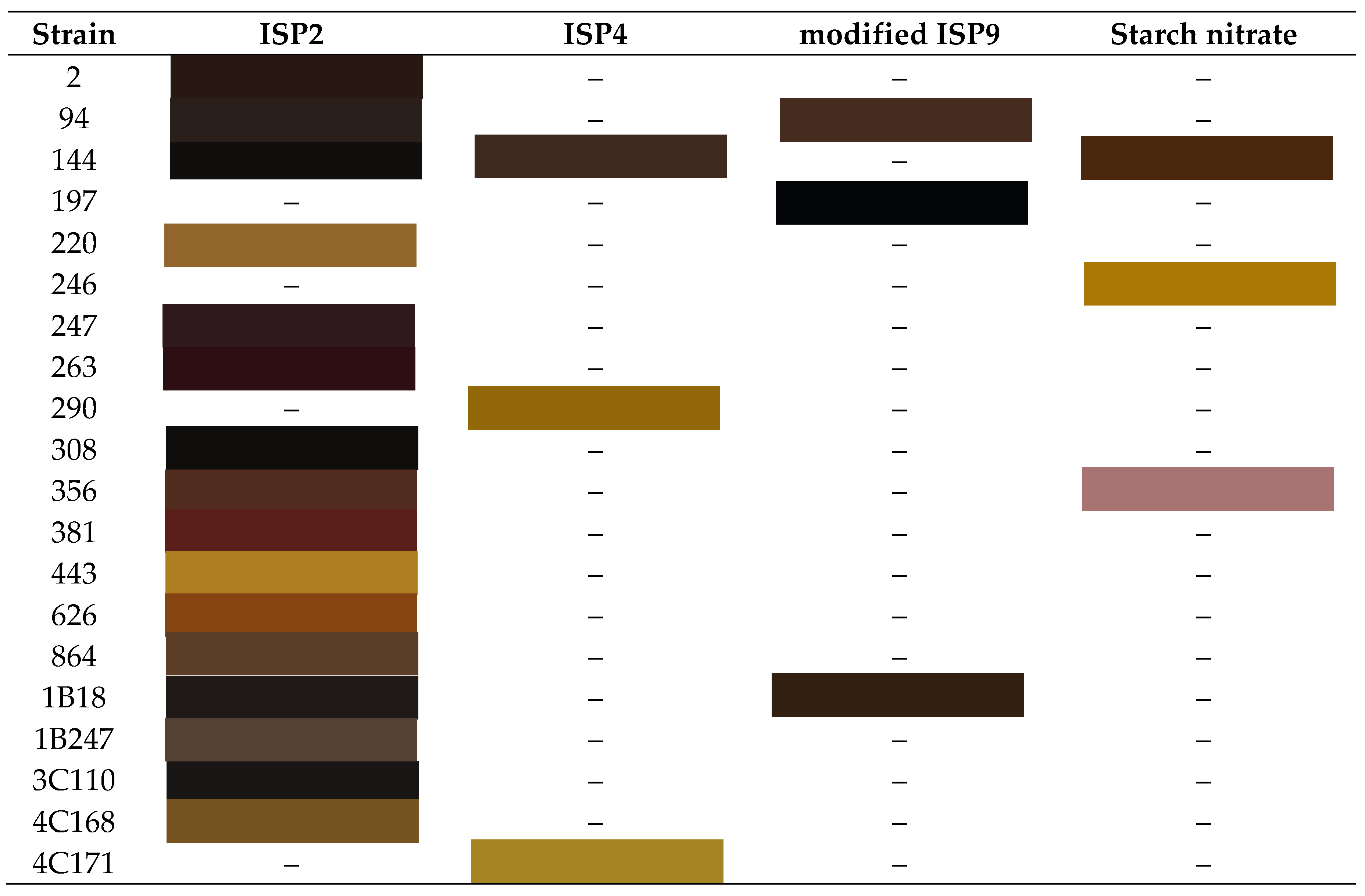 |
3.2. Molecular Identification
3.3. Biological Activity Assessment
3.3.1. Antioxidant Capacity
3.3.2. Antibacterial Activity
| Pigmented Extracts |
Tested bacterial strains | Pigmented Extracts |
Tested bacterial strains | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 4 | 5 | 6 | ||
| 2-A | – | – | – | – | – | – | 356-A | – | – | – | – | ++ | – |
| 94-A | – | – | – | – | – | – | 356-D | – | – | – | – | – | – |
| 94-C | – | – | – | – | – | – | 381-A | – | – | – | – | – | – |
| 144-A | – | – | – | – | – | – | 443-A | – | – | – | – | – | – |
| 144-B | – | – | – | – | – | – | 626-A | – | – | – | – | – | – |
| 144-D | – | – | – | – | – | – | 864-A | – | – | – | – | – | – |
| 197-C | – | – | – | – | – | – | 1B18-A | – | – | – | – | – | – |
| 220-A | – | – | – | – | – | – | 1B18-C | – | – | – | – | – | – |
| 246-D | – | – | – | + | ++ | ++ | 1B247-A | – | – | – | – | ++ | +++ |
| 246-A | – | – | – | – | +++ | ++ | 3C110-A | – | – | – | – | ++ | – |
| 263-A | – | – | – | – | – | – | 4C168-A | – | – | – | – | – | – |
| 290-B | – | – | – | +++ | ++ | ++ | 4C171-B | – | – | – | ++ | +++ | +++ |
| 308-A | – | – | – | – | – | – | |||||||
| Strain code | Identification | % Similarity |
|---|---|---|
| 2 | Streptomyces griseochromogenes | 99.93% |
| 94 | Streptomyces tendae | 99.86% |
| 144 | Streptomyces humi | 99.55% |
| 197 | Streptomyces misionensis | 99.72% |
| 220 | Streptomyces argillaceus | 100% |
| 246 | Streptomyces murinus | 99.93% |
| 290 | Streptomyces noursei | 99.24% |
| 308 | Streptomyces humi | 99.64% |
| 356 | Streptomyces hygroscopicus | 99.93% |
| 381 | Streptomyces albospinus | 99.18% |
| 443 | Streptomyces murinus | 98.96% |
| 626 | Streptomyces albospinus | 99.85% |
| 864 | Streptomyces lacticiproducens | 99.17% |
| 1B18 | Streptomyces lactacystinicus | 99.35% |
| 1B247 | Streptomyces mediolani | 99.64% |
| 3C110 | Streptomyces fodineus | 99.86% |
| 4C168 | Streptomyces murinus | 99.93% |
| 4C171 | Streptomyces murinus | 99.86% |
3.3.3. Cytotoxic Activity
3.4. HPLC-MS-Based Annotation (Level 3) of Metabolites Possibly Responsible for the Observed Activities by the Different Streptomyces-Derived Extracts
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marcano, D. Introducción a La Química de Los Colorantes; 2018. ISBN 9789806195592.
- Lyu, X.; Lyu, Y.; Yu, H.; Chen, W.; Ye, L.; Yang, R. Biotechnological Advances for Improving Natural Pigment Production: A State-of-the-Art Review. Bioresour. Bioprocess. 2022, 9, 8. [Google Scholar] [CrossRef]
- Ahmad, W.A.; Yusof, N.Z.; Nordin, N.; Zakaria, Z.A.; Rezali, M.F. Production and Characterization of Violacein by Locally Isolated Chromobacterium Violaceum Grown in Agricultural Wastes. Appl. Biochem. Biotechnol. 2012, 167, 1220–1234. [Google Scholar] [CrossRef] [PubMed]
- Sajda. S. Affat Classifications, Advantages, Disadvantages, Toxicity Effects of Natural and Synthetic Dyes: A Review. Univ. Thi-Qar J. Sci. 2021, 8, 130–135. [Google Scholar]
- Venil, C.K.; Zakaria, Z.A.; Ahmad, W.A. Bacterial Pigments and Their Applications. Process Biochem. 2013, 48, 1065–1079. [Google Scholar] [CrossRef]
- Paul, T.; Bandyopadhyay, T.K.; Mondal, A.; Tiwari, O.N.; Muthuraj, M.; Bhunia, B. A Comprehensive Review on Recent Trends in Production, Purification, and Applications of Prodigiosin. Biomass Convers. Biorefinery 2020. [Google Scholar] [CrossRef]
- Husain, Q. Potential Applications of the Oxidoreductive Enzymes in the Decolorization and Detoxification of Textile and Other Synthetic Dyes from Polluted Water: A Review. Crit. Rev. Biotechnol. 2006, 26, 201–221. [Google Scholar] [CrossRef]
- Khattab, T.A.; Abdelrahman, M.S.; Rehan, M. Textile Dyeing Industry: Environmental Impacts and Remediation. Environ. Sci. Pollut. Res. 2020, 27, 3803–3818. [Google Scholar] [CrossRef]
- Sarayu, K.; Sandhya, S. Current Technologies for Biological Treatment of Textile Wastewater–A Review. Appl. Biochem. Biotechnol. 2012, 167, 645–661. [Google Scholar] [CrossRef]
- Lellis, B.; Fávaro-Polonio, C.Z.; Pamphile, J.A.; Polonio, J.C. Effects of Textile Dyes on Health and the Environment and Bioremediation Potential of Living Organisms. Biotechnol. Res. Innov. 2019, 3, 275–290. [Google Scholar] [CrossRef]
- Sanchez-Marroquín, A.; Zapata, M. Observations on the Pigment of Streptomyces Coelicolor. 1954, 2.
- Manivasagan, P.; Venkatesan, J.; Sivakumar, K.; Kim, S.-K. Actinobacterial Melanins: Current Status and Perspective for the Future. World J. Microbiol. Biotechnol. 2013, 29, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Dharmaraj, S. Marine Streptomyces as a Novel Source of Bioactive Substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Mann, J. Natural Products as Immunosuppressive Agents. Nat. Prod. Rep. 2001, 18, 417–430. [Google Scholar] [CrossRef]
- Olano, C.; Méndez, C.; Salas, J.A. Antitumor Compounds from Marine Actinomycetes. Mar. Drugs 2009, 7, 210–248. [Google Scholar] [CrossRef]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary Metabolites and Biodiversity of Actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19. [Google Scholar] [CrossRef]
- Pathom-aree, W.; Stach, J.E.M.; Ward, A.C.; Horikoshi, K.; Bull, A.T.; Goodfellow, M. Diversity of Actinomycetes Isolated from Challenger Deep Sediment (10,898 m) from the Mariana Trench. Extremophiles 2006, 10, 181–189. [Google Scholar] [CrossRef]
- Bull, A.T. Microbial Diversity and Bioprospecting; Bull, A.T., Ed.; ASM Press: Washington, DC, USA, 2003. ISBN 9781683672173.
- Berdi, J. Bioactive Microbial Metabolites. J. Antibiot. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Devi Salam, M. Antimicrobial Potential of Actinomycetes Isolated from Soil Samples of Punjab, India. J. Microbiol. Exp. 2014, 1. [Google Scholar] [CrossRef]
- Darshan, N.; Manonmani, H.K. Prodigiosin and Its Potential Applications. J. Food Sci. Technol. 2015, 52, 5393–5407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.C.; Zhan, J.X.; Su, K.M.; Zhang, Y.X. A Kind of Potential Food Additive Produced by Streptomyces Coelicolor: Characteristics of Blue Pigment and Identification of a Novel Compound, Lambda-Actinorhodin. Food Chem. 2006, 95, 186–192. [Google Scholar] [CrossRef]
- Petinate, S.D.; Martins, R.M.; Coelho, R.R.; Meirelles, M.N.L.; Branquinha, M.H.; Vermelho, A.B. Influence of Growth Medium in Proteinase and Pigment Production by Streptomyces Cyaneus. Mem. Inst. Oswaldo Cruz 1999, 94, 173–177. [Google Scholar] [CrossRef]
- Zhu, H.-H.; Guo, J.; Yao, Q.; Yang, S.-Z.; Deng, M.-R.; Le Phuong, T.B.; Hanh, V.T.; Ryan, M.J. Streptomyces Vietnamensis Sp. Nov., a Streptomycete with Violet Blue Diffusible Pigment Isolated from Soil in Vietnam. Int. J. Syst. Evol. Microbiol. 2007, 57, 1770–1774. [Google Scholar] [CrossRef]
- Gupta, C.; Garg, A.P.; Prakash, D.; Goyal, S. Microbes as Potential Sources of Biocolours. Pharmacologyonline 2011, 1309–1318. [Google Scholar]
- Lin, Y.B.; Wang, X.Y.; Fang, H.; Ma, Y.N.; Tang, J.; Tang, M.; Wei, G.H. Streptomyces Shaanxiensis Sp. Nov., a Blue Pigment-Producing Streptomycete from Sewage Irrigation Soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 1725–1730. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.-H.; Guo, J.; Yao, Q.; Yang, S.-Z.; Deng, M.-R.; Li, T.-H. Streptomyces Caeruleatus Sp. Nov., with Dark Blue Diffusible Pigment. Int. J. Syst. Evol. Microbiol. 2011, 61, 507–511. [Google Scholar] [CrossRef]
- Malik, K.; Tokkas, J.; Goyal, S. Microbial Pigments : A Review. Int. J. Microb. Resour. Technol. 2012, 361–365. [Google Scholar]
- Morales-Oyervides, L.; Oliveira, J.; Sousa-Gallagher, M.; Méndez-Zavala, A.; Montañez, J.C. Assessment of the Dyeing Properties of the Pigments Produced by Talaromyces Spp. J. Fungi 2017, 3. [Google Scholar] [CrossRef]
- Narsing Rao, M.P.; Xiao, M.; Li, W. Fungal and Bacterial Pigments : Secondary Metabolites with Wide Applications. Front. Microbiol. 2017, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pombeiro-Sponchiado, S.R.; Sousa, G.S.; Andrade, J.C.R.; Lisboa, H.F.; Gonçalves, R.C.R. Production of Melanin Pigment by Fungi and Its Biotechnological Applications. Melanin 2017. [Google Scholar] [CrossRef]
- Pailliè-Jiménez, M.E.; Stincone, P.; Brandelli, A. Natural Pigments of Microbial Origin. Front. Sustain. Food Syst. 2020, 4, 1–8. [Google Scholar] [CrossRef]
- Ramesh, C.; Vinithkumar, N.V.; Kirubagaran, R.; Venil, C.K.; Dufossé, L. Multifaceted Applications of Microbial Pigments: Current Knowledge, Challenges and Future Directions for Public Health Implications; 2019; Vol. 7; ISBN 3326221754.
- Uribe Vélez, D. Metagenómica ¿Una Oportunidad Para El Estudio de La Diversidad Microbiana En Colombia? Rev. Colomb. Biotecnol. 2009, XI, 4–7. [Google Scholar]
- Shirling, E.B.; Gottlieb, D. Methods for Characterization of Streptomyces Species. Int. J. Syst. Bacteriol. 1966, 16, 313–340. [Google Scholar] [CrossRef]
- El-Naggar, N.E.-A.; El-Ewasy, S.M. Bioproduction, Characterization, Anticancer and Antioxidant Activities of Extracellular Melanin Pigment Produced by Newly Isolated Microbial Cell Factories Streptomyces Glaucescens NEAE-H. Sci. Rep. 2017, 7, 42129. [Google Scholar] [CrossRef] [PubMed]
- Benouagueni, S.; Ranque, S.; Gacemi Kirane, D. A Non-Polyenic Antifungal Produced by a Streptomyces Yatensis Strain Isolated from Mellah Lake in El Kala, North-East of Algeria. J. Mycol. Med. 2015, 25, 2–10. [Google Scholar] [CrossRef]
- Siddharth, S.; Vittal, R.R. Evaluation of Antimicrobial, Enzyme Inhibitory, Antioxidant and Cytotoxic Activities of Partially Purified Volatile Metabolites of Marine Streptomyces Sp.S2A. Microorganisms 2018, 6, 72. [Google Scholar] [CrossRef]
- Francenia Santos-Sánchez, N.; Salas-Coronado, R.; Villanueva-Cañongo, C.; Hernández-Carlos, B. Antioxidant Compounds and Their Antioxidant Mechanism. In Antioxidants; IntechOpen, 2019; pp. 1–29.
- Taroco, R.; Seija, V.; Vignoli, R. Métodos de Estudio de La Sensibilidad Antibiótica. In Temas de Bacterología y Virología Médica; 2008; pp. 663–671.
- Prashanthi, K.; Suryan, S.; Varalakshmi, K.N. In Vitro Anticancer Property of Yellow Pigment from Streptomyces Griseoaurantiacus JUACT 01. Brazilian Arch. Biol. Technol. 2015, 58, 869–876. [Google Scholar] [CrossRef]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the Human-Ape Splitting by a Molecular Clock of Mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Ramirez-Rodriguez, L.; Stepanian-Martinez, B.; Morales-Gonzalez, M.; Diaz, L. Optimization of the Cytotoxic Activity of Three Streptomyces Strains Isolated from Guaviare River Sediments (Colombia, South America). Biomed Res. Int. 2018, 2018, 1–14. [Google Scholar] [CrossRef]
- Bernal, F.A.; Orduz-Díaz, L.L.; Coy-Barrera, E. Application of Parafac and OPLS-DA Analyses on HPLC Fingerprints for the Characterization of Hibiscus Sabdariffa Calyxes. Quim. Nova 2016. [Google Scholar] [CrossRef]
- Schmid, R.; Heuckeroth, S.; Korf, A.; Smirnov, A.; Myers, O.; Dyrlund, T.S.; Bushuiev, R.; Murray, K.J.; Hoffmann, N.; Lu, M.; et al. Integrative Analysis of Multimodal Mass Spectrometry Data in MZmine 3. Nat. Biotechnol. 2023, 41, 447–449. [Google Scholar] [CrossRef]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C. V; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An Updated Compendium of Streptomycetes Natural Products. Nucleic Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2023 Update. Nucleic Acids Res. 2023, 51, D1373–D1380. [Google Scholar] [CrossRef] [PubMed]
- Rutz, A.; Sorokina, M.; Galgonek, J.; Mietchen, D.; Willighagen, E.; Gaudry, A.; Graham, J.G.; Stephan, R.; Page, R.; Vondrášek, J.; et al. The LOTUS Initiative for Open Knowledge Management in Natural Products Research. Elife 2022, 11. [Google Scholar] [CrossRef]
- van Santen, J.A.; Poynton, E.F.; Iskakova, D.; McMann, E.; Alsup, T.A.; Clark, T.N.; Fergusson, C.H.; Fewer, D.P.; Hughes, A.H.; McCadden, C.A.; et al. The Natural Products Atlas 2.0: A Database of Microbially-Derived Natural Products. Nucleic Acids Res. 2022, 50, D1317–D1323. [Google Scholar] [CrossRef]
- van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef]
- Wink, J.; Fatemeh, M.; Javad, H. Biology and Biotechnology of Actinobacteria; Wink, J. , Mohammadipanah, F., Hamedi, J., Eds.; Springer International Publishing: Cham, 2017; ISBN 978-3-319-60338-4. [Google Scholar]
- Anzai, K.; Ohno, M.; Nakashima, T.; Kuwahara, N.; Suzuki, R.; Tamura, T.; Komaki, H.; Miyadoh, S.; Harayama, S.; Ando, K. Taxonomic Distribution of Streptomyces Species Capable of Producing Bioactive Compounds among Strains Preserved at NITE/NBRC. Appl. Microbiol. Biotechnol. 2008, 80, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Ryu, Y.-G.; Butler, M.J.; Chater, K.F.; Lee, K.J. Engineering of Primary Carbohydrate Metabolism for Increased Production of Actinorhodin in Streptomyces Coelicolor. Appl. Environ. Microbiol. 2006, 72, 7132–7139. [Google Scholar] [CrossRef]
- Demain, A.L.; Sanchez, S. Microbial Drug Discovery: 80 Years of Progress. J. Antibiot. (Tokyo). 2009, 62, 5–16. [Google Scholar] [CrossRef]
- Romero-Rodríguez, A.; Maldonado-Carmona, N.; Ruiz-Villafán, B.; Koirala, N.; Rocha, D.; Sánchez, S. Interplay between Carbon, Nitrogen and Phosphate Utilization in the Control of Secondary Metabolite Production in Streptomyces. Antonie Van Leeuwenhoek 2018, 111, 761–781. [Google Scholar] [CrossRef]
- Görke, B.; Stülke, J. Carbon Catabolite Repression in Bacteria: Many Ways to Make the Most out of Nutrients. Nat. Rev. Microbiol. 2008, 6, 613–624. [Google Scholar] [CrossRef]
- Omura, S.; Tanaka, Y.; Mamada, H.; Masuma, R. Effect of Ammonium Ion, Inorganic Phosphate and Amino Acids on the Biosynthesis of Protylonolide, a Precursor of Tylosin Aglycone. J. Antibiot. (Tokyo). 1984, 37, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Tomé, D. Yeast Extracts: Nutritional and Flavoring Food Ingredients. ACS Food Sci. Technol. 2021, 1, 487–494. [Google Scholar] [CrossRef]
- Narayana, K.J.P.; Kumar, K.G.; Vijayalakshmi, M. L-Asparaginase Production by Streptomyces Albidoflavus. Indian J. Microbiol. 2008, 48, 331–336. [Google Scholar] [CrossRef] [PubMed]
- Khopade, A.; Ren, B.; Liu, X.-Y.; Mahadik, K.; Zhang, L.; Kokare, C. Production and Characterization of Biosurfactant from Marine Streptomyces Species B3. J. Colloid Interface Sci. 2012, 367, 311–318. [Google Scholar] [CrossRef]
- Langheinrich, W.; Ring, K. Regulation of Amino Acid Transport in Growing Cells of Streptomyces Hydrogenans. Arch. Microbiol. 1976, 109, 227–235. [Google Scholar] [CrossRef]
- Van Wezel, G.P.; Mahr, K.; König, M.; Traag, B.A.; Pimentel-Schmitt, E.F.; Willimek, A.; Titgemeyer, F. GlcP Constitutes the Major Glucose Uptake System of Streptomyces Coelicolor A3(2). Mol. Microbiol. 2004, 55, 624–636. [Google Scholar] [CrossRef] [PubMed]
- Čihák, M.; Kameník, Z.; Šmídová, K.; Bergman, N.; Benada, O.; Kofroňová, O.; Petříčková, K.; Bobek, J. Secondary Metabolites Produced during the Germination of Streptomyces Coelicolor. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Saengkhae, C.; Srivibool, R.; Watanadilok, R.; Enomoto, K. Partially Purified Pigment Extract from Streptomyces A 16-1 Induces Apoptosis of Human Carcinoma of Nasopharynx Cell (KB Cells) via the Mitochondrial and Caspase-3 Pathway. Walailak J. Sci. Technol. 2017, 14, 51–63. [Google Scholar] [CrossRef]
- Karuppiah, V.; Aarthi, C.; Ramohan, *!!! REPLACE !!!*; Sivakumar, K.; Kannan, L. Statistical Optimization and Anticancer Activity of a Red Pigment Isolated from Streptomyces Sp. PM4. Asian Pac. J. Trop. Biomed. 2013, 3, 650–656. [Google Scholar] [CrossRef]
- Vaishnavi, M.; Manigundan, K.; Smalia, T.; N, *!!! REPLACE !!!*; Hini, S.U.; Gopikrishnan, V.; Kumar, A.; Hanna, L.E.; Radhakrishnan, M.; Aruni, W. Antibacterial and Anti-HIV Activity of Extracellular Pigment from Streptomyces Sp. S45 Isolated from Sabarimala Forest Soil, India. Indian J. Exp. Biol. 2020, 58, 861–868. [Google Scholar] [CrossRef]
- Bayram, S.; Dengiz, C.; Gerçek, Y.C.; Cetin, I.; Topcul, M.R. Bioproduction, Structure Elucidation and in Vitro Antiproliferative Effect of Eumelanin Pigment from Streptomyces Parvus BSB49. Arch. Microbiol. 2020, 202, 2401–2409. [Google Scholar] [CrossRef] [PubMed]
- Bayram, S. Production, Purification, and Characterization of Streptomyces Sp. Strain MPPS2 Extracellular Pyomelanin Pigment. Arch. Microbiol. 2021. [Google Scholar] [CrossRef]
- Dastager, S.G.; Wen-Jun, L.; Dayanand, A.; Shu-Kun, T.; Xin-Peng, T.; Xiao-Yang, Z.; Li-Hua, X.; Cheng-Lin, J. Seperation, Identification and Analysis of Pigment (Melanin) Production in Streptomyces. African J. Biotechnol. 2006, 5, 1131–1134. [Google Scholar]
- Tang-um, J.; Niamsup, H. Extracellular Amylase Activity from Endophytic Streptomyces Griseoflavus P4. Chiang Mai J. Sci. 2012, 39, 346–350. [Google Scholar]
- Singh, R.; Tiwari, M.; Singh, R.; Lee, J.-K. From Protein Engineering to Immobilization: Promising Strategies for the Upgrade of Industrial Enzymes. Int. J. Mol. Sci. 2013, 14, 1232–1277. [Google Scholar] [CrossRef] [PubMed]
- Elibol, M. Optimization of Medium Composition for Actinorhodin Production by Streptomyces Coelicolor A3(2) with Response Surface Methodology. Process Biochem. 2004, 39, 1057–1062. [Google Scholar] [CrossRef]
- Elyasi Far, B.; Ahmadi, Y.; Yari Khosroshahi, A.; Dilmaghani, A. Microbial Alpha-Amylase Production: Progress, Challenges and Perspectives. Adv. Pharm. Bull. 2020, 10, 350–358. [Google Scholar] [CrossRef]
- Abraham, J.; Chauhan, R. Profiling of Red Pigment Produced by Streptomyces Sp. JAR6 and Its Bioactivity. 3 Biotech 2018, 8, 22. [Google Scholar] [CrossRef]
- Azimi, S.; Baserisalehi, M.; Bahador, N. Evaluation of Antimicrobial Pigment Produced by Streptomyces Coeruleorubidus. Nat. Environ. Pollut. Technol. 2014, 13, 641–644. [Google Scholar]
- Soundari, A. Parimala Gnana Mani, V.M.; Bose, V.S.C.; Jabastin, J.; Priyadarisini, V.B. A Preliminary Assessment of Yellow Pigment from Streptomyces Parvulus C5-5Y. J. Pure Appl. Microbiol. 2017, 11, 197–203. [Google Scholar] [CrossRef]
- Vijayabharathi, R.; Bruheim, P.; Andreassen, T.; Raja, D.S.; Devi, P.B.; Sathyabama, S.; Priyadarisini, V.B. Assessment of Resistomycin, as an Anticancer Compound Isolated and Characterized from Streptomyces Aurantiacus AAA5. J. Microbiol. 2011, 49, 920–926. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution (N. Y). 1985, 39, 783. [Google Scholar] [CrossRef]
- Abellan-Schneyder, I.; Matchado, M.S.; Reitmeier, S.; Sommer, A.; Sewald, Z.; Baumbach, J.; List, M.; Neuhaus, K. Primer, Pipelines, Parameters: Issues in 16S RRNA Gene Sequencing. mSphere 2021, 6, 10–1128. [Google Scholar] [CrossRef]
- Labeda, D.P.; Goodfellow, M.; Brown, R.; Ward, A.C.; Lanoot, B.; Vanncanneyt, M.; Swings, J.; Kim, S.-B.; Liu, Z.; Chun, J.; et al. Phylogenetic Study of the Species within the Family Streptomycetaceae. Antonie Van Leeuwenhoek 2012, 101, 73–104. [Google Scholar] [CrossRef] [PubMed]
- Nicault, M.; Tidjani, A.-R.; Gauthier, A.; Dumarcay, S.; Gelhaye, E.; Bontemps, C.; Leblond, P. Mining the Biosynthetic Potential for Specialized Metabolism of a Streptomyces Soil Community. Antibiotics 2020, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Xi, W.; Hu, Y.; Nie, C.; Zhou, Z. Antioxidant Activity of Citrus Fruits. Food Chem. 2016, 196, 885–896. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A. The Role of Antioxidants in the Chemistry of Oxidative Stress: A Review. Eur. J. Med. Chem. 2015, 97, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.N.; Bristi, N.J.; Rafiquzzaman, M. Review on in Vivo and in Vitro Methods Evaluation of Antioxidant Activity. Saudi Pharm. J. 2013, 21, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-O.; Lee, K.W.; Lee, H.J.; Lee, C.Y. Vitamin C Equivalent Antioxidant Capacity (VCEAC) of Phenolic Phytochemicals. J. Agric. Food Chem. 2002, 50, 3713–3717. [Google Scholar] [CrossRef]
- Arnao, M.B.; Cano, A.; Acosta, M. The Hydrophilic and Lipophilic Contribution to Total Antioxidant Activity. Food Chem. 2001, 73, 239–244. [Google Scholar] [CrossRef]
- Hyun, H.B.; Shrestha, S.; Boo, K.H.; Cho, S.K. Evaluation of Antioxidant Potential of Ethyl Acetate Fraction of Rosmarinus Officinalis L. and Its Major Components. J. Korean Soc. Appl. Biol. Chem. 2015, 58, 715–722. [Google Scholar] [CrossRef]
- Floegel, A.; Kim, D.-O.; Chung, S.-J.; Koo, S.I.; Chun, O.K. Comparison of ABTS/DPPH Assays to Measure Antioxidant Capacity in Popular Antioxidant-Rich US Foods. J. Food Compos. Anal. 2011, 24, 1043–1048. [Google Scholar] [CrossRef]
- Arnao, M.B. Some Methodological Problems in the Determination of Antioxidant Activity Using Chromogen Radicals: A Practical Case. Trends Food Sci. Technol. 2000, 11, 419–421. [Google Scholar] [CrossRef]
- Floegel, A.; Kim, D.-O.; Chung, S.-J.; Chun, O.K. Comparison of ABTS/DPPH Assays for the Detection of Antioxidant Capacity in Foods. FASEB J. 2010, 24, 535–9. [Google Scholar] [CrossRef]
- Kuskoski, E.M.; Asuero, A.G.; Troncoso, A.M.; Mancini-filho, J.; Fett, R. Aplicación de Diversos Métodos Químicos Para Determinar Actividad Antioxidante En Pulpa de Frutos. Ciência e Tecnol. Aliment. 2005, 25, 726–732. [Google Scholar] [CrossRef]
- Thirumalai, D.; Manikkam, R.; Ramasamy, B. Antioxidant Activity of Melanin Pigment from Streptomyces Species D5 Isolated from Desert Soil, Rajasthan, India. Drug Invent. Today 2010, 2, 12–13. [Google Scholar]
- Miethke, M.; Pieroni, M.; Weber, T.; Brönstrup, M.; Hammann, P.; Halby, L.; Arimondo, P.B.; Glaser, P.; Aigle, B.; Bode, H.B.; et al. Towards the Sustainable Discovery and Development of New Antibiotics. Nat. Rev. Chem. 2021, 5, 726–749. [Google Scholar] [CrossRef]
- Byrne, M.K.; Miellet, S.; McGlinn, A.; Fish, J.; Meedya, S.; Reynolds, N.; van Oijen, A.M. The Drivers of Antibiotic Use and Misuse: The Development and Investigation of a Theory Driven Community Measure. BMC Public Health 2019, 19, 1425. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Liu, Z.; Chen, Y.; Xiao, Y.; Huang, X.; Fan, X.-G. Bacterial and Fungal Infections in COVID-19 Patients: A Matter of Concern. Infect. Control Hosp. Epidemiol. 2020, 41, 1124–1125. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; So, M.; Raybardhan, S.; Leung, V.; Westwood, D.; MacFadden, D.R.; Soucy, J.-P.R.; Daneman, N. Bacterial Co-Infection and Secondary Infection in Patients with COVID-19: A Living Rapid Review and Meta-Analysis. Clin. Microbiol. Infect. 2020, 26, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, V.M.; Gandhi, T.N.; Petty, L.A.; Patel, P.K.; Prescott, H.C.; Malani, A.N.; Ratz, D.; McLaughlin, E.; Chopra, V.; Flanders, S.A. Empiric Antibacterial Therapy and Community-Onset Bacterial Coinfection in Patients Hospitalized With Coronavirus Disease 2019 (COVID-19): A Multi-Hospital Cohort Study. Clin. Infect. Dis. 2021, 72, e533–e541. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J. How Covid-19 Is Accelerating the Threat of Antimicrobial Resistance. BMJ 2020, m1983. [Google Scholar] [CrossRef] [PubMed]
- Bassetti, M.; Giacobbe, D.R. A Look at the Clinical, Economic, and Societal Impact of Antimicrobial Resistance in 2020. Expert Opin. Pharmacother. 2020, 21, 2067–2071. [Google Scholar] [CrossRef] [PubMed]
- Nieuwlaat, R.; Mbuagbaw, L.; Mertz, D.; Burrows, L.L.; Bowdish, D.M.E.; Moja, L.; Wright, G.D.; Schünemann, H.J. Coronavirus Disease 2019 and Antimicrobial Resistance: Parallel and Interacting Health Emergencies. Clin. Infect. Dis. 2021, 72, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Rupp, M.E.; Fey, P.D.; Heilmann, C.; Götz, F. Characterization of the Importance of Staphylococcus Epidermidis Autolysin and Polysaccharide Intercellular Adhesin in the Pathogenesis of Intravascular Catheter–Associated Infection in a Rat Model. J. Infect. Dis. 2001, 183, 1038–1042. [Google Scholar] [CrossRef]
- Otto, M. Staphylococcus Epidermidis — the “accidental” Pathogen. Nat. Rev. Microbiol. 2009, 7, 555–567. [Google Scholar] [CrossRef]
- Forbes, B.A.; Schaberg, D.R. Transfer of Resistance Plasmids from Staphylococcus Epidermidis to Staphylococcus Aureus: Evidence for Conjugative Exchange of Resistance. J. Bacteriol. 1983, 153, 627–634. [Google Scholar] [CrossRef]
- Loggenberg, S.R.; Twilley, D.; De Canha, M.N.; Lall, N. Chapter 4 - Medicinal Plants Used in South Africa as Antibacterial Agents for Wound Healing. In Medicinal Plants as Anti-Infectives; Chassagne, F.B.T., Ed.; Academic Press, 2022; pp. 139–182. ISBN 978-0-323-90999-0.
- Khan, T.M.; Kok, Y.L.; Bukhsh, A.; Lee, L.-H.; Chan, K.-G.; Goh, B.-H. Incidence of Methicillin Resistant Staphylococcus Aureus (MRSA) in Burn Intensive Care Unit: A Systematic Review. Germs 2018, 8, 113–125. [Google Scholar] [CrossRef]
- Clebak, K.T.; Malone, M.A. Skin Infections. Prim. Care Clin. Off. Pract. 2018, 45, 433–454. [Google Scholar] [CrossRef]
- Baddour, L.M.; Wilson, W.R.; Bayer, A.S.; Fowler, V.G.; Tleyjeh, I.M.; Rybak, M.J.; Barsic, B.; Lockhart, P.B.; Gewitz, M.H.; Levison, M.E.; et al. Infective Endocarditis in Adults: Diagnosis, Antimicrobial Therapy, and Management of Complications. Circulation 2015, 132, 1435–1486. [Google Scholar] [CrossRef]
- Selvameenal, L.; Radhakrishnan, M.; Balagurunathan, R. Antibiotic Pigment from Desert Soil Actinomycetes; Biological Activity, Purification and Chemical Screening. Indian J. Pharm. Sci. 2009, 71, 499–504. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, R.; Venugopal, G.; Ramasamy, B.; Kumar, V. Effect of Critical Medium Components and Culture Conditions on Antitubercular Pigment Production from Novel Streptomyces Sp D25 Isolated from Thar Desert, Rajasthan. J. Appl. Pharm. Sci. 2015, 5, 15–19. [Google Scholar] [CrossRef]
- Ohnishi, Y.; Furusho, Y.; Higashi, T.; Chun, H.K.; Furihata, K.; Sakuda, S.; Horinouchi, S. Structures of Grixazone A and B, A-Factor-Dependent Yellow Pigments Produced under Phosphate Depletion by Streptomyces Griseus. J. Antibiot. (Tokyo). 2004, 57, 218–223. [Google Scholar] [CrossRef]
- Lu, J.-J.; Wang, Y.-T. Identification of Anti-Cancer Compounds from Natural Products. Chin. J. Nat. Med. 2020, 18, 481–482. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Rangappa, S.; Nayak, S.C.; Jadimurthy, R.; Wang, L.; Sethi, G.; Garg, M.; Rangappa, K.S. Bacteria as a Treasure House of Secondary Metabolites with Anticancer Potential. Semin. Cancer Biol. 2022, 86, 998–1013. [Google Scholar] [CrossRef]
- Schabel, F.M.; Pittillo, R.F. Screening for and Biological Characterization of Antitumor Agents Using Microorganisms. In; 1961; pp. 223–256.
- Tan, L.T.H.; Chan, K.G.; Pusparajah, P.; Yin, W.F.; Khan, T.M.; Lee, L.H.; Goh, B.H. Mangrove Derived Streptomyces Sp. MUM265 as a Potential Source of Antioxidant and Anticolon-Cancer Agents. BMC Microbiol. 2019, 19. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Cui, J.; Wang, P.; Wang, X.; Wen, J. Enhancement of Bleomycin Production in Streptomyces Verticillus through Global Metabolic Regulation of N-Acetylglucosamine and Assisted Metabolic Profiling Analysis. Microb. Cell Fact. 2020, 19, 32. [Google Scholar] [CrossRef]
- Sajjad, W.; Ahmad, S.; Aziz, I.; Azam, S.S.; Hasan, F.; Shah, A.A. Antiproliferative, Antioxidant and Binding Mechanism Analysis of Prodigiosin from Newly Isolated Radio-Resistant Streptomyces Sp. Strain WMA-LM31. Mol. Biol. Rep. 2018, 45, 1787–1798. [Google Scholar] [CrossRef]
- Sarmiento-Tovar, A.A.; Silva, L.; Sánchez-Suárez, J.; Diaz, L. Streptomyces-Derived Bioactive Pigments: Ecofriendly Source of Bioactive Compounds. Coatings 2022, 12, 1858. [Google Scholar] [CrossRef]
- Salla, T.D.; Astarita, L. V.; Santarém, E.R. Defense Responses in Plants of Eucalyptus Elicited by Streptomyces and Challenged with Botrytis Cinerea. Planta 2016, 243, 1055–1070. [Google Scholar] [CrossRef]
- Kim, S.S.; Lee, C.K.; Kang, S.S.; Jung, H.A.; Choi, J.S. Chlorogenic Acid, an Antioxidant Principle from the Aerial Parts of Artemisia Iwayomogi That Acts on 1,1-Diphenyl-2-Picrylhydrazyl Radical. Arch. Pharm. Res. 1997, 20, 148–154. [Google Scholar] [CrossRef]
- Saqib, M.; Iqbal, S.; Mahmood, A.; Akram, R. Theoretical Investigation for Exploring the Antioxidant Potential of Chlorogenic Acid: A Density Functional Theory Study. Int. J. Food Prop. 2016, 19, 745–751. [Google Scholar] [CrossRef]
- Keudell, K.C.; Huang, J.K.; Wen, L.; Klopfenstein, W.E.; Bagby, M.O.; Lanser, A.C.; Plattner, R.D.; Peterson, R.E.; Weisleder, D. Fatty Acids Enhanced Tubermycin Production by Pseudomonas Strain 2HS. Microbios 2000, 102, 27–38. [Google Scholar] [PubMed]
- Weber, W.; Zähner, H.; Siebers, J.; Schröder, K.; Zeeck, A. [Metabolic products of microorganisms. 175. Tetracenomycin C (author’s transl)]. Arch. Microbiol. 1979, 121, 111–116. [Google Scholar] [CrossRef]
- Remali, J.; Sahidin, I.; Aizat, W.M. Xanthone Biosynthetic Pathway in Plants: A Review. Front. Plant Sci. 2022, 13. [Google Scholar] [CrossRef] [PubMed]
- Leetanasaksakul, K.; Koomsiri, W.; Suga, T.; Matsuo, H.; Hokari, R.; Wattana-Amorn, P.; Takahashi, Y.; Shiomi, K.; Nakashima, T.; Inahashi, Y.; et al. Sattahipmycin, a Hexacyclic Xanthone Produced by a Marine-Derived Streptomyces. J. Nat. Prod. 2022, 85, 1211–1217. [Google Scholar] [CrossRef]
- Matselyukh, B.P.; Matselyukh, D.Y.; Golembiovska, S.L.; Polishchuk, L. V; Lavrinchuk, V.Y. Isolation of Streptomyces Globisporus and Blakeslea Trispora Mutants with Increased Carotenoid Content. Mikrobiol. Z. 2013, 75, 10–16. [Google Scholar]
- Han, Z.; Zhao, X.; Zhang, E.; Ma, J.; Zhang, H.; Li, J.; Xie, W.; Li, X. Resistomycin Induced Apoptosis and Cycle Arrest in Human Hepatocellular Carcinoma Cells by Activating P38 MAPK Pathway In Vitro and In Vivo. Pharmaceuticals 2021, 14. [Google Scholar] [CrossRef]
- Shaaban, K.A.; Singh, S.; Elshahawi, S.I.; Wang, X.; Ponomareva, L. V; Sunkara, M.; Copley, G.C.; Hower, J.C.; Morris, A.J.; Kharel, M.K.; et al. Venturicidin C, a New 20-Membered Macrolide Produced by Streptomyces Sp. TS-2-2. J. Antibiot. (Tokyo). 2014, 67, 223–230. [Google Scholar] [CrossRef]
- Tan, L.T.; Mahendra, C.K.; Yow, Y.; Chan, K.; Khan, T.M.; Lee, L.; Goh, B. Streptomyces Sp. MUM273b: A Mangrove-derived Potential Source for Antioxidant and UVB Radiation Protectants. Microbiologyopen 2019, 8. [Google Scholar] [CrossRef]
- Berger, H.; Bacher, M.; Labuda, R.; Eppel, I.M.; Bayer, F.; Sulyok, M.; Gasparotto, E.; Zehetbauer, F.; Doppler, M.; Gratzl, H.; et al. Polaramycin B, and Not Physical Interaction, Is the Signal That Rewires Fungal Metabolism in the Streptomyces–Aspergillus Interaction. Environ. Microbiol. 2022, 24, 4899–4914. [Google Scholar] [CrossRef] [PubMed]
- Ebrahim, W.; El-Neketi, M.; Lewald, L.-I.; Orfali, R.S.; Lin, W.; Rehberg, N.; Kalscheuer, R.; Daletos, G.; Proksch, P. Metabolites from the Fungal Endophyte Aspergillus Austroafricanus in Axenic Culture and in Fungal–Bacterial Mixed Cultures. J. Nat. Prod. 2016, 79, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Kemung, H.M.; Tan, L.T.-H.; Chan, K.-G.; Ser, H.-L.; Law, J.W.-F.; Lee, L.-H.; Goh, B.-H. Antioxidant Activities of Streptomyces Sp. Strain MUSC 14 from Mangrove Forest Soil in Malaysia. Biomed Res. Int. 2020, 2020, 6402607. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.M.A.; Pereira, P.S.; Silva, B.I.M. da; Ribeiro, N.E.; Borba, E.F. de O.; Oliveira Tintino, C.D. de M.; Randau, K.P.; Coutinho, H.D.M.; Lima-Gomes, G.M. de S.; Chávez-González, M.L.; et al. Antioxidant, Antimicrobial and Cytotoxic Activities of Secondary Metabolites from Streptomyces Sp. Isolated of the Amazon-Brazil Region. Res. Soc. Dev. 2021, 10, e366101018974. [Google Scholar] [CrossRef]
- de Moura, P.A.; de Albuquerque Lima, T.; Ferreira, M.R.A.; Soares, L.A.L.; de Souza Lima, G.M.; Napoleão, T.H.; da Silva, M.V.; de Oliveira, A.P.S.; Paiva, P.M.G. The Relevance of Actinobacteria as Sources of Antioxidant Compounds: Evaluation of Streptomyces Isolates from Rhizosphere Collected at Brazilian Caatinga. In Microbial and Natural Macromolecules - Synthesis and Applications; Das, S., Dash, H.R.B.T.-M. and N.M., Eds.; Academic Press: London UK, 2021; pp. 401–418. ISBN 978-0-12-820084-1. [Google Scholar]
| Composition/Medium | ISP2 | ISP4 | modified ISP9 | Starch nitrate |
|---|---|---|---|---|
| agar | 15 | 15 | 15 | 15 |
| starch | 0 | 10 | 0 | 20 |
| CaCO3 | 0 | 2 | 0 | 3 |
| CuSO4·5H2O | 0 | 0 | 0.00064 | 0 |
| yeast extract | 4 | 0 | 0 | 0 |
| malt extract | 10 | 0 | 0 | 0 |
| FeSO4·7H2O | 0 | 0.01 | 0.00011 | 0.01 |
| glucose | 4 | 0 | 10 | 0 |
| K2HPO4 | 0 | 1 | 5.65 | 1 |
| KH2PO4 | 0 | 0 | 2.38 | 0 |
| KNO3 | 0 | 0 | 0 | 2 |
| MgSO4·7H2O | 0 | 1 | 1 | 0.5 |
| MnCl2·4H2O | 0 | 0.001 | 0.00079 | 0 |
| NaCl | 0 | 0.5 | 0 | 0 |
| (NH4)2SO4 | 0 | 2 | 2.64 | 0 |
| ZnSO4·7H2O | 0 | 0.001 | 0.00015 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





