Submitted:
13 August 2024
Posted:
13 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Experimental Methods
2.2.1. Behavioral Observations of Scallops
2.2.2. Crab-Scallop Experiments
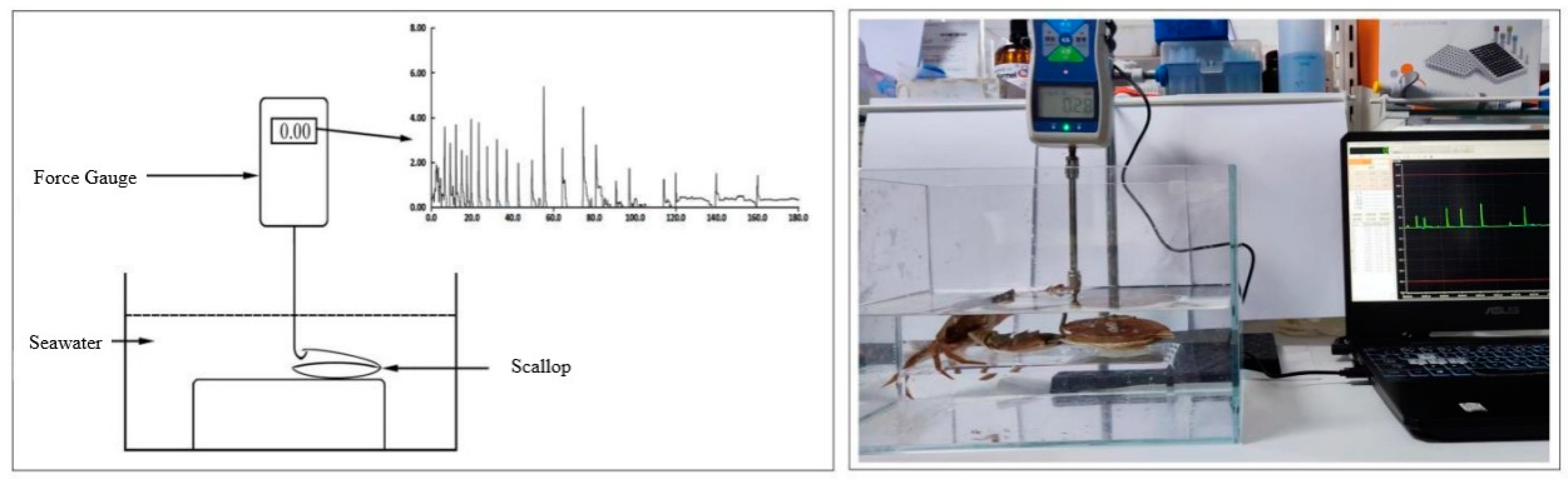
2.2.3 Force of Clap Measurement
- Experimental design
- Force recordings
2.2.4 Enzyme Activity Assay
2.3 Transcriptome Analysis
2.3.1 RNA Extraction and Transcriptome Sequencing
2.3.2 Bioinformatics Analysis
2.3.3 qRT-PCR
2.4 Statistical Analysis
3. Results
3.1. Scallop Behavior
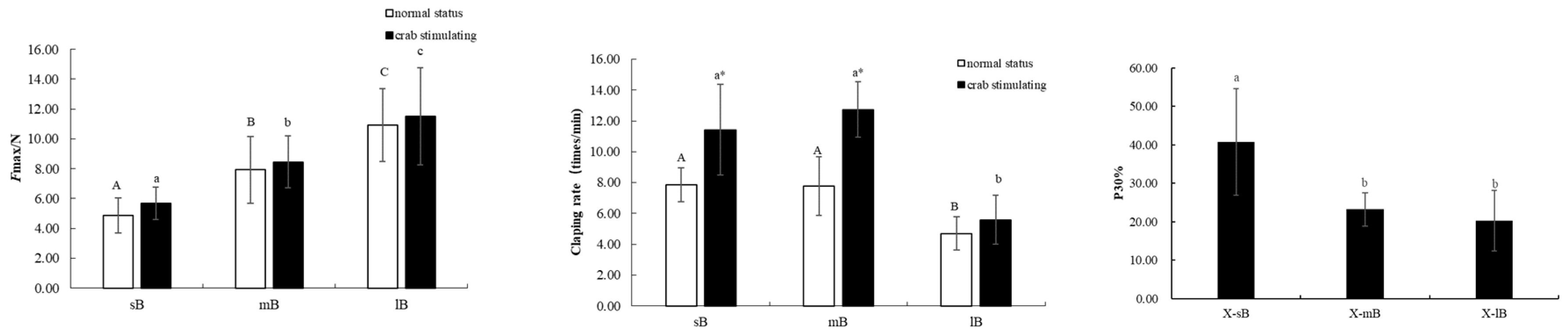
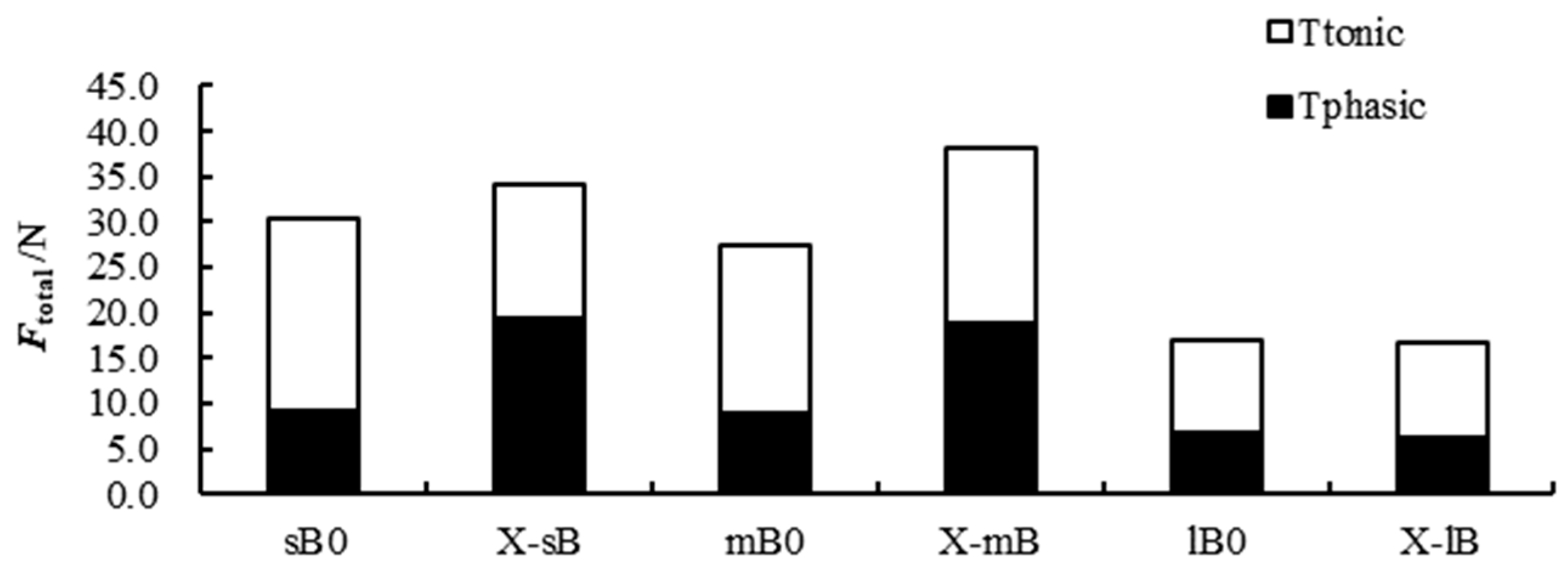
3.2 Force of Clap
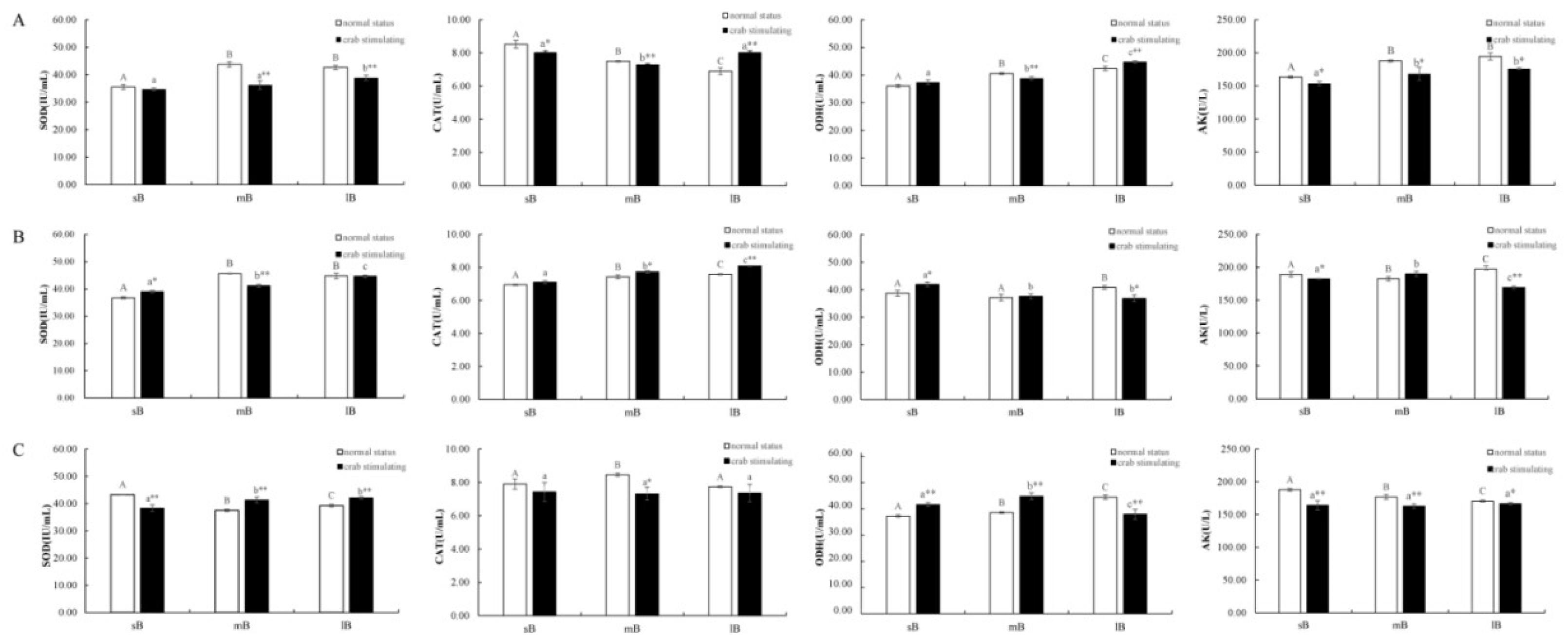
3.3 Changes in Enzyme Activity of Scallops due to Crab Predation
3.4 Transcriptome Results
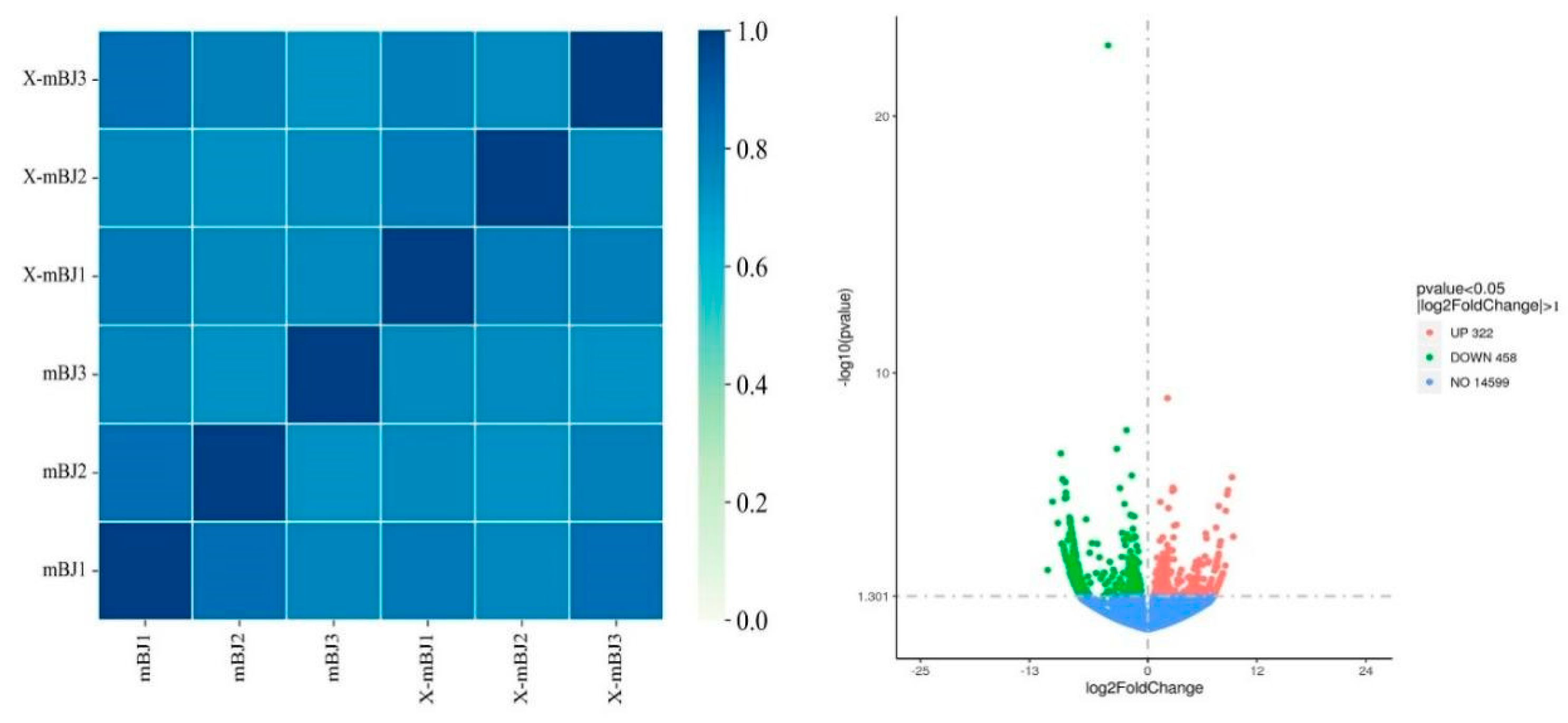
3.4.1 Transcriptome Sequencing
3.4.2 Analysis of Differentially Expressed Genes (DEGs)
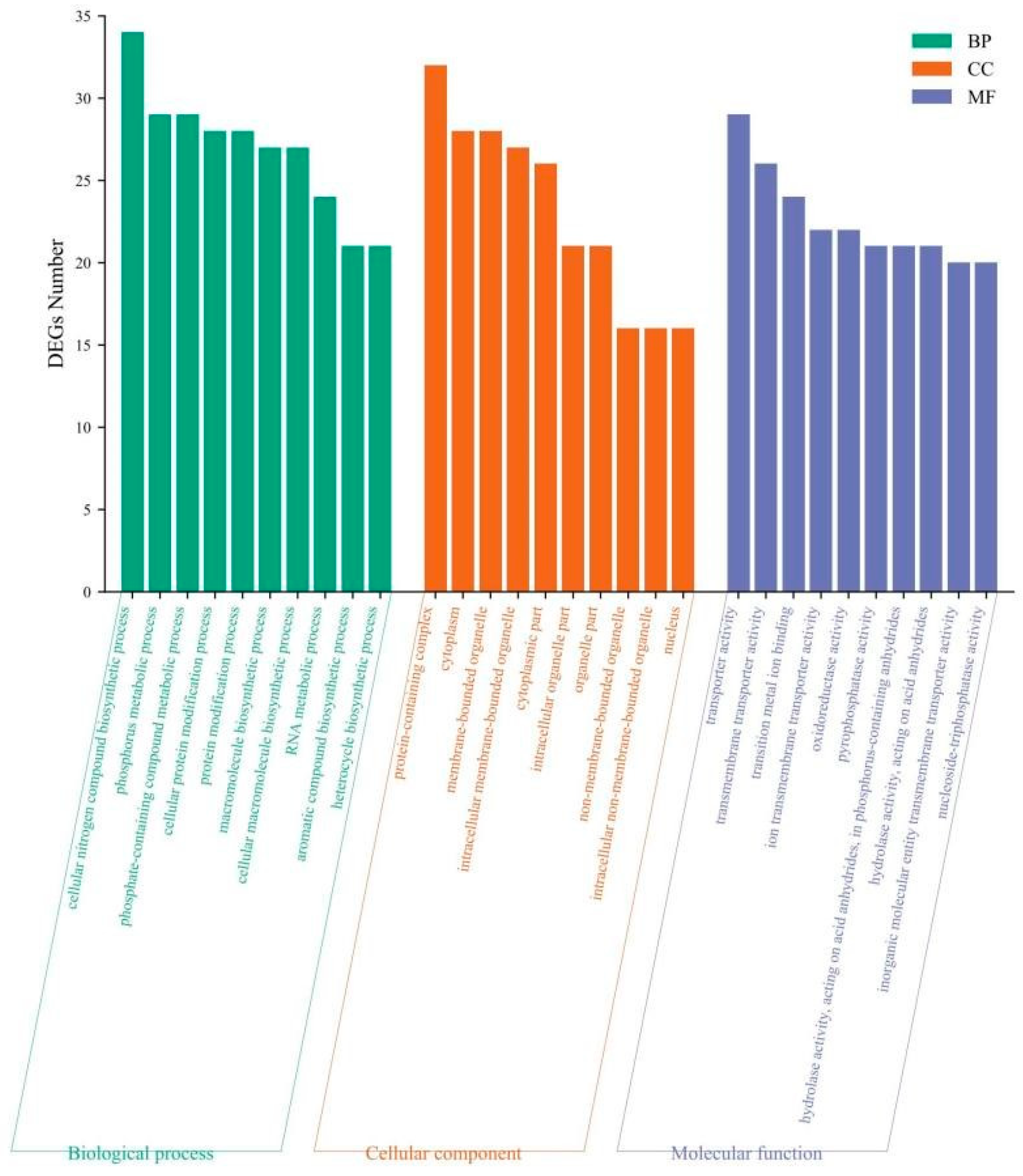
3.4.3 Gene Ontology (GO) Functional Enrichment Analysis of DEGs
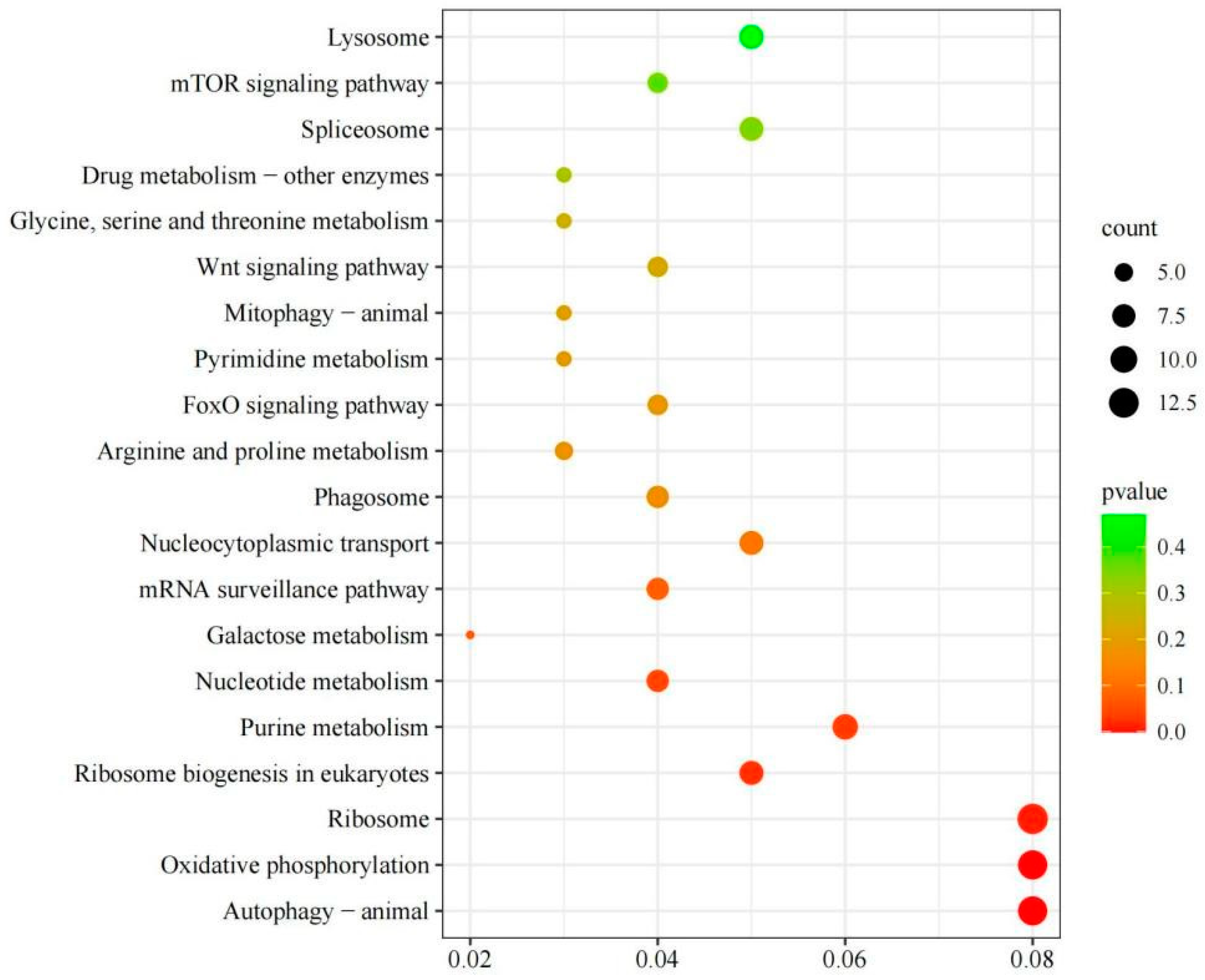
3.4.4 Kyoto Encyclopedia of Genes and Genomes (KEGG) Enrichment Analysis of DEGs
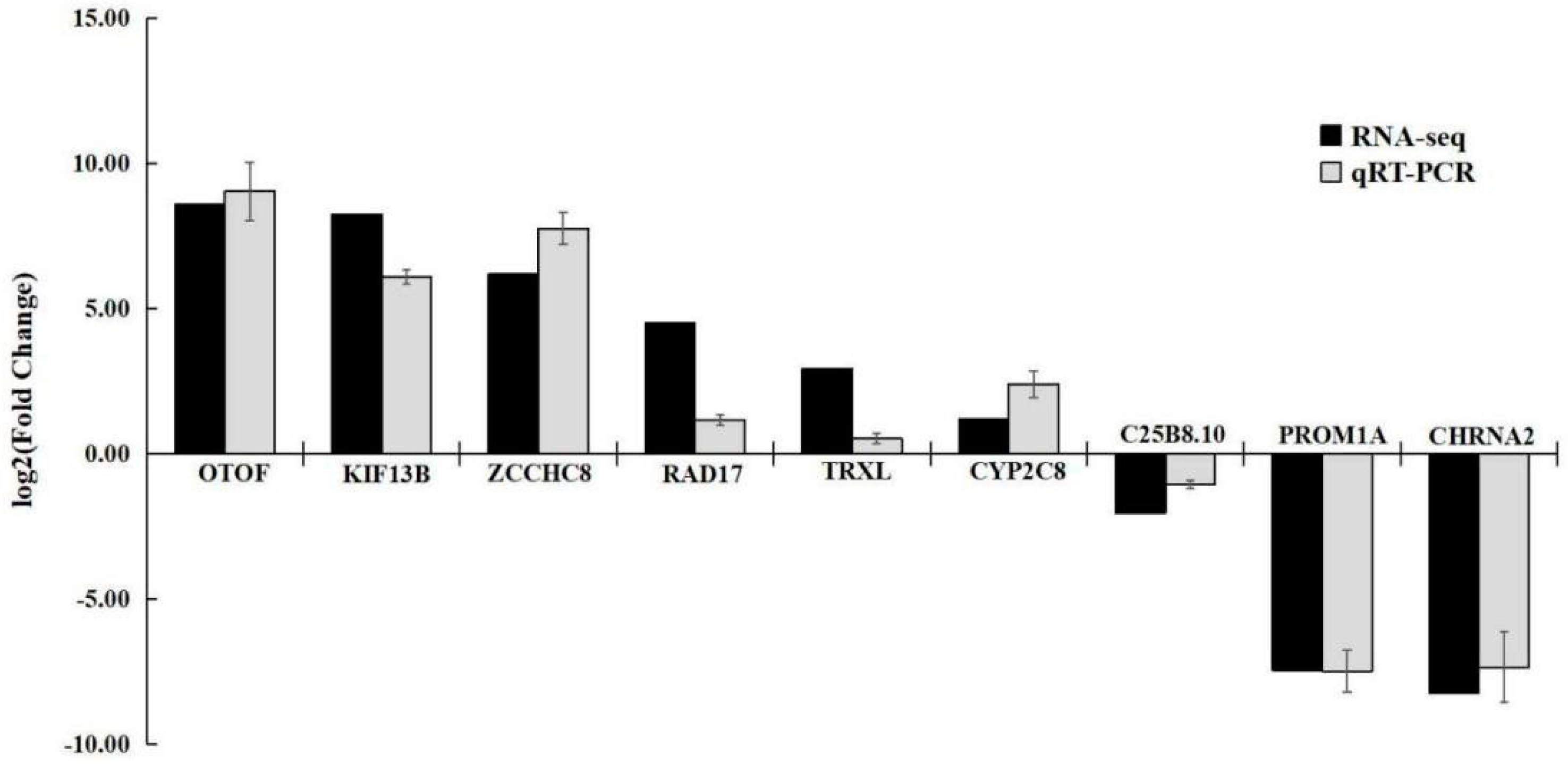
3.4.5 qRT-PCR Validation Analysis
4. Discussion
4.1 Effects of Crab Predation on the Closed-Shell Force of Scallops
4.2 Effects of Crab Predation on the Enzyme Activities of Scallops
4.3 Effects of Crab Predation on the Transcriptome of Scallops
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.X., Zhang, J.H., & Wu, W.G. Ecological carrying capacity assessment of bottom-culture Yesso scallops, Patinopecten yessoensis, in Zhangzi Island. Journal of Fishery Sciences of China. 2021,28(07):878-887.
- Cong, H.H., Geng, W.H., & Zhu, J.W. Patinopecten yessoensis mantle polypeptides enhanced the structural stability of myofibrillar proteins from silver carp. Meat Research. 2022, 36(09): 13-19.
- Wang, Y., & Zhou, L. Bottom sowing of proliferation of Patinopecten yessoensis yield research; case in Zhangzidao. Chinese Fisheries Economics. 2014, 32(01): 104-109.
- Gao, Z. K. Effects of environmental stresses on physiological, immunological parameters and behavioral characteristics of Patinopecten yessoensis. M.A. Thesis. Shanghai Ocean University. Shanghai. 2016.
- Shank, B.V., Hart, D.R., & Friedland, K.D. Post-settlement predation by sea stars and crabs on the sea scallop in the mid-Atlantic bight. Marine Ecology Progress. 2012, 468, 161–177. [CrossRef]
- Barbeau, M.A., Scheibling, R.E. Behavioral mechanisms of prey size selection by sea stars (Asterias vulgaris verrill) and crabs (Cancer irroratus say) preying on juvenile sea scallops (placopecten magellanicus (Gmelin)). Journal of Experimental Marine Biology and Ecology. 1994, 180(1), 103–136. [CrossRef]
- Yu, Z.H., Yang, H.S., & Liu, B.Z. Predation of scallop Chlamys farreri by crab Charybdis japonica. Marine Science. 2010,34(12), 62-66.
- Sclafani, M., Bopp, J., & Havelin, J. Predation on planted and wild bay scallops (Argopecten Irradians Irradians) by busyconine whelks: studies of behavior incorporating acoustic telemetry.2021, 169(66). [CrossRef]
- Zhang, J.H., Xia, Y.Y., & Gao, Z.K. Force production during shell clap of scallop Pationopecten yessoensis and its response to predator starfish. Journal of Fishery Sciences of China. 2021,28(07), 871–877.
- Guderley, H.E., Himmelman, J.H., & Nadeau, M. Effect of different predators on the escape response of placopecten magellanicus. Marine Biology: International Journal on Life in Oceans and Coastal Waters. 2015, 162(7), 1407–1415. [CrossRef]
- Xia, Y.Y. Effects of Environmental Stress on the Survival, Behavior Metabolism and Immunity of Scallops. M.A. Thesis. Shanghai Ocean University. Shanghai. 2010.
- Duan L Z., Huang X Y., & Hu W B. Comparison of three coelomic fluid enzymes activities in scallop Patinopecten yesoensis. Chinese Agricultural Science Bulletin. 2015, 31(02), 112–117.
- Tremblay, I., Guderley, H.E., & Himmelman, J.H. Swimming away or clamming up: The use of phasic and tonic adductor muscles during escape responses varies with shell morphology in scallops. The Jounrnal of Experimental Biology. 2012, 215(Pt23), 4131–4143. [CrossRef]
- Bolger, A.M., Lohse, M., & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014, 30(15), 2114–2120. [CrossRef]
- Kim, D., Langmead, B., & Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nature Methods. 2015,12(4), 357–360. [CrossRef]
- Trapnell, C., Williams, B.A., & Pertea, G. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010,28(5), 511–515. [CrossRef]
- Anders, S., Pyl, P.T., & Huber, W. HTSeq—A python framework to work with high-throughput sequencing data. Bioinformatics. 2015,31(2),166–169. [CrossRef]
- Anders, S., & Huber, W. Differential expression analysis for sequence count data. Nature Precedings. 2010, 1–1. [CrossRef]
- Kanehisa, M., Araki, M., & Goto, S., Hattori, M. KEGG for linking genomes to life and the environment. Nucleic Acids Research, 2007, 36(suppl-1), D480–D484. [CrossRef]
- Livak, K. J., & Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods. 2001, 25(4), 402–408.
- Xia, Y.Y., Zhang, J.H., & Liu, Y. Behavioral characteristics and physiological responses to hypoxic stress in Patinopecten yessoensis. Journal of Fishery Sciences of China. 2021,28(10), 1319–1328.
- Schalkhausser, B., Bock, C., & Partner, H-O. Escape performance of temperate king scallop, Pecten maximus under ocean warming and acidification. Marine Biology. 2014, 161, 2819–2829. [CrossRef]
- Schmidt, M., Philipp, E.E.R., & Abele, D. Size and age-dependent changes of escape response to predator attack in the Queen scallop Aequipecten opercularis. Marine Biology Research. 2008, 4(6), 442–450. [CrossRef]
- Greenway, S.C., & Storey, K.B. The effect of prolonged anoxia on enzyme activities in oysters (Crassostrea virginica) at different seasons. Journal of Experimental Marine Biology and Ecology. 1999, 242(2), 259–272. [CrossRef]
- Liu, Z.H., Mou, H.J., & Wang, Q.Y. Research progress of immune related enzymes in Mollusca. Marine Fisheries Research. 2003, 024(003), 86–90.
- Yao, C.L., Wang, W.N., & Wang, A.L. Progess of studies on superoxide dismutase in the body of aquatic animals. Marine Sciences. 2003, 27(10), 18–21.
- Yang, X.L., & Zhou, J.G. Influence of age size and nutrition of trionyx sinensis on the immune response. Journal of Fishery Sciences of China. 1999, 23(4), 5.
- Mourente, G., & Daz-Salvago, E. Characterization of antioxidant systems, oxidation status and lipids in brain of wild-caught size-class distributed Aristeus antennatus (Risso,1816) Crustacea, Decapoda. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 1999, 124(4), 405–416.
- Hao, Z.L., Tang, X.J., & Ding, J. Survival rate,oxygen consumption rate and immune enzymetic activity of Mizuhopecten yessoensis at high temperature. Chinese Journal of Ecology. 2014, 33(6), 7.
- Ding, R.X., Huang, X.M., & Zhao, W. Effects of pH acute stress on the behavior and immune enzyme activity of Babylonia Areolata. Fishery modernization. 2022, 49(6), 7.
- Thoai, N.V., Huc, C., & Pho, D.B. Octopine déshydrogénase Purification et propriétés catalytiques. Biochimica et Biophysica Acta (BBA)-Enzymology. 1969, 191(1), 46–57.
- Zheng Y. Octopine dehydrogenase in the adductor muscle of live scallop and its changes during postharvest. M.A. Thesis. Dalian Ocean University. Dalian. 2018.
- Smits, S.H.J., Meyer, T., & Mueller, A. Insights into the Mechanism of Ligand Binding to Octopine Dehydrogenase from Pecten maximus by NMR and Crystallography. Plos One. 2010, 5(8), e12312. [CrossRef]
- Strahl, J., Dringen, R., & Schmidt, M. M. Metabolic and physiological responses in tissues of the long-lived bivalve Arctica islandica to oxygen deficiency. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2011, 158(4), 513–519. [CrossRef]
- Guerra, C., Zenteno-Savín, T., & Maeda-Martínez, A.N. The effect of predator exposure and reproduction on oxidative stress parameters in the Catarina scallop Argopecten ventricosus. Comparative Biochemistry and physiology. Part A: Molecular & Integrative Physiology. 2013, 165(1), 89–96. [CrossRef]
- Morris, S., Van Aardt, W.J., & Ahern, M.D. The effect of lead on the metabolic and energetic status of the Yabby, Cherax destructor, during environmental hypoxia. Aquatic Toxicology. 2005, 75(1), 16–31. [CrossRef]
- Yao, C.L., Wang, Z.Y., & Xiang, J.H. Structure and function of arginine kinase in crustacean. Chinese Journal of Biochemistry and Molecular Biology. 2008, 24(3), 203–208.
- Huang, L.N., Liu, N., & Zhao, J. The research prospect of Arginine Kinase. Life Science Research. 2015, 19(05), 452–456.
- Voncken, F., Gao, F., & Wadforth, C. The phosphoarginine energy-buffering system of Trypanosoma brucei involves multiple arginine kinase isoforms with different subcellular locations. PLoS One. 2013, 8(6), e65908. [CrossRef]
- Gäde, G., Weeda, E., & Gabbott, P.A. Changes in the level of octopine during the escape responses of the scallop, Pecten maximus (L.). Journal of comparative physiology. 1978, 124, 121–127. [CrossRef]
- Qin, Y.L., Peng, H.L.Y., & Fu, S.J. Effects of food deprivation on fast-start swimming and predator-prey interaction between a predator and prey fish species. Chinese Journal of Ecology. 2016, 35(09), 2429–2434.
- Hirai, H., Pang, Z., & Bao, D. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nature Neuroscience. 2005, 8(11), 1534–1541. [CrossRef]
- Matsuda, K., Yuzaki, M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. European Journal of Neuroscience. 2011, 33(8), 1447–1461. [CrossRef]
- Stevens, B., Allen, N.J., & Vazquez, L.E. The classical complement cascade mediates CNS synapse elimination. Cell. 2007, 131(6), 1164–1178. [CrossRef]
- Uemura T., Lee S J., & Yasumura M. Trans-synaptic interaction of GluRδ2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010, 141(6), 1068–1079. [CrossRef]
- Reid, K.B.M., Gagnon, J., & Frampton, J. Completion of the amino acid sequences of the A and B chains of subcomponent C1q of the first component of human complement. Biochemical Journal. 1982, 203(3), 559–569. [CrossRef]
- Yamauchi T., Kamon J., Waki H., Imai, Y., Shimozawa, N., Hioki, K., Uchida, S., Ito, Y., Takakuwa, K., Matsui, J., Takata, M., Eto, K., Terauchi, Y., Komeda, K., Tsunoda., M., Murakami, K., Ohnishi, Y., Naitoh, T., Yamamura, K., Ueyama, Y., Froguel, P., Kimura, S., Nagai, R., Kadowaki, T. Globular adiponectin protected ob/ob mice from diabetes and ApoE-deficient mice from atherosclerosis. Journal of Biological Chemistry. 2003, 278(4), 2461–2468. [CrossRef]
- Bolliger, M.F., Martinelli, D.C., & Südhof, T.C. The cell-adhesion G protein-coupled receptor BAI3 is a high-affinity receptor for C1q-like proteins. Proceedings of the National Academy of Sciences. 2011, 108(6), 2534–2539. [CrossRef]
- Ressl, S., Vu, B.K., & Vivona, S. Structures of C1q-like proteins reveal unique features among the C1q/TNF superfamily. Structure. 2015, 23(4), 688–699. [CrossRef]
- Gao, Z. Identification, expression and functional characterization of complement components C1q-like and C3a molecules in amphioxus. M.A. Thesis. Ocean University Of China. Qingdao. 2015.
- Nayak, A., Ferluga, J., & Tsolaki, A.G. The non-classical functions of the classical complement pathway recognition subcomponent C1q. Immunology Letters. 2010, 131(2), 139–150. [CrossRef]
- Nauta, A.J., Trouw, L.A., & Daha, M.R. Direct binding of C1q to apoptotic cells and cell blebs induces complement activation. European Journal of Immunology. 2002, 32(6), 1726–1736. [CrossRef]
- Korb, L.C., & Ahearn, J.M. C1q binds directly and specifically to surface blebs of apoptotic human keratinocytes: complement deficiency and systemic lupus erythematosus revisited. Journal of Immunology (Baltimore, Md.: 1950). 1997, 158(10), 4525–4528. [CrossRef] [PubMed]
- Young Jr, K.R., Ambrus Jr, J.L., & Malbran, A. Complement subcomponent C1q stimulates Ig production by human B lymphocytes. Journal of Immunology (Baltimore, Md.: 1950). 1991, 146(10), 3356–3364.
- Ferry, H., Potter, P.K., & Crockford, T.L. Increased positive selection of B1 cells and reduced B cell tolerance to intracellular antigens in c1q-deficient mice. The Journal of Immunology. 2007, 178(5), 2916–2922. [CrossRef]
- Kobayashi, H., Hirashima, Y., & Terao, T. Human myometrial cells in culture express specific binding sites for urinary trypsin inhibitor. Molecular Human Reproduction. 2000, 6(8), 735–742. [CrossRef]
- Bordin, S., Ghebrehiwet, B., & Page, R.C. Participation of C1q and its receptor in adherence of human diploid fibroblast. Journal of immunology. 1990, 145(8), 2520–2526. [CrossRef]
- Naito, A.T., Sumida, T., & Nomura, S. Complement C1q activates canonical Wnt signaling and promotes aging-related phenotypes. Cell. 2012, 149(6), 1298–1313. [CrossRef]
- Bossi, F., Tripodo, C., & Rizzi, L. C1q as a unique player in angiogenesis with therapeutic implication in wound healing. Proceedings of the National Academy of Sciences. 2014, 111(11), 4209–4214. [CrossRef]
- Tan, A., Ke, S.Y., & Chen, Y. Expression patterns of C1ql4 and its cell-adhesion GPCR Bai3 in the murine testis and functional roles in steroidogenesis. The FASEB Journal. 2019, 33(4), 4893–4906. [CrossRef]
- Siddiqui, N., & Straube, A. (2020). The Kinesin–3 Family: Long-Distance Transporters. The Kinesin Superfamily Handbook.
- Venkateswarlu, K., Hanada, T., & Chishti, A. H. Centaurin-α1 interacts directly with kinesin motor protein KIF13B. Journal of Cell Science. 2005, 118(11), 2471–2484. [CrossRef]
- Willemsen, M.H., Ba, W., & Wissink-Lindhout, W.M. Involvement of the kinesin family members KIF4A and KIF5C in intellectual disability and synaptic function. Journal of Medical Genetics. 2014, 51(7).
- Kaori, H., Yamada, Y., & Geyer. KIF13B regulates angiogenesis through Golgi to plasma membrane trafficking of VEGFR2. Journal of cell science. 2014, 127(20), 4518-4530.
- Amiri, M., Conserva, F., & Panayiotou, C. The human adenylate kinase 9 is a nucleoside mono-and diphosphate kinase. The international journal of biochemistry & cell biology. 2013, 45(5), 925–931. [CrossRef]
- Dzeja, P., & Terzic, A. Adenylate Kinase and AMP signaling networks: metabolic monitoring, signal communication and body energy sensing. International Journal of Molecular Sciences. 2009, 10(4), 1729–1772. [CrossRef]
- Yang, A.L. Effects of adenylate kinase ak4 knockout on germ cell apoptosis of male zebrafish. M.A. Thesis. ShanDong University. Shandong. 2017.
- Vogel, B.E., Muriel, J.M., & Dong, C. Hemicentins: what have we learned from worms. Cell Research. 2006, 16(11), 872–878. [CrossRef]
- Vogel, B.E, Hedgecock, E.M. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development. 2001, 128(6), 883-894.
- Chowdhury, A., Herzog, C., & Hasselbach, L. Expression of fibulin-6 in failing hearts and its role for cardiac fibroblast migration. Cardiovasc Research. 2014, 103(4), 509–520. [CrossRef]
- Carney, T.J., Natália M.F., & Sonntag, C. (2010) Genetic Analysis of Fin Development in Zebrafish Identifies Furin and Hemicentin1 as Potential Novel Fraser Syndrome Disease Genes. PLoS Genetics. [CrossRef]
- Lin, M.H., Pope III, B.D., & Sasaki, T. Mammalian hemicentin 1 is assembled into tracks in the extracellular matrix of multiple tissues. Developmental Dynamics. 2020, 249(6), 775–788.
- Yang, L.D., Jiang, H.F., & He, S.P. Comparative genomics reveals accelerated evolution of fright reaction genes in ostariophysan fishes. Frontiers in Genetics. 2019, 10, 1283. [CrossRef]


| Large size (l) | Middle size (m) | Small size (s) | |
|---|---|---|---|
| Shell length/mm | 119.85±3.23 | 89.24±3.77 | 60.10±3.23 |
| Shell height/mm | 116.92±6.02 | 87.46±3.55 | 61.39±6.02 |
| Shell width/mm | 26.98±2.98 | 26.98±2.98 | 16.21±2.63 |
| Total wet weight/g | 176.50±28.57 | 83.70±13.97 | 30.16±5.29 |
| Gene name | Forword Primer (5’-3’) | Reverse Primer (5’-3’) |
|---|---|---|
| Gapdh | TGGTATGGCTTTCCGTGTGC | TCCTCTGTGTAACCAAGGAACC |
| KIF13B | GCAGCCAACCTCAGTCCTAACAG | TCGTGCTCGTCCTCTACCATCAT |
| CYP2C8 | GTTGCTCCTCTTGGCGTTCCT | GGCGACCGACAGAGAATGCT |
| ZCCHC8 | ACCACCACTGCCAATCAACACTC | CCATCACCTGTAGCTCCACCTCT |
| TRXL | TGTCTACAACACCCGCCAGAAT | ACACCACGAAGCATGGAAGTC |
| RAD17 | ACGAGTCGGAGTTGTGGTCTG | TGCCTGTGCCTTGAGATGTGT |
| OTOF | GTTGACGGACTCGGACGACATC | GCCTTCAGCACTCGCACAGT |
| C25B8.10 | GTTGAGCTTGGAGCTGGAACAG | GCCACCACAGTCCTAACAGAGT |
| CHRNA2 | GCCGTGCTCAGAATCCACAACT | TCCCGACGACACGCCACAATA |
| PROM1A | GGTTTGGCTTGGGATGGTGTCT | GCGTGGCTGACCTTGTTGCT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





