Submitted:
29 July 2024
Posted:
01 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
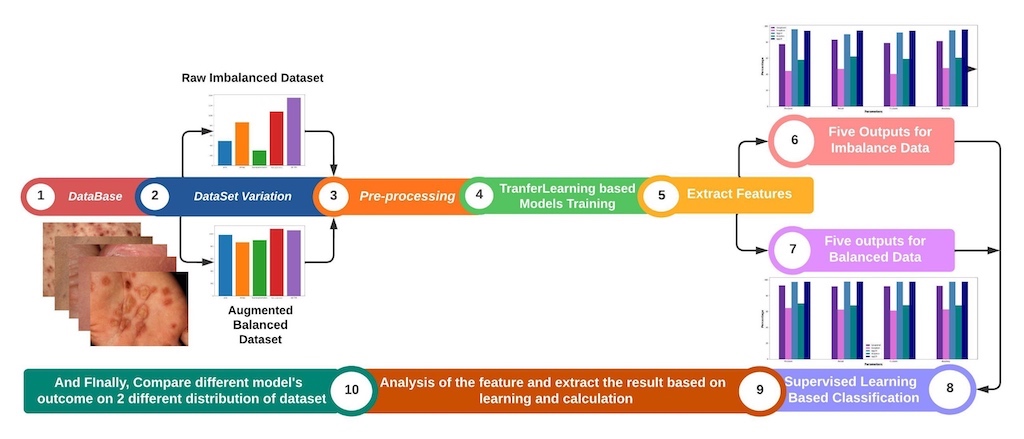
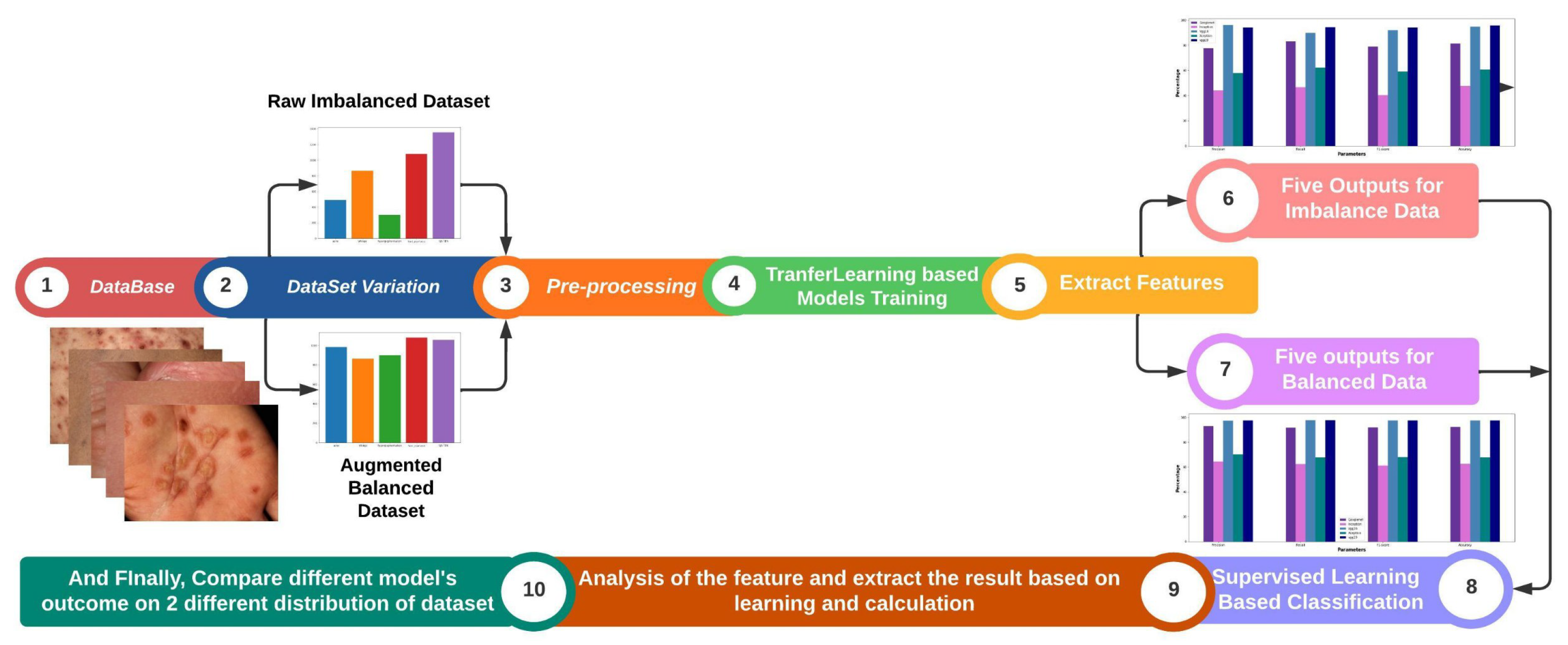
- Evaluation of Deep Learning Models: The study comprehensively evaluated five deep learning models, including GoogleNet, Inception, VGG-16, VGG-19, and Xception, for the identification of skin diseases.
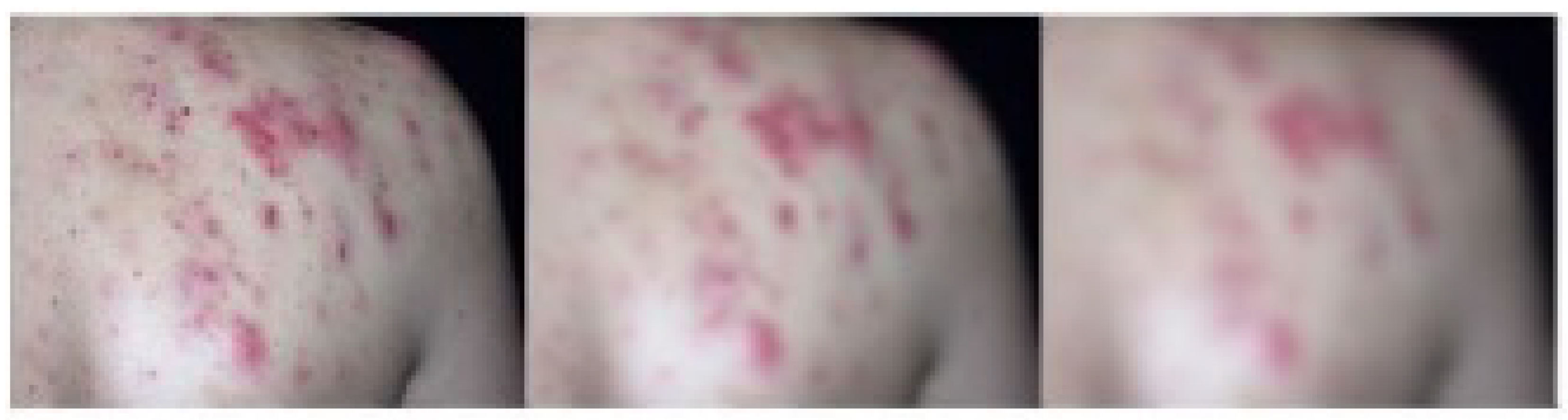
- Creation of a Self-Collected Dataset: In this study, a dataset consisting of approximately 5,185 images of rare skin diseases was created.
- Importance of Data Balancing and Augmentation: The study highlighted the significance of data balancing techniques, specifically through augmentation, in improving the accuracy of skin disease identification models. By applying augmentation on the imbalanced dataset, the study successfully enhanced the performance of the models, with VGG-16-Aug achieving the highest accuracy of 97.07% on the balanced data. This emphasizes the importance of addressing imbalanced data distributions for accurate disease prediction.
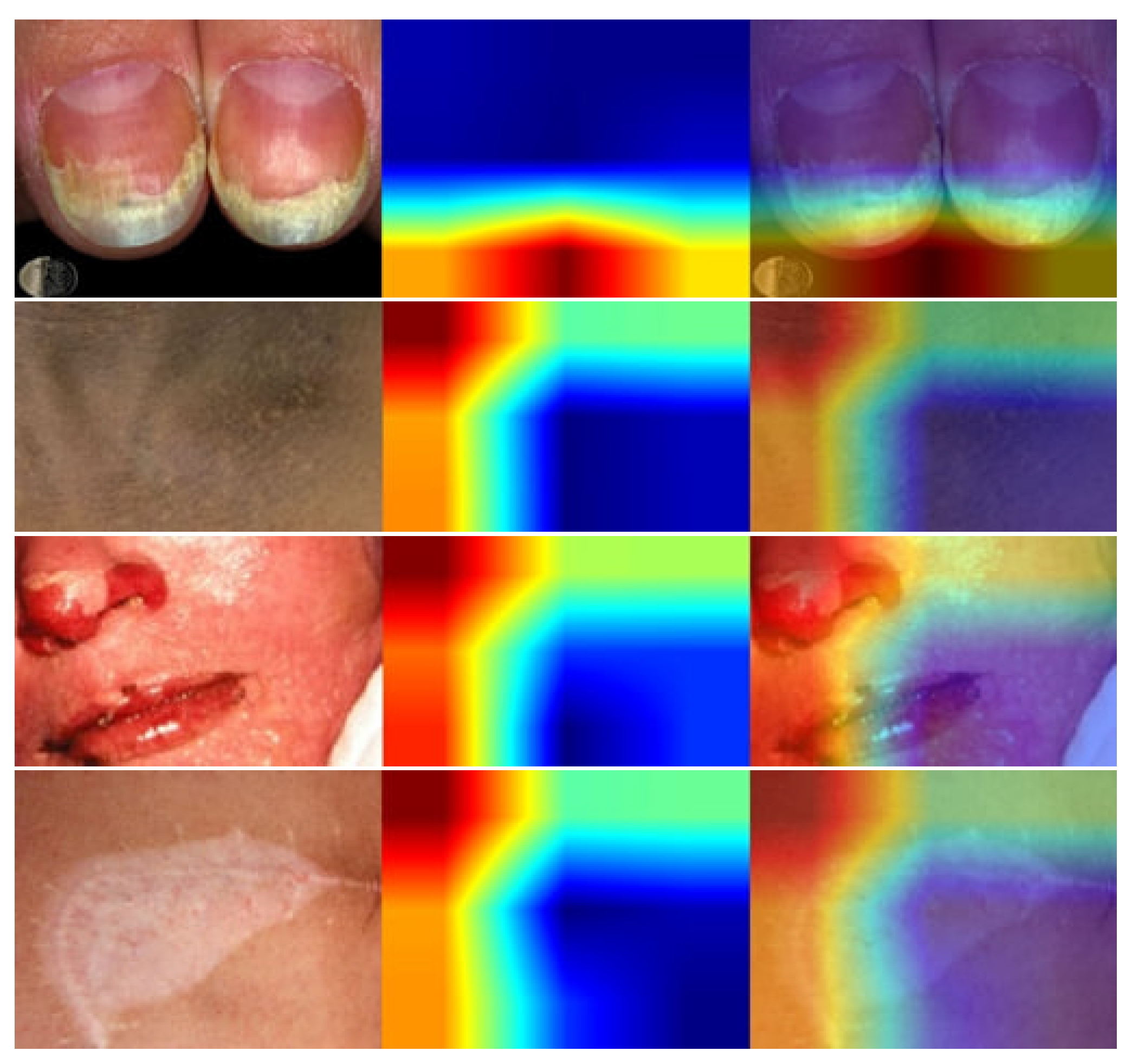
- We have used lime to explain how the model performs in the hidden layer and also identify the disease areas by using the deep color contrast.
2. Literature Review
3. Materials and Methods
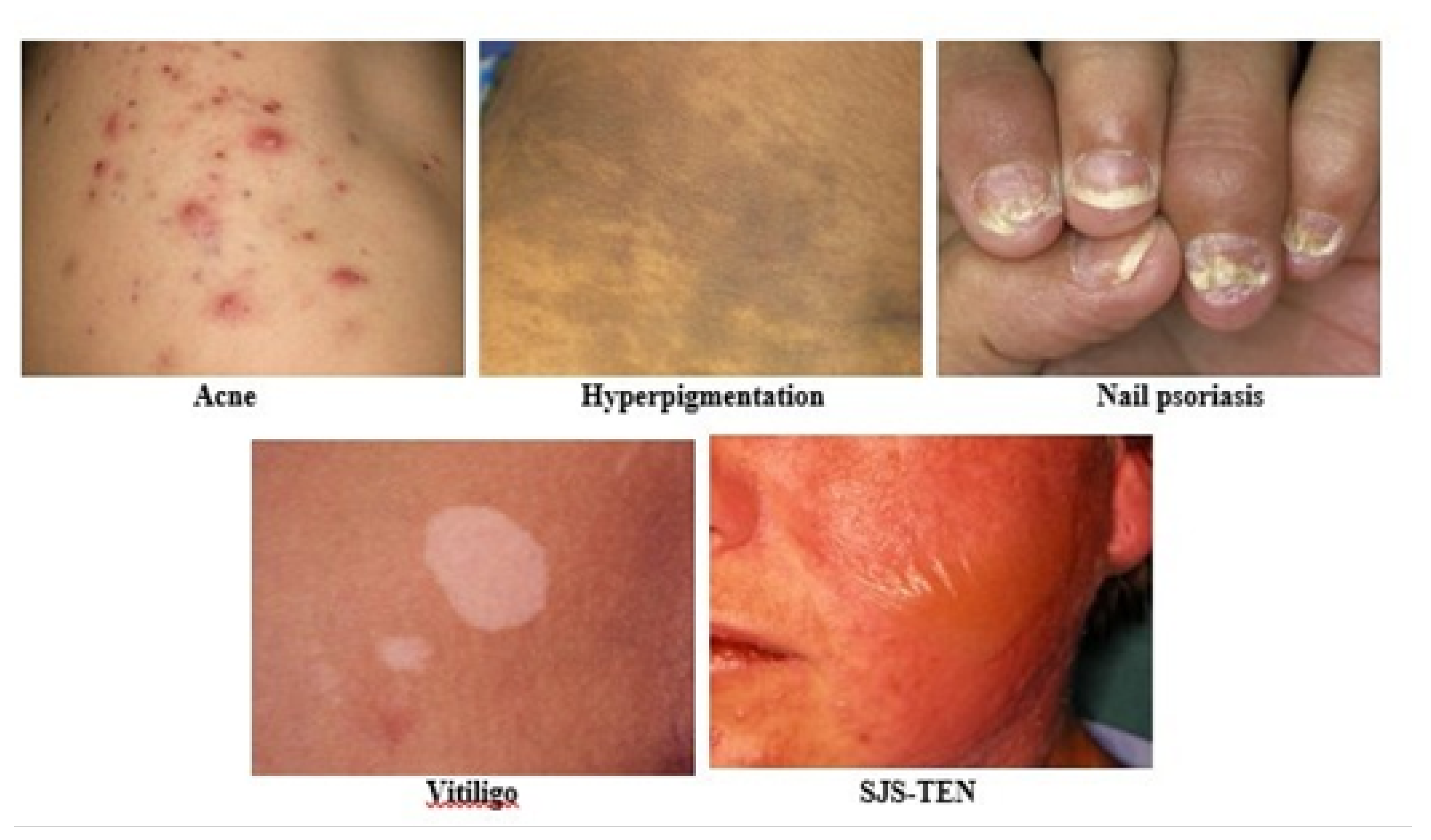
3.1. Dataset Description
3.2. Dataset Preparation
3.3. Random Oversampling
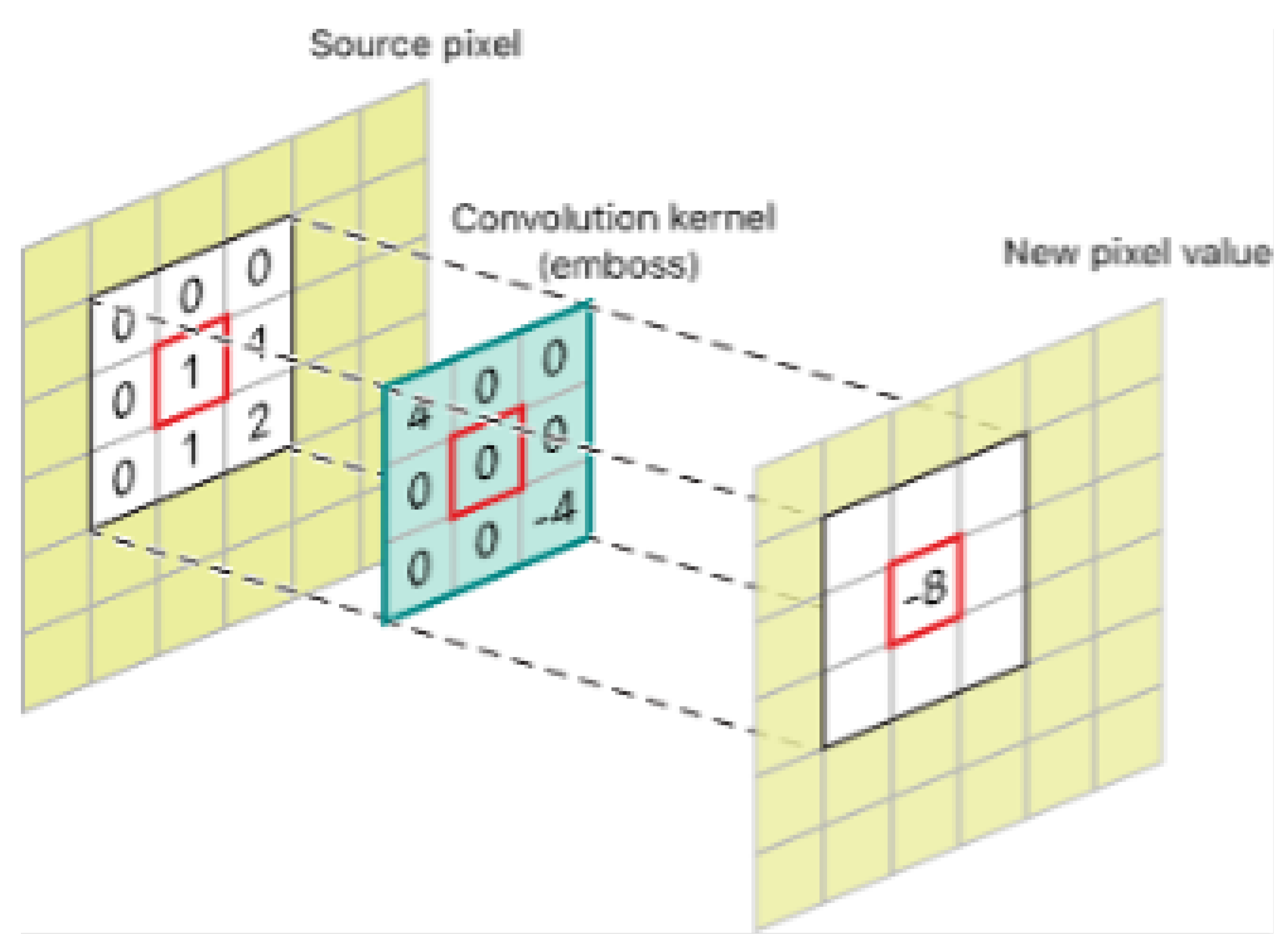
3.4. Image Processing
3.5. Image Resizing
3.6. Noise Removal
3.7. Augmentation
3.8. Blur Techniques
3.8.1. Gaussian Blur:
3.8.2. Averaging Blur:
3.8.3. Median Blur:
3.8.4. Bilateral Blur:
3.9. Statistical Features
3.9.1. Energy:
3.9.2. Contrast:
3.9.3. Homogeneity:
3.9.4. Entropy:
3.9.5. Mean:
3.9.6. Variance:
3.9.7. Standard Deviation:
3.9.8. Root Mean Square (Rms):
3.10. Transfer Learning Based Models
3.10.1. Custom Cnn Architecture:
3.11. Experimental Setup
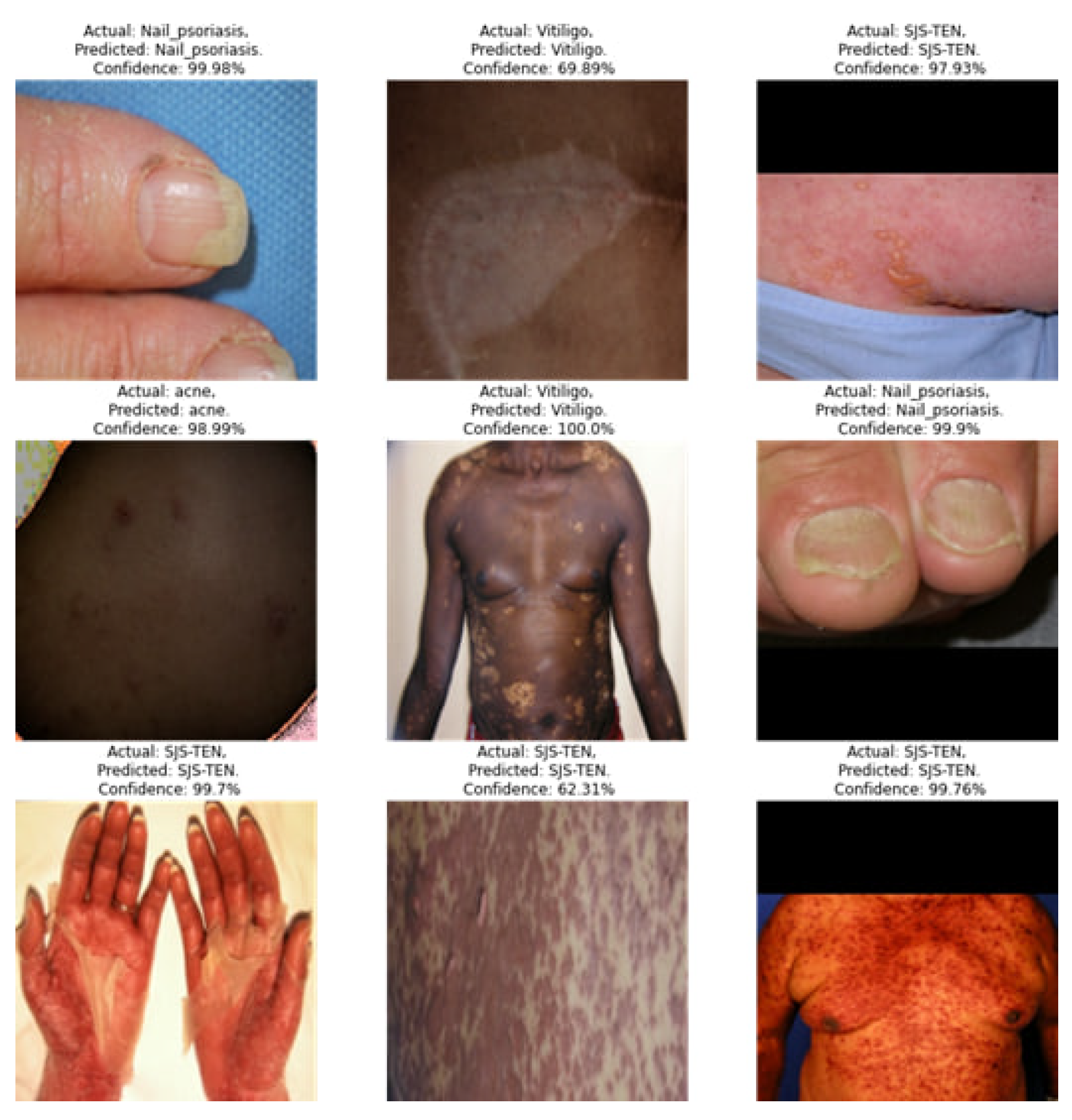
4. Result Analysis & Discussion
5. Conclusion and Future Work
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DL | Deep Learning |
| Grad-CAM | Gradient-weighted Class Activation Mapping |
| VGG-16 | Visual Geometry Group 16 |
| SJS-TEN | Stevens-Johnson syndrome and toxic epidermal necrolysis |
References
- G. Martin, S. Gu´erard, M.-M. R. Fortin, D. Rusu, J. Soucy, P. E. Poubelle, and R. Pouliot, Pathologicalcrosstalk in vitro between t lymphocytes and lesional keratinocytes in psoriasis: necessity of direct cell-to-cell contact, Laboratory investigation, 2012, vol. 92, no. 7, pp. 1058–1070.
- R. J. Hay, N. E. Johns, H. C. Williams, I. W. Bolliger, R. P. Dellavalle, D. J. Margolis, R. Marks, L. Naldi, M. A. Weinstock, S. K. Wulf, et al., The global burden of skin disease in 2010: an analysis of the prevalence and impact of skin conditions, Journal of Investigative Dermatology, 2014, vol. 134, no. 6, pp. 1527–1534.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal, and F. Bray, Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: a cancer journal for clinicians, 2021, vol. 71, no. 3, pp. 209–249.
- M. F. Akter, S. S. Sathi, S. Mitu, and M. O. Ullah, Lifestyle and heritability effects on cancer in bangladesh: an application of cox proportional hazards model, Asian Journal of Medical and Biological Research, 2021, vol. 7, no. 1, pp. 82–89.
- J. D. Khoury, E. Solary, O. Abla, Y. Akkari, R. Alaggio, J. F. Apperley, R. Bejar, E. Berti, L. Busque, J. K. Chan, et al., The 5th edition of the world health organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms, Leukemia, 2022, vol. 36, no. 7, pp. 1703–1719.
- W. Roberts, Air pollution and skin disorders, International Journal of Women’s Dermatology, 2021, vol. 7, no. 1, pp. 91–97.
- S. S. Han, M. S. Kim, W. Lim, G. H. Park, I. Park, and S. E. Chang, Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm, Journal of Investigative Dermatology, 2018, vol. 138, no. 7, pp. 1529–1538.
- P. Tschandl, C. Rosendahl, and H. Kittler, The ham10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions, Scientific data, 2018, vol. 5, no. 1, pp. 1–9.
- M. H. Jafari, E. Nasr-Esfahani, N. Karimi, S. R. Soroushmehr, S. Samavi, and K. Najarian, Extraction of skin lesions from non-dermoscopic images for surgical excision of melanoma, International journal of computer assisted radiology and surgery, 2017, vol. 12, pp. 1021–1030.
- X. Zhang, S. Wang, J. Liu, and C. Tao, Towards improving diagnosis of skin diseases by combining deep neural network and human knowledge, BMC medical informatics and decision making, 2018, vol. 18, no. 2, pp. 69–76.
- B. Harangi, A. Baran, and A. Hajdu, Classification of skin lesions using an ensemble of deep neural networks, In Proceedings of the 2018 40th annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp. 2575–2578, IEEE, 2018.
- Z. Ge, S. Demyanov, R. Chakravorty, A. Bowling, and R. Garnavi, Skin disease recognition using deep saliency features and multimodal learning of dermoscopy and clinical images, In Proceedings of the International conference on medical image computing and computer-assisted intervention, pp. 250–258, Springer, 2017.
- S. Inthiyaz, B. R. Altahan, S. H. Ahammad, V. Rajesh, R. R. Kalangi, L. K. Smirani, M. A. Hossain, and A. N. Z. Rashed, Skin disease detection using deep learning, Advances in Engineering Software, 2023, vol. 175, p. 103361.
- Y. Liu, A. Jain, C. Eng, D. H. Way, K. Lee, P. Bui, K. Kanada, G. de Oliveira Marinho, J. Gallegos, S. Gabriele, et al., A deep learning system for differential diagnosis of skin diseases,Nature medicine, 2020, vol. 26, no. 6, pp. 900–908.
- P. N. Srinivasu, J. G. SivaSai, M. F. Ijaz, A. K. Bhoi, W. Kim, and J. J. Kang, Classification of skin disease using deep learning neural networks with mobilenet v2 and lstm, Sensors, 2021, vol. 21, no. 8, p. 2852.
- T. Shanthi, R. Sabeenian, and R. Anand, Automatic diagnosis of skin diseases using convolution neural network, Microprocessors and Microsystems, 2020, vol. 76, p. 103074.
- M. Chen, P. Zhou, D. Wu, L. Hu, M. M. Hassan, and A. Alamri, Ai-skin: Skin disease recognition based on self-learning and wide data collection through a closed-loop framework, Information Fusion, 2020, vol. 54, pp. 1–9.
- E. Goceri, Diagnosis of skin diseases in the era of deep learning and mobile technology, Computers in Biology and Medicine, 2021, vol. 134, p. 104458.
- A. Esteva, K. Chou, S. Yeung, N. Naik, A. Madani, A. Mottaghi, Y. Liu, E. Topol, J. Dean, and R. Socher, Deep learning-enabled medical computer vision, NPJ digital medicine, 2021, vol. 4, no. 1, pp. 1–9.
- V. R. Allugunti, A machine learning model for skin disease classification using convolution neural network, International Journal of Computing, Programming and Database Management, 2022, vol. 3, no. 1, pp. 141–147.
- E. Goceri, Deep learning based classification of facial dermatological disorders, Computers in Biology and Medicine, 2021, vol. 128, p. 104118.
- M. Groh, C. Harris, L. Soenksen, F. Lau, R. Han, A. Kim, A. Koochek, and O. Badri, Evaluating deep neural networks trained on clinical images in dermatology with the fitzpatrick 17k dataset, In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 1820– 1828, 2021.
- L. R. Soenksen, T. Kassis, S. T. Conover, B. Marti-Fuster, J. S. Birkenfeld, J. Tucker-Schwartz, A. Naseem, R. R. Stavert, C. C. Kim, M. M. Senna, et al., Using deep learning for dermatologistlevel detection of suspicious pigmented skin lesions from wide-field images, Science Translational Medicine, 2021, vol. 13, no. 581, p. eabb3652.
- P. M. Burlina, N. J. Joshi, E. Ng, S. D. Billings, A. W. Rebman, and J. N. Aucott, Automated detection of erythema migrans and other confounding skin lesions via deep learning, Science Translational Medicine, 2019, vol. 105, pp. 151–156.
- M. A. Al-Masni, D.-H. Kim, and T.-S. Kim, Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification, Computer methods and programs in biomedicine, 2020, vol. 190, p. 105351.
- N. Janbi, R. Mehmood, I. Katib, A. Albeshri, J. M. Corchado, and T. Yigitcanlar, Imtidad: A reference architecture and a case study on developing distributed ai services for skin disease diagnosis over cloud, fog and edge, Sensors, 2022, vol. 22, no. 5, p. 1854.
- M. Abbas, M. Imran, A. Majid, and N. Ahmad, Skin diseases diagnosis system based on machine learning, Journal of Computing & Biomedical Informatics, 2022, vol. 4, no. 01, pp. 37–53.
- F. Weng, Y. Xu, Y. Ma, J. Sun, S. Shan, Q. Li, J. Zhu, and Y. Wang, An interpretable imbalanced semi-supervised deep learning framework for improving differential diagnosis of skin diseases, arXiv preprint arXiv:2211.10858, 2022.
- W. Gouda, N. U. Sama, G. Al-Waakid, M. Humayun, and N. Z. Jhanjhi, Detection of skin cancer based on skin lesion images using deep learning, in Healthcare MDPI 2022, vol. 10, p. 1183.
- B. Shetty, R. Fernandes, A. P. Rodrigues, R. Chengoden, S. Bhattacharya, and K. Lakshmanna, Skin lesion classification of dermoscopic images using machine learning and convolutional neural network, Scientific Reports 2022, vol. 12, no. 1, p. 18134.
- V. D. P. Jasti, A. S. Zamani, K. Arumugam, M. Naved, H. Pallathadka, F. Sammy, A. Raghuvanshi, and K. Kaliyaperumal, Computational technique based on machine learning and image processing for medical image analysis of breast cancer diagnosis, Security and communication networks 2022, vol. 2022, pp. 1–7.
- S. N. Almuayqil, S. Abd El-Ghany, and M. Elmogy, Computer-aided diagnosis for early signs of skin diseases using multi types feature fusion based on a hybrid deep learning model, Electronics 2022, vol. 11, no. 23, p. 4009.
- R. Karthik, T. S. Vaichole, S. K. Kulkarni, O. Yadav, and F. Khan, Eff2net: An efficient channel attention-based convolutional neural network for skin disease classification, Biomedical Signal Processing and Control 2022, vol. 73, p. 103406.
- A. C. Foahom Gouabou, J. Collenne, J. Monnier, R. Iguernaissi, J.-L. Damoiseaux, A. Moudafi, and D. Merad, Computer aided diagnosis of melanoma using deep neural networks and game theory: Application on dermoscopic images of skin lesions, International Journal of Molecular Sciences 2022, vol. 23, no. 22, p. 13838.
- K. Sreekala, N. Rajkumar, R. Sugumar, K. Sagar, R. Shobarani, K. P. Krishnamoorthy, A. Saini, H. Palivela, and A. Yeshitla, Skin diseases classification using hybrid ai based localization approach, Computational Intelligence and Neuroscience, 2022, vol. 2022.
- P. R. Kshirsagar, H. Manoharan, S. Shitharth, A. M. Alshareef, N. Albishry, and P. K. Balachandran, Deep learning approaches for prognosis of automated skin disease, Life, 2022, vol. 12, no. 3, p. 426.
- B. Cassidy, C. Kendrick, A. Brodzicki, J. Jaworek-Korjakowska, and M. H. Yap, Analysis of the isic image datasets: Usage, benchmarks and recommendations, Medical image analysis, 2022, vol. 75, p. 102305.
- F. Alenezi, A. Armghan, and K. Polat, Wavelet transform based deep residual neural network and relu based extreme learning machine for skin lesion classification, Expert Systems with Applications, 2023, vol. 213, p. 119064.
- S. Singh et al., Image filtration in python using opencv, Turkish Journal of Computer and Math- ematics Education (TURCOMAT), 2021, vol. 12, no. 6, pp. 5136–5143.
- M. Xu, S. Yoon, A. Fuentes, and D. S. Park, A comprehensive survey of image augmentation techniques for deep learning, Pattern Recognition, 2023, p. 109347.
- P. Chlap, H. Min, N. Vandenberg, J. Dowling, L. Holloway, and A. Haworth, A review of medical image data augmentation techniques for deep learning applications, Journal of Medical Imaging and Radiation Oncology, 2021, vol. 65, no. 5, pp. 545–563.
- K. Maharana, S. Mondal, and B. Nemade, A review: Data pre-processing and data augmentation techniques, Global Transitions Proceedings, 2022.
- S. Bang, F. Baek, S. Park, W. Kim, and H. Kim, Image augmentation to improve construction resource detection using generative adversarial networks, cut-and-paste, and image transformation techniques, Automation in Construction, 2020, vol. 115, p. 103198.
- A. Singla, L. Yuan, and T. Ebrahimi, Food/non-food image classification and food categorization using pre-trained googlenet model, In Proceedings of the 2nd International Workshop on Multimedia Assisted Dietary Management, pp. 3–11, 2016.
- Q. Mi, J. Keung, Y. Xiao, S. Mensah, and X. Mei, An inception architecture-based model for improving code readability classification, In Proceedings of the 22nd International Conference on Evaluation and Assessment in Software Engineering 2018, pp. 139–144, 2018.
- A. Younis, L. Qiang, C. O. Nyatega, M. J. Adamu, and H. B. Kawuwa, Brain tumor analysis using deep learning and vgg-16 ensembling learning approaches, Applied Sciences, 2022, vol. 12, no. 14, p. 7282.
- T. Subetha, R. Khilar, and M. S. Christo, Withdrawn: A comparative analysis on plant pathology classification using deep learning architecture–resnet and vgg19, 2021.
- F. Chollet, Xception: Deep learning with depthwise separable convolutions, In Proceedings of the IEEE conference on computer vision and pattern recognition, pp. 1251–1258, 2017.
- khushbu, sharun akter, “Skin Disease Classification Dataset”, Mendeley Data, V1, 2024. [CrossRef]
| Name | Description |
|---|---|
| Total number of images | 5184 |
| Dimension | 224*224 |
| Image format | JPG |
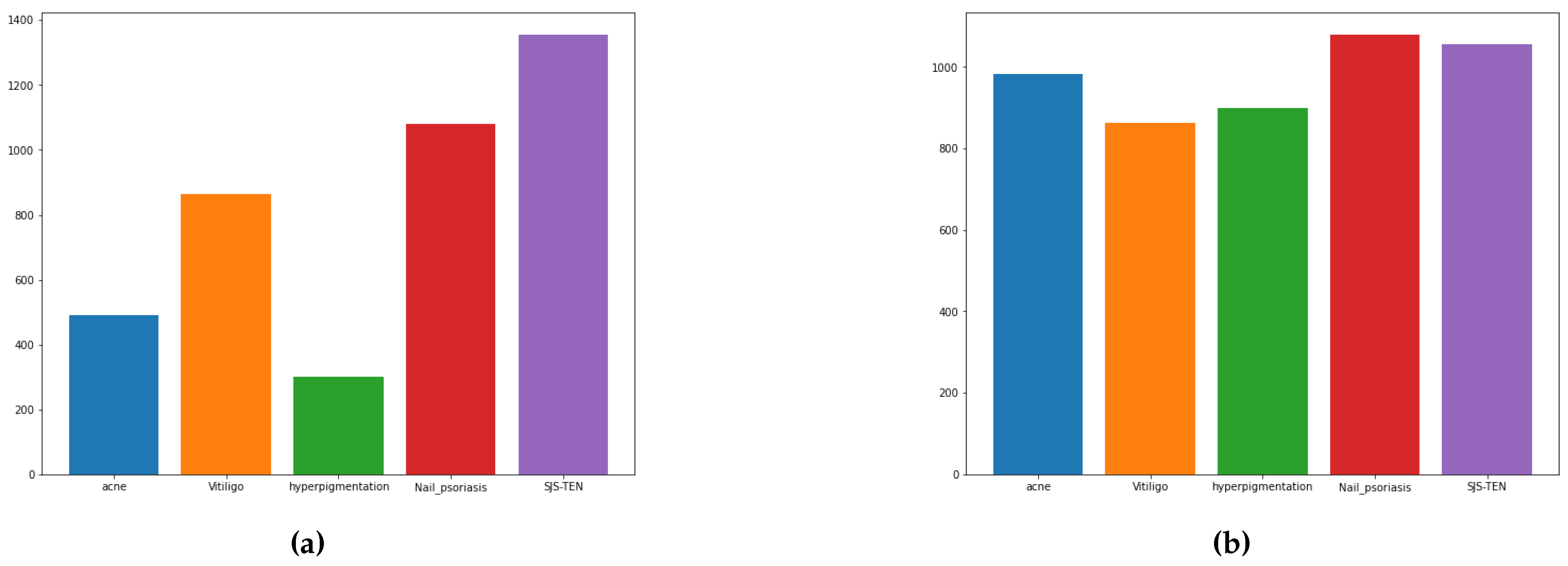
| Acne | 984 |
| Hyperpigmentation | 900 |
| Nail psoriasis | 1080 |
| SJS-TEN | 1356 |
| Vitiligo | 864 |
| Image | Image_1 | Image_2 | Image_3 | Image_4 | Image_5 | Image_6 | Image_7 | Image_8 | Image_9 | Image_10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Energy | 0.0485 | 0.0290 | 0.0286 | 0.0286 | 0.0325 | 0.0284 | 0.0348 | 0.0636 | 0.0392 | 0.0478 |
| Correlation | 0.8838 | 0.7944 | 0.9763 | 0.9711 | 0.9838 | 0.9029 | 0.9471 | 0.9964 | 0.9752 | 0.9938 |
| Contrast | 1112.3901 | 546.9877 | 45.1985 | 64.6160 | 22.8364 | 559.1883 | 244.3821 | 1.8534 | 27.5155 | 4.4025 |
| Homogeneity | 0.3357 | 0.2438 | 0.2451 | 0.3491 | 0.3242 | 0.2446 | 0.4573 | 0.5935 | 0.3337 | 0.4511 |
| Entropy | 10.8958 | 11.5756 | 11.1457 | 10.9050 | 10.7034 | 11.2315 | 10.4978 | 8.3734 | 10.3353 | 9.2798 |
| Mean | 0.3171 | 0.3760 | 0.5281 | 0.2792 | 0.4288 | 0.2504 | 0.3817 | 0.3769 | 0.5094 | 0.5319 |
| Variance | 0.0735 | 0.0204 | 0.0146 | 0.0172 | 0.0109 | 0.0443 | 0.0358 | 0.0039 | 0.0086 | 0.0055 |
| SD | 0.2711 | 0.1429 | 0.1209 | 0.1313 | 0.1046 | 0.2104 | 0.1892 | 0.0628 | 0.0927 | 0.0738 |
| RMS | 0.4171 | 0.4023 | 0.5418 | 0.3085 | 0.4413 | 0.3270 | 0.4260 | 0.3821 | 0.5177 | 0.5370 |
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| GoogleNet | 82.91 | 90.69 | 96.61 | 91.40 |
| Inception | 47.40 | 67.74 | 88.59 | 57.87 |
| VGG-16 | 93.97 | 92.81 | 99.30 | 97.87 |
| VGG-19 | 95.00 | 97.72 | 98.30 | 99.14 |
| Xception | 60.30 | 68.51 | 77.60 | 69.78 |
| Model | Accuracy (%) | Precision (%) | Recall (%) | F1-Score (%) |
|---|---|---|---|---|
| GoogleNet | 91.11 | 97.56 | 98.87 | 96.00 |
| Inception | 58.20 | 78.72 | 85.83 | 76.05 |
| VGG-16 | 97.07 | 98.52 | 98.34 | 99.16 |
| VGG-19 | 96.29 | 97.12 | 97.82 | 98.37 |
| Xception | 68.36 | 73.58 | 76.47 | 76.57 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





