Submitted:
02 March 2024
Posted:
05 March 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- SkinLiTE is designed to leverage the strengths of both supervised learning and contrastive learning for skin lesion detection and disease typification from dermoscopic images. This hybrid approach is particularly potent for tasks that involve complex medical datasets.
- SkinLiTE addresses the challenge of imbalanced datasets we encountered in skin lesions by focusing on discerning relative similarities and differences among various classes, which is often the case with rare skin diseases.
- The model uses labeled data to form pairs or groups based on class labels, which is essential for supervised contrastive learning. The representations derived from SkinLiTE are designed to be strong and adaptable, enabling the model to discover features of various skin conditions effectively.
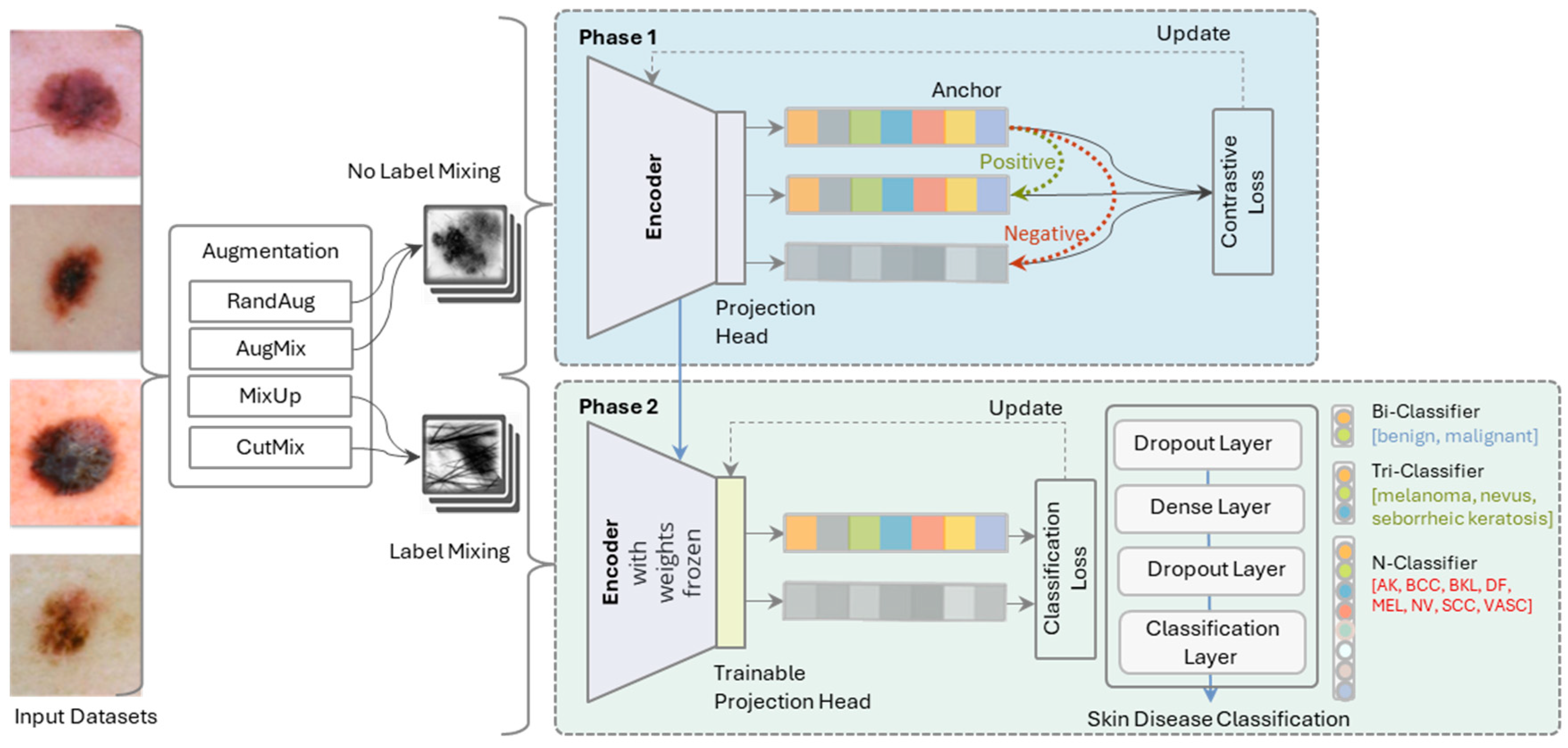
- The two-phase architecture enhances the learning process in SkinLiTE and yields a light-weight model for the application in the world of the internet of medical things (IoMT). In the first phase, the model utilizes an encoder and a projection head to map the input dermoscopic images into a representation space where contrastive loss is applied. Here, the model is trained to update its weights to minimize this loss, effectively learning to anchor data points of the same class closely together and push different classes apart. The second phase involves the encoder with its frozen weights and a trainable projection head, meaning the representations learned in the earlier phase are kept stable. The focus here is on classification, where a series of layers, including dropout and dense layers, lead to a classification layer.
- SkinLiTE is trained for multi-tier classification to handle different classification scenarios: Bi-Classifier: Differentiates between benign and malignant lesions., Tri-Classifier: Categorizes lesions into melanoma, nevus, or seborrheic keratosis., and N-Classifier: A more granular classification that includes multiple types of skin conditions like Actinic Keratosis (AK), Basal Cell Carcinoma (BCC), Benign Keratosis-like Lesions (BKL), Dermatofibroma (DF), Melanoma (MEL), Nevus (NV), Squamous Cell Carcinoma (SCC), and Vascular Lesions (VASC).
2. Related Work
2.1. The Glance of Machine Learning
2.2. The Move to Deep Learning
2.2.1. Use of Pre-Trained Models and Transfer Learning
2.2.2. Innovative Approaches and Combination Strategies
2.2.3. Focus on Specific Challenges
2.2.4. Advanced Optimization Techniques
2.2.5. Dataset Utilization
2.2.6. Performance and Interpretability
2.3. The Challenge of Imbalanced Data
2.4. Attention to Skin Lesions
2.5. The Trends of Internet of Medical Things and Remote Patient Monitoring
2.6. The Gap
3. Methodology
3.1. Problem Formulation
3.2. SkinLiTE Model Architecture
3.3. Augmentation
3.4. Supervised Contrastive Learning
- Input Images: Dermoscopic images are input to the encoder.
- Encoding: We used ResNet50V2 for the encoder neural network to produce embeddings.
- Projection: The embeddings are then mapped to a projection space using a projection head, which is typically a shallow neural network.
- Contrastive Loss Calculation: The model computes the supervised contrastive loss for the anchor, positive, and negative samples. Given an anchor image embedding , and a set of positive examples where constitutes examples from the same label plus augmentations of anchor , and a set of negative examples from other classes, the supervised contrastive loss used in this study was defined as:
3.5. Supervised Classification Learning
-
Inputs:
- o
- Dermoscopic images are input to the encoder.
- o
- Frozen Encoder which is the encoder from phase (1) with its frozen weights provides the feature embeddings.
- Trainable Projection Head: A new projection head, which can be trained, is used for fine-tuning to the classification task.
- Classification Layers: A series of layers including dropout and dense layers process the embeddings.
- Cross-Entropy Loss Calculation: The model calculates the cross-entropy loss based on the output of the classification layers and the true labels. Given a set of true labels and the predicted probabilities by the model, the cross-entropy loss for classification is defined as:
4. Experimental Setup
4.1. Datasets
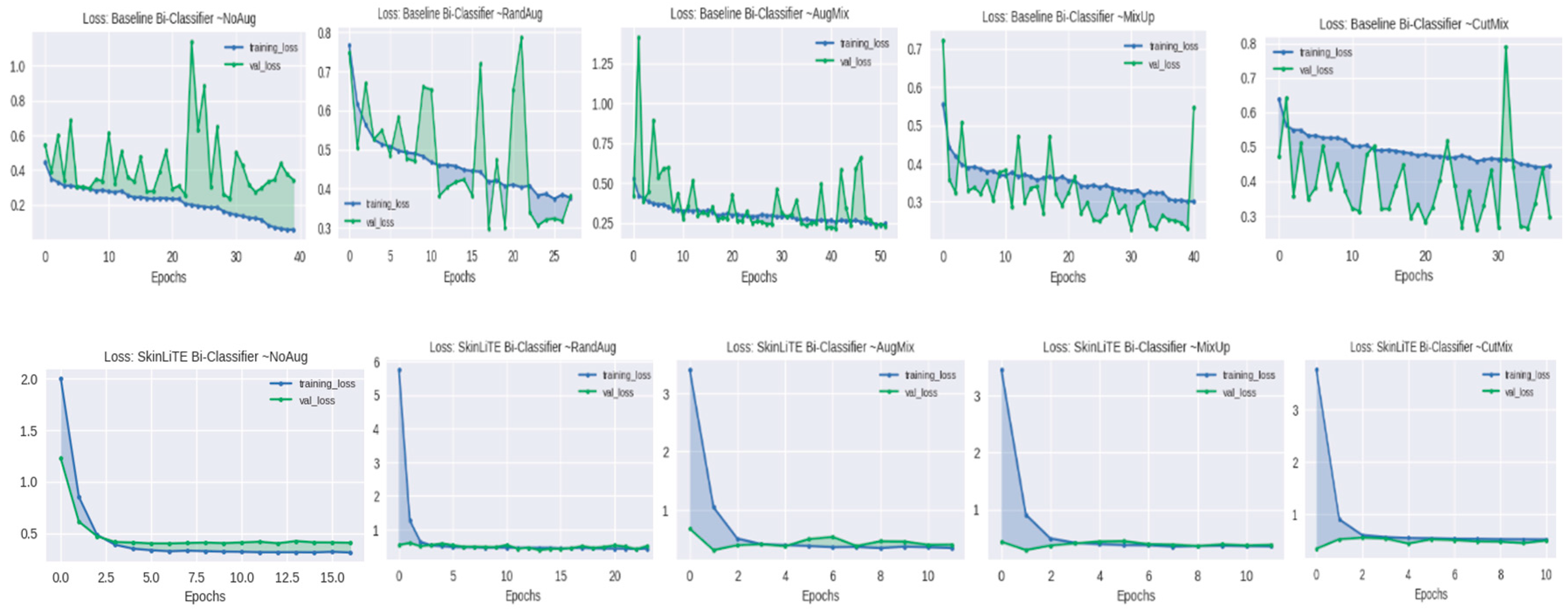
4.2. Augmentation Results
4.3. Evaluation and Computing Resources
5. Results and Discussion
6. Conclusion and Future Work
Data Availability Statement
Acknowledgments
Declaration of Competing Interest
References
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Medical Image Analysis 2022, 75, 102305. [Google Scholar] [CrossRef]
- Mohanty, A.; Sutherland, A.; Bezbradica, M.; Javidnia, H. Skin Disease Analysis With Limited Data in Particular Rosacea: A Review and Recommended Framework. IEEE Access 2022, 10, 39045–39068. [Google Scholar] [CrossRef]
- Kumar, Y.; Koul, A.; Singla, R.; Ijaz, M.F. Artificial intelligence in disease diagnosis: a systematic literature review, synthesizing framework and future research agenda. Journal of Ambient Intelligence and Humanized Computing 2022. [Google Scholar] [CrossRef]
- Vatiwutipong, P.; Vachmanus, S.; Noraset, T.; Tuarob, S. Artificial Intelligence in Cosmetic Dermatology: A Systematic Literature Review. IEEE Access 2023, 11, 71407–71425. [Google Scholar] [CrossRef]
- Li, X.H.; Zhao, X.Y.; Ma, H.R.; Xie, B. Image Analysis and Diagnosis of Skin Diseases-A Review. Current Medical Imaging 2023, 19, 199–242. [Google Scholar] [CrossRef]
- Kaur, B.; Goyal, B.; Daniel, E. A Survey on Machine Learning Based Medical Assistive Systems in Current Oncological Sciences. Current Medical Imaging 2022, 18, 445–459. [Google Scholar] [CrossRef]
- Huérfano-Maldonado, Y.; Mora, M.; Vilches, K.; Hernández-García, R.; Gutiérrez, R.; Vera, M. A comprehensive review of extreme learning machine on medical imaging. Neurocomputing 2023, 556. [Google Scholar] [CrossRef]
- Mangotra, H.; Srivastava, S.; Jaiswal, G.; Rani, R.; Sharma, A. Hyperspectral imaging for early diagnosis of diseases: A review. Expert Systems 2023, 40. [Google Scholar] [CrossRef]
- Singh, M.; Singh, M.; De, D.; Handa, S.; Mahajan, R.; Chatterjee, D. Towards Diagnosis of Autoimmune Blistering Skin Diseases Using Deep Neural Network. Archives of Computational Methods in Engineering 2023, 30, 3529–3557. [Google Scholar] [CrossRef]
- Hosny, K.M.; Elshoura, D.; Mohamed, E.R.; Vrochidou, E.; Papakostas, G.A. Deep Learning and Optimization-Based Methods for Skin Lesions Segmentation: A Review. IEEE Access 2023, 11, 85467–85488. [Google Scholar] [CrossRef]
- Jiang, H.Y.; Diao, Z.S.; Shi, T.Y.; Zhou, Y.; Wang, F.Y.; Hu, W.R.; Zhu, X.L.; Luo, S.J.; Tong, G.Y.; Yao, Y.D. A review of deep learning-based multiple-lesion recognition from medical images: classification, detection and segmentation. Computers in Biology and Medicine 2023, 157. [Google Scholar] [CrossRef]
- Noronha, S.S.; Mehta, M.A.; Garg, D.; Kotecha, K.; Abraham, A. Deep Learning-Based Dermatological Condition Detection: A Systematic Review With Recent Methods, Datasets, Challenges, and Future Directions. IEEE Access 2023, 11, 140348–140381. [Google Scholar] [CrossRef]
- Painuli, D.; Bhardwaj, S.; Kose, U. Recent advancement in cancer diagnosis using machine learning and deep learning techniques: A comprehensive review. Computers in Biology and Medicine 2022, 146. [Google Scholar] [CrossRef] [PubMed]
- Rai, H.M. Cancer detection and segmentation using machine learning and deep learning techniques: a review. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Nawaz, M.; Uvaliyev, A.; Bibi, K.; Wei, H.; Abaxi, S.M.D.; Masood, A.; Shi, P.L.; Ho, H.P.; Yuan, W. Unraveling the complexity of Optical Coherence Tomography image segmentation using machine and deep learning techniques: A review. Computerized Medical Imaging and Graphics 2023, 108. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Mohamed, H.K.; Maher, A.; Zhang, B.C. A Survey on Human Cancer Categorization Based on Deep Learning. Frontiers in Artificial Intelligence 2022, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.-C.; Ahn, E.; Feng, D.; Kim, J. A Review of Predictive and Contrastive Self-supervised Learning for Medical Images. Machine Intelligence Research 2023, 20, 483–513. [Google Scholar] [CrossRef]
- Khosla, P.; Teterwak, P.; Wang, C.; Sarna, A.; Tian, Y.; Isola, P.; Maschinot, A.; Liu, C.; Krishnan, D. Supervised Contrastive Learning. 2020, arXiv:2004.11362. [CrossRef]
- AlDera, S.A.; Ben Othman, M.T. A Model for Classification and Diagnosis of Skin Disease using Machine Learning and Image Processing Techniques. International Journal of Advanced Computer Science and Applications 2022, 13, 252–259. [Google Scholar] [CrossRef]
- Hatem, M.Q. Skin lesion classification system using a K-nearest neighbor algorithm. Visual Computing for Industry Biomedicine and Art 2022, 5. [Google Scholar] [CrossRef]
- Saghir, U.; Hasan, M. Skin cancer detection and classification based on differential analyzer algorithm. Multimedia Tools and Applications 2023, 82, 41129–41157. [Google Scholar] [CrossRef]
- Mustafa, S.; Jaffar, A.; Iqbal, M.W.; Abubakar, A.; Alshahrani, A.S.; Alghamdi, A. Hybrid Color Texture Features Classification Through ANN for Melanoma. Intelligent Automation and Soft Computing 2023, 35. [Google Scholar] [CrossRef]
- Huong, L.H.; Khang, N.H.; Quynh, L.; Thang, L.; Canh, D.M.; Sang, H.P. A Proposed Approach for Monkeypox Classification. International Journal of Advanced Computer Science and Applications 2023, 14, 643–651. [Google Scholar] [CrossRef]
- MunishKhanna; Singh, L.K.; Garg, H. A novel approach for human diseases prediction using nature inspired computing & machine learning approach. Multimedia Tools and Applications 2023. [CrossRef]
- Meena, K.; Veni, N.N.K.; Deepapriya, B.S.; Vardhini, P.A.H.; Kalyani, B.J.D.; Sharmila, L. A novel method for prediction of skin disease through supervised classification techniques. Soft Computing 2022, 26, 10527–10533. [Google Scholar] [CrossRef]
- Zou, Q.X.; Cheng, J.Y.; Liang, Z.L. Automatic Diagnosis of Melanoma Based on EfficientNet and Patch Strategy. International Journal of Computational Intelligence Systems 2023, 16. [Google Scholar] [CrossRef]
- Spolaôr, N.; Lee, H.D.; Mendes, A.I.; Nogueira, C.V.; Parmezan, A.R.S.; Takaki, W.S.R.; Coy, C.S.R.; Wu, F.C.; Fonseca-Pinto, R. Fine-tuning pre-trained neural networks for medical image classification in small clinical datasets. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Patel, M.B. Multi Class Skin Diseases Classification Based On Dermoscopic Skin Images Using Deep Learning. International Journal of Next-Generation Computing 2022, 13, 151–161. [Google Scholar] [CrossRef]
- Shaheen, H.; Singh, M.P. Multiclass skin cancer classification using particle swarm optimization and convolutional neural network with information security. Journal of Electronic Imaging 2023, 32. [Google Scholar] [CrossRef]
- Alghieth, M. Skin Disease Detection for Kids at School Using Deep Learning Techniques. International Journal of Online and Biomedical Engineering 2022, 18, 114–128. [Google Scholar] [CrossRef]
- Sivakumar, M.S.; Leo, L.M.; Gurumekala, T.; Sindhu, V.; Priyadharshini, A.S. Deep learning in skin lesion analysis for malignant melanoma cancer identification. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Alrusaini, O.A. Deep Learning Models for the Detection of Monkeypox Skin Lesion on Digital Skin Images. International Journal of Advanced Computer Science and Applications 2023, 14, 637–644. [Google Scholar] [CrossRef]
- Tripathi, A.; Singh, A.K.; Singh, A.; Choudhary, A.; Pareek, K.; Mishra, K.K. Analyzing Skin Disease Using XCNN (eXtended Convolutional Neural Network). International Journal of Software Science and Computational Intelligence-Ijssci 2022, 14. [Google Scholar] [CrossRef]
- Bala, D.; Hossain, M.S.; Hossain, M.A.; Abdullah, M.I.; Rahman, M.M.; Manavalan, B.; Gu, N.J.; Islam, M.S.; Huang, Z.J. MonkeyNet: A robust deep convolutional neural network for monkeypox disease detection and classification. Neural Networks 2023, 161, 757–775. [Google Scholar] [CrossRef]
- Wei, M.J.; Wu, Q.W.; Ji, H.Y.; Wang, J.K.; Lyu, T.; Liu, J.Y.; Zhao, L. A Skin Disease Classification Model Based on DenseNet and ConvNeXt Fusion. Electronics 2023, 12. [Google Scholar] [CrossRef]
- El Gannour, O.; Hamida, S.; Lamalem, Y.; Cherradi, B.; Saleh, S.; Raihani, A. Enhancing Skin Diseases Classification Through Dual Ensemble Learning and Pre-trained CNNs. International Journal of Advanced Computer Science and Applications 2023, 14, 436–445. [Google Scholar] [CrossRef]
- Aboulmira, A.; Hrimech, H.; Lachgar, M. Comparative Study of Multiple CNN Models for Classification of 23 Skin Diseases. International Journal of Online and Biomedical Engineering 2022, 18, 127–142. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Koundal, D.; Nayak, S.R.; Nayak, J.; Vimal, S. Multi-class Skin Disease Classification Using Transfer Learning Model. International Journal on Artificial Intelligence Tools 2022, 31. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Nayak, S.R.; Koundal, D.; Prakash, D.; Verma, K.D. An automated deep learning models for classification of skin disease using Dermoscopy images: a comprehensive study. Multimedia Tools and Applications 2022, 81, 37379–37401. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Koundal, D.; Mahajan, S.; Pandit, A.K.; Zaguia, A. Deep Learning Based Automated Diagnosis of Skin Diseases Using Dermoscopy. Cmc-Computers Materials & Continua 2022, 71, 3145–3160. [Google Scholar] [CrossRef]
- Nath, S.; Das Gupta, S.; Saha, S. Deep learning-based common skin disease image classification. Journal of Intelligent & Fuzzy Systems 2023, 44, 7483–7499. [Google Scholar] [CrossRef]
- Arora, G.; Dubey, A.K.; Jaffery, Z.A.; Rocha, A. A comparative study of fourteen deep learning networks for multi skin lesion classification (MSLC) on unbalanced data. Neural Computing & Applications 2023, 35, 7989–8015. [Google Scholar] [CrossRef]
- Al-Tuwaijari, J.M.; Yousir, N.T.; Alhammad, N.A.M.; Mostafa, S. Deep Residual Learning Image Recognition Model for Skin Cancer Disease Detection and Classification. Acta Informatica Pragensia 2023, 12, 19–31. [Google Scholar] [CrossRef]
- Tasar, B. SkinCancerNet: Automated Classification of Skin Lesion Using Deep Transfer Learning Method. Traitement Du Signal 2023, 40, 285–295. [Google Scholar] [CrossRef]
- Magdy, A.; Hussein, H.; Abdel-Kader, R.F.; Abd El Salam, K. Performance Enhancement of Skin Cancer Classification Using Computer Vision. IEEE Access 2023, 11, 72120–72133. [Google Scholar] [CrossRef]
- Pramila, R.P.; Subhashini, R. Automated skin lesion detection and classification using fused deep convolutional neural network on dermoscopic images. Computational Intelligence 2023. [Google Scholar] [CrossRef]
- Arshed, M.A.; Mumtaz, S.; Ibrahim, M.; Ahmed, S.; Tahir, M.; Shafi, M. Multi-Class Skin Cancer Classification Using Vision Transformer Networks and Convolutional Neural Network-Based Pre-Trained Models. Information 2023, 14. [Google Scholar] [CrossRef]
- Raghavendra, P.; Charitha, C.; Begum, K.G.; Prasath, V.B.S. Deep Learning-Based Skin Lesion Multi-class Classification with Global Average Pooling Improvement. Journal of Digital Imaging 2023, 36, 2227–2248. [Google Scholar] [CrossRef] [PubMed]
- Kani, M.; Parvathy, M.S.; Banu, S.M.; Kareem, M.S.A. Classification of skin lesion images using modified Inception V3 model with transfer learning and augmentation techniques. Journal of Intelligent & Fuzzy Systems 2023, 44, 4627–4641. [Google Scholar] [CrossRef]
- Aloraini, M. Two-stream convolutional networks for skin cancer classification. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Riaz, L.; Qadir, H.M.; Ali, G.; Ali, M.; Raza, M.A.; Jurcut, A.D.; Ali, J. A Comprehensive Joint Learning System to Detect Skin Cancer. IEEE Access 2023, 11, 79434–79444. [Google Scholar] [CrossRef]
- Kaur, R.; Kaur, N. Ti-FCNet: Triple fused convolutional neural network-based automated skin lesion classification. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Li, H.Y.; Zhang, P.; Wei, Z.K.; Qian, T.; Tang, Y.Q.; Hu, K.; Huang, X.Q.; Xia, X.X.; Zhang, Y.S.; Cheng, H.X.; et al. Deep skin diseases diagnostic system with Dual-channel Image and Extracted Text. Frontiers in Artificial Intelligence 2023, 6. [Google Scholar] [CrossRef]
- Li, S.H.; Li, X.M.; Xu, X.W.; Cheng, K.T. Dynamic Subcluster-Aware Network for Few-Shot Skin Disease Classification. IEEE Transactions on Neural Networks and Learning Systems 2023. [Google Scholar] [CrossRef]
- Reddy, D.A.; Roy, S.; Kumar, S.; Tripathi, R. Enhanced U-Net segmentation with ensemble convolutional neural network for automated skin disease classification. Knowledge and Information Systems 2023, 65, 4111–4156. [Google Scholar] [CrossRef]
- Josphineleela, R.; Rao, P.; Shaikh, A.; Sudhakar, K. A Multi-Stage Faster RCNN-Based iSPLInception for Skin Disease Classification Using Novel Optimization. Journal of Digital Imaging 2023, 36, 2210–2226. [Google Scholar] [CrossRef] [PubMed]
- Sreekala, K.; Rajkumar, N.; Sugumar, R.; Sagar, K.V.D.; Shobarani, R.; Krishnamoorthy, K.P.; Saini, A.K.; Palivela, H.; Yeshitla, A. Skin Diseases Classification Using Hybrid AI Based Localization Approach. Computational Intelligence and Neuroscience 2022, 2022. [Google Scholar] [CrossRef] [PubMed]
- He, X.Y.; Wang, Y.; Zhao, S.; Chen, X. Joint segmentation and classification of skin lesions via a multi-task learning convolutional neural network. Expert Systems with Applications 2023, 230. [Google Scholar] [CrossRef]
- Alam, M.J.; Mohammad, M.S.; Hossain, M.A.F.; Showmik, I.A.; Raihan, M.S.; Ahmed, S.; Ibn Mahmud, T. S-2-C-DeLeNet: A parameter transfer based segmentation-classification integration for detecting skin cancer lesions from dermoscopic images. Computers in Biology and Medicine 2022, 150. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.X.; Dai, D.W.; Zhang, Y.Z.; Zhang, C.Y.; Li, Z.F.; Xu, S.H. Learning from dermoscopic images in association with clinical metadata for skin lesion segmentation and classification. Computers in Biology and Medicine 2023, 152. [Google Scholar] [CrossRef]
- Yadav, A.K.; Mehta, R.; Kumar, V.; Medikondu, N.R. An optimized boosting framework for skin lesion segmentation and classification. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Duman, E.; Tolan, Z. Ensemble the recent architectures of deep convolutional networks for skin diseases diagnosis. International Journal of Imaging Systems and Technology 2023, 33, 1293–1305. [Google Scholar] [CrossRef]
- Kalaivani, A.; Karpagavalli, S. Advanced domain adaptation for skin disease segmentation and classification using bootstrapping of fine-tuned deep learner. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Ramamurthy, K.; Muthuswamy, A.; Mathimariappan, N.; Kathiresan, G.S. A novel two-staged network for skin disease detection using atrous residual convolutional networks. Concurrency and Computation-Practice & Experience 2023, 35. [Google Scholar] [CrossRef]
- Rao, G.M.; Ramesh, D.; Gantela, P.; Srinivas, K. A hybrid deep learning strategy for image based automated prognosis of skin disease. Soft Computing 2023. [Google Scholar] [CrossRef]
- Manoj, S.O.; Abirami, K.R.; Victor, A.; Arya, M. Automatic Detection and Categorization of Skin Lesions for Early Diagnosis of Skin Cancer Using YOLO-v3 - DCNN Architecture. Image Analysis & Stereology 2023, 42, 101–117. [Google Scholar] [CrossRef]
- Hao, S.N.; Zhang, L.G.; Jiang, Y.Y.; Wang, J.K.; Ji, Z.L.; Zhao, L.; Ganchev, I. ConvNeXt-ST-AFF: A Novel Skin Disease Classification Model Based on Fusion of ConvNeXt and Swin Transformer. IEEE Access 2023, 11, 117460–117473. [Google Scholar] [CrossRef]
- Kalyani, K.; Althubiti, S.A.; Ahmed, M.A.; Lydia, E.L.; Kadry, S.; Han, N.; Nam, Y. Arithmetic Optimization with Ensemble Deep Transfer Learning Based Melanoma Classification. Cmc-Computers Materials & Continua 2023, 75, 149–164. [Google Scholar] [CrossRef]
- Seeja, R.D.; Geetha, A. Effective melanoma classification using inter neighbour mean order interleaved pattern on dermoscopy images. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Albraikan, A.A.; Nemri, N.; Alkhonaini, M.A.; Hilal, A.M.; Yaseen, I.; Motwakel, A. Automated Deep Learning Based Melanoma Detection and Classification Using Biomedical Dermoscopic Images. Cmc-Computers Materials & Continua 2023, 74, 2443–2459. [Google Scholar] [CrossRef]
- Meswal, H.; Kumar, D.; Gupta, A.; Roy, S. A weighted ensemble transfer learning approach for melanoma classification from skin lesion images. Multimedia Tools and Applications 2023. [Google Scholar] [CrossRef]
- Asif, S.; Zhao, M.; Tang, F.X.; Zhu, Y.S.; Zhao, B.K. Metaheuristics optimization-based ensemble of deep neural networks for Mpox disease detection. Neural Networks 2023, 167, 342–359. [Google Scholar] [CrossRef]
- Aloraini, M. An effective human monkeypox classification using vision transformer. International Journal of Imaging Systems and Technology 2023. [Google Scholar] [CrossRef]
- Ren, G.Y. Monkeypox Disease Detection with Pretrained Deep Learning Models. Information Technology and Control 2023, 52, 288–296. [Google Scholar] [CrossRef]
- Ahsan, M.M.; Uddin, M.R.; Ali, M.S.; Islam, M.K.; Farjana, M.; Sakib, A.N.; Al Momin, K.; Luna, S.A. Deep transfer learning approaches for Monkeypox disease diagnosis. Expert Systems with Applications 2023, 216. [Google Scholar] [CrossRef]
- Bian, X.F.; Pan, H.W.; Zhang, K.J.; Li, P.Y.; Li, J.B.; Chen, C.L. Skin lesion image classification method based on extension theory and deep learning. Multimedia Tools and Applications 2022, 81, 16389–16409. [Google Scholar] [CrossRef]
- Radhika, V.; Chandana, B.S. MSCDNet-based multi-class classification of skin cancer using dermoscopy images. Peerj Computer Science 2023, 9. [Google Scholar] [CrossRef]
- Ghosh, A.; Jana, N.D.; Das, S.; Mallipeddi, R. Two-Phase Evolutionary Convolutional Neural Network Architecture Search for Medical Image Classification. IEEE Access 2023, 11, 115280–115305. [Google Scholar] [CrossRef]
- Rajeshwari, J.; Sughasiny, M. Modified Filter Based Feature Selection Technique for Dermatology Dataset Using Beetle Swarm Optimization. Eai Endorsed Transactions on Scalable Information Systems 2023, 10. [Google Scholar] [CrossRef]
- Mostafa, R.R.; Khedr, A.M.; Al Aghbari, Z.; Afyouni, I.; Kamel, I.; Ahmed, N. An adaptive hybrid mutated differential evolution feature selection method for low and high-dimensional medical datasets. Knowledge-Based Systems 2024, 283. [Google Scholar] [CrossRef]
- Zhou, J.C.; Wu, Z.; Jiang, Z.X.; Huang, K.; Guo, K.H.; Zhao, S. Background selection schema on deep learning-based classification of dermatological disease. Computers in Biology and Medicine 2022, 149. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Rao, A.C.S.; Jain, P.K.; Abraham, A. Multi-type skin diseases classification using OP-DNN based feature extraction approach. Multimedia Tools and Applications 2022, 81, 6451–6476. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.N.; Rejeesh, M.R.; Sundararaj, V.; Gunasundari, B. ACO-KELM: Anti Coronavirus Optimized Kernel-based Softplus Extreme Learning Machine for classification of skin cancer. Expert Systems with Applications 2023, 232. [Google Scholar] [CrossRef]
- Vidhyalakshmi, A.M.; Kanchana, M. Skin cancer classification using improved transfer learning model-based random forest classifier and golden search optimization. International Journal of Imaging Systems and Technology 2023. [Google Scholar] [CrossRef]
- Renith, G.; Senthilselvi, A. An efficient skin cancer detection and classification using Improved Adaboost Aphid-Ant Mutualism model. International Journal of Imaging Systems and Technology 2023, 33, 1957–1972. [Google Scholar] [CrossRef]
- Desale, R.P.; Patil, P.S. An efficient multi-class classification of skin cancer using optimized vision transformer. Medical & Biological Engineering & Computing 2023. [Google Scholar] [CrossRef]
- Villa-Pulgarin, J.P.; Ruales-Torres, A.A.; Arias-Garzón, D.; Bravo-Ortiz, M.A.; Arteaga-Arteaga, H.B.; Mora-Rubio, A.; Alzate-Grisales, J.A.; Mercado-Ruiz, E.; Hassaballah, M.; Orozco-Arias, S.; et al. Optimized Convolutional Neural Network Models for Skin Lesion Classification. Cmc-Computers Materials & Continua 2022, 70, 2131–2148. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Koundal, D.; Singh, K. Fusion of U-Net and CNN model for segmentation and classification of skin lesion from dermoscopy images. Expert Systems with Applications 2023, 213. [Google Scholar] [CrossRef]
- Adepu, A.K.; Sahayam, S.; Jayaraman, U.; Arramraju, R. Melanoma classification from dermatoscopy images using knowledge distillation for highly imbalanced data. Computers in Biology and Medicine 2023, 154. [Google Scholar] [CrossRef] [PubMed]
- El Saleh, R.; Chantaf, S.; Nait-ali, A. Identification of facial skin diseases from face phenotypes using FSDNet in uncontrolled environment. Machine Vision and Applications 2022, 33. [Google Scholar] [CrossRef]
- Bozkurt, F. Skin lesion classification on dermatoscopic images using effective data augmentation and pre-trained deep learning approach. Multimedia Tools and Applications 2023, 82, 18985–19003. [Google Scholar] [CrossRef]
- Pokhrel, K.; Sanin, C.; Sakib, M.K.H.; Islam, M.R.; Szczerbicki, E. Improved Skin Disease Classification with Mask R-CNN and Augmented Dataset. Cybernetics and Systems 2023. [Google Scholar] [CrossRef]
- Holmes, K.; Sharma, P.; Fernandes, S. Facial skin disease prediction using StarGAN v2 and transfer learning. Intelligent Decision Technologies-Netherlands 2023, 17, 55–66. [Google Scholar] [CrossRef]
- Rezk, E.; Eltorki, M.; El-Dakhakhni, W. Interpretable Skin Cancer Classification based on Incremental Domain Knowledge Learning. Journal of Healthcare Informatics Research 2023, 7, 59–83. [Google Scholar] [CrossRef] [PubMed]
- Hoang, V.; Vo, X.T.; Jo, K.H. Categorical Weighting Domination for Imbalanced Classification With Skin Cancer in Intelligent Healthcare Systems. IEEE Access 2023, 11, 105170–105181. [Google Scholar] [CrossRef]
- Yu, L.S.; Wang, Y.F.; Zhou, L.Y.; Wu, J.S.; Wang, Z.H. Residual neural network-assisted one-class classification algorithm for melanoma recognition with imbalanced data. Computational Intelligence 2023. [Google Scholar] [CrossRef]
- Chen, K.H.; Lei, W.X.; Zhao, S.; Zheng, W.S.; Wang, R.X. PCCT: Progressive Class-Center Triplet Loss for Imbalanced Medical Image Classification. IEEE Journal of Biomedical and Health Informatics 2023, 27, 2026–2036. [Google Scholar] [CrossRef] [PubMed]
- Okuboyejo, D.A.; Olugbara, O.O. Classification of Skin Lesions Using Weighted Majority Voting Ensemble Deep Learning. Algorithms 2022, 15. [Google Scholar] [CrossRef]
- Vidhyalakshmi, A.M.; Kanchana, M. Classification of skin disease using a novel hybrid flash butterfly optimization from dermoscopic images. Neural Computing & Applications 2023. [Google Scholar] [CrossRef]
- Huang, Z.Z.; Wu, J.W.; Wang, T.; Li, Z.Y.; Ioannou, A. Class-Specific Distribution Alignment for semi-supervised medical image classification. Computers in Biology and Medicine 2023, 164. [Google Scholar] [CrossRef]
- Zhuang, J.X.; Cai, J.B.; Zhang, J.G.; Zheng, W.S.; Wang, R.X. Class attention to regions of lesion for imbalanced medical image recognition. Neurocomputing 2023, 555. [Google Scholar] [CrossRef]
- Yao, P.; Shen, S.W.; Xu, M.J.; Liu, P.; Zhang, F.; Xing, J.Y.; Shao, P.F.; Kaffenberger, B.; Xu, R.X. Single Model Deep Learning on Imbalanced Small Datasets for Skin Lesion Classification. IEEE Transactions on Medical Imaging 2022, 41, 1242–1254. [Google Scholar] [CrossRef]
- Alshawi, S.A.; Musawi, G. Skin Cancer Image Detection and Classification by CNN based Ensemble Learning. International Journal of Advanced Computer Science and Applications 2023, 14, 710–717. [Google Scholar] [CrossRef]
- Yue, G.H.; Wei, P.S.; Zhou, T.W.; Jiang, Q.P.; Yan, W.Q.; Wang, T.F. Toward Multicenter Skin Lesion Classification Using Deep Neural Network With Adaptively Weighted Balance Loss. IEEE Transactions on Medical Imaging 2023, 42, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Omeroglu, A.N.; Mohammed, H.M.A.; Oral, E.A.; Aydin, S. A novel soft attention-based multi-modal deep learning framework for multi-label skin lesion classification. Engineering Applications of Artificial Intelligence 2023, 120. [Google Scholar] [CrossRef]
- Zhang, R.Z.; Wang, L.J.; Cheng, S.L.; Song, S.J. MLP-based classification of COVID-19 and skin diseases. Expert Systems with Applications 2023, 228. [Google Scholar] [CrossRef]
- Desale, R.P.; Patil, P.S. An automated hybrid attention based deep convolutional capsule with weighted autoencoder approach for skin cancer classification. Imaging Science Journal 2023. [Google Scholar] [CrossRef]
- Bao, Q.W.; Han, H.; Huang, L.; Muzahid, A.A.M. A Convolutional Neural Network Based on Soft Attention Mechanism and Multi-Scale Fusion for Skin Cancer Classification. International Journal of Pattern Recognition and Artificial Intelligence 2023, 37. [Google Scholar] [CrossRef]
- To, H.D.; Nguyen, H.G.; Le, H.T.T.; Le, H.M.; Quan, T.T. MetaAttention model: a new approach for skin lesion diagnosis using AB features and attention mechanism. Biomedical Physics & Engineering Express 2023, 9. [Google Scholar] [CrossRef]
- Surati, S.; Trivedi, H.; Shrimali, B.; Bhatt, C.; Travieso-González, C.M. An Enhanced Diagnosis of Monkeypox Disease Using Deep Learning and a Novel Attention Model Senet on Diversified Dataset. Multimodal Technologies and Interaction 2023, 7. [Google Scholar] [CrossRef]
- Damineni, D.H.; Sekharamantry, P.K.; Badugu, R. An Adaptable Model for Medical Image Classification Using the Streamlined Attention Mechanism. International Journal of Online and Biomedical Engineering 2023, 19, 93–110. [Google Scholar] [CrossRef]
- Ashwath, V.A.; Sikha, O.K.; Benitez, R. TS-CNN: A Three-Tier Self-Interpretable CNN for Multi-Region Medical Image Classification. IEEE Access 2023, 11, 78402–78418. [Google Scholar] [CrossRef]
- Ren, Y.G.; Xu, W.Q.; Mao, Y.X.; Wu, Y.C.; Fu, B.; Thanh, D.N.H. Few-shot learning for dermatological conditions with Lesion Area Aware Swin Transformer. International Journal of Imaging Systems and Technology 2023, 33, 1549–1560. [Google Scholar] [CrossRef]
- Zhang, D.Z.; Li, A.L.; Wu, W.D.; Yu, L.; Kang, X.J.; Huo, X.Z. CR-Conformer: a fusion network for clinical skin lesion classification. Medical & Biological Engineering & Computing 2023. [Google Scholar] [CrossRef]
- Shaik, T.; Tao, X.H.; Higgins, N.; Li, L.; Gururajan, R.; Zhou, X.J.; Acharya, U.R. Remote patient monitoring using artificial intelligence: Current state, applications, and challenges. Wiley Interdisciplinary Reviews-Data Mining and Knowledge Discovery 2023, 13. [Google Scholar] [CrossRef]
- Oztel, I.; Oztel, G.Y.; Sahin, V.H. Deep Learning-Based Skin Diseases Classification using Smartphones. Advanced Intelligent Systems 2023. [Google Scholar] [CrossRef]
- Yang, Y.G.; Xie, F.Y.; Zhang, H.P.; Wang, J.C.; Liu, J.; Zhang, Y.L.; Ding, H.D. Skin lesion classification based on two-modal images using a multi-scale fully-shared fusion network. Computer Methods and Programs in Biomedicine 2023, 229. [Google Scholar] [CrossRef]
- Manzoor, K.; Majeed, F.; Siddique, A.; Meraj, T.; Rauf, H.T.; El-Meligy, M.A.; Sharaf, M.; Abd Elgawad, A.E. A Lightweight Approach for Skin Lesion Detection Through Optimal Features Fusion. Cmc-Computers Materials & Continua 2022, 70, 1617–1630. [Google Scholar] [CrossRef]
- Vivekananda, G.N.; Almufti, S.M.; Suresh, C.; Samsudeen, S.; Devarajan, M.V.; Srikanth, R.; Jayashree, S. Retracing-efficient IoT model for identifying the skin-related tags using automatic lumen detection. Intelligent Data Analysis 2023, 27, S161–S180. [Google Scholar] [CrossRef]
- Shahin, M.; Chen, F.F.; Hosseinzadeh, A.; Koodiani, H.K.; Shahin, A.; Nafi, O.A. A smartphone-based application for an early skin disease prognosis: Towards a lean healthcare system via computer-based vision. Advanced Engineering Informatics 2023, 57. [Google Scholar] [CrossRef]
- Kumar, K.A.; Satheesha, T.Y.; Salvador, B.B.L.; Mithileysh, S.; Ahmed, S.T. Augmented Intelligence enabled Deep Neural Networking (AuDNN) framework for skin cancer classification and prediction using multi-dimensional datasets on industrial IoT standards. Microprocessors and Microsystems 2023, 97. [Google Scholar] [CrossRef]
- Shi, Y.H.; Li, X.; Chen, S.G. Skin Lesion Intelligent Diagnosis in Edge Computing Networks: An FCL Approach. Acm Transactions on Intelligent Systems and Technology 2023, 14. [Google Scholar] [CrossRef]
- Gupta, A.; Bhagat, M.; Jain, V. Blockchain-enabled healthcare monitoring system for early Monkeypox detection. Journal of Supercomputing 2023, 79, 15675–15699. [Google Scholar] [CrossRef] [PubMed]
- Hossen, M.N.; Panneerselvam, V.; Koundal, D.; Ahmed, K.; Bui, F.M.; Ibrahim, S.M. Federated Machine Learning for Detection of Skin Diseases and Enhancement of Internet of Medical Things (IoMT) Security. IEEE Journal of Biomedical and Health Informatics 2023, 27, 835–841. [Google Scholar] [CrossRef] [PubMed]
- Hendrycks, D.; Mu, N.; Cubuk, E.D.; Zoph, B.; Gilmer, J.; Lakshminarayanan, B. AugMix: A Simple Data Processing Method to Improve Robustness and Uncertainty. 2019, arXiv:1912.02781. [CrossRef]
- Zhang, H.; Cisse, M.; Yann; Lopez-Paz, D. mixup: Beyond Empirical Risk Minimization. arXiv pre-print server 2018, doi:arxiv:1710.09412. [CrossRef]
- Yun, S.; Han, D.; Chun, S.; Oh, S.J.; Yoo, Y.; Choe, J. CutMix: Regularization Strategy to Train Strong Classifiers With Localizable Features. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV); 2019; pp. 6022–6031. [Google Scholar]
- Rotemberg, V.; Kurtansky, N.; Betz-Stablein, B.; Caffery, L.; Chousakos, E.; Codella, N.; Combalia, M.; Dusza, S.; Guitera, P.; Gutman, D.; et al. A patient-centric dataset of images and metadata for identifying melanomas using clinical context. Scientific Data 2021, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Codella, N.C.F.; Gutman, D.; Emre Celebi, M.; Helba, B.; Marchetti, M.A.; Dusza, S.W.; Kalloo, A.; Liopyris, K.; Mishra, N.; Kittler, H.; Halpern, A. Skin Lesion Analysis Toward Melanoma Detection: A Challenge at the 2017 International Symposium on Biomedical Imaging (ISBI), Hosted by the International Skin Imaging Collaboration (ISIC). 2017, arXiv:1710.05006. 0500. [Google Scholar] [CrossRef]
- Tschandl, P.; Rosendahl, C.; Kittler, H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific Data 2018, 5, 180161. [Google Scholar] [CrossRef]
- Combalia, M.; Codella, N.C.F.; Rotemberg, V.; Helba, B.; Vilaplana, V.; Reiter, O.; Carrera, C.; Barreiro, A.; Halpern, A.C.; Puig, S.; Malvehy, J. BCN20000: Dermoscopic Lesions in the Wild. 2019, arXiv:1908.02288. [CrossRef]
- Singh, P.; Sizikova, E.; Cirrone, J. CASS: Cross Architectural Self-Supervision for Medical Image Analysis. 2022, arXiv:2206.04170. [CrossRef]


| Datasets | Desc. | No. class | Weighting per class | No. train | No. val | Total |
|---|---|---|---|---|---|---|
| Skin Cancer ISIC 2019 & 2020 Malignant or Benign [128] | The dataset was compiled using images from the ISIC 2019 and ISIC 2020 collections, which were sourced from the International Society for Digital Imaging of the Skin. It comprises a total of 11,396 images, with 5,096 categorized as benign tumors and 6,300 categorized as malignant tumors. | 2 | {0: 0.9044, 1: 1.1181} |
9,117 | 2,279 | 11,396 images |
| Melanoma Detection Dataset [129] | The objective of this dataset is to facilitate the research and creation of automated systems for the diagnosis of melanoma, which is the deadliest form of skin cancer. The collection of labeled images includes 2,750 images in total, of which 521 are identified as melanoma, 1,843 as nevus, and 386 as seborrheic keratosis. | 3 | {0: 1.7594, 1: 0.4973, 2: 2.3747} |
2200 | 550 | 2,750 |
|
ISIC 2019 Skin Lesion images for classification [129,130,131] |
The dataset encompasses the training data for the ISIC 2019 competition, which also incorporates data from the 2018 and 2017 challenges. The ISIC 2019 dataset offers a total of 25,331 dermoscopic images for classification across eight distinct diagnostic categories: - Actinic keratosis (AK): 867 images. - Basal cell carcinoma (BCC): 3,323 images. - Benign keratosis group (BKL): 2,624 images. - Dermatofibroma (DF).239 images. - Melanoma (MEL): 4,522 images. - Melanocytic nevus (NV): 12,875 images. - Squamous cell carcinoma (SCC): 628 images. - Vascular lesion (VASC): 253 images. |
8 | {0: 3.6521, 1: 0.9528, 2: 1.2066, 3: 13.2484, 4: 0.7002, 5: 0.2459, 6: 5.0419, 7: 12.515} |
20265 | 5066 | 25,331 images |
| Skin Lesions Model | Aug. Method | Traditional classifier (Supervised Learning) | SkinLiTE (Supervised Contrastive Learning) | |||||||
| Cross-entropy | Accuracy | AUC | F1 score |
Contrastive loss | Cross-entropy | Accuracy | AUC | F1 score |
||
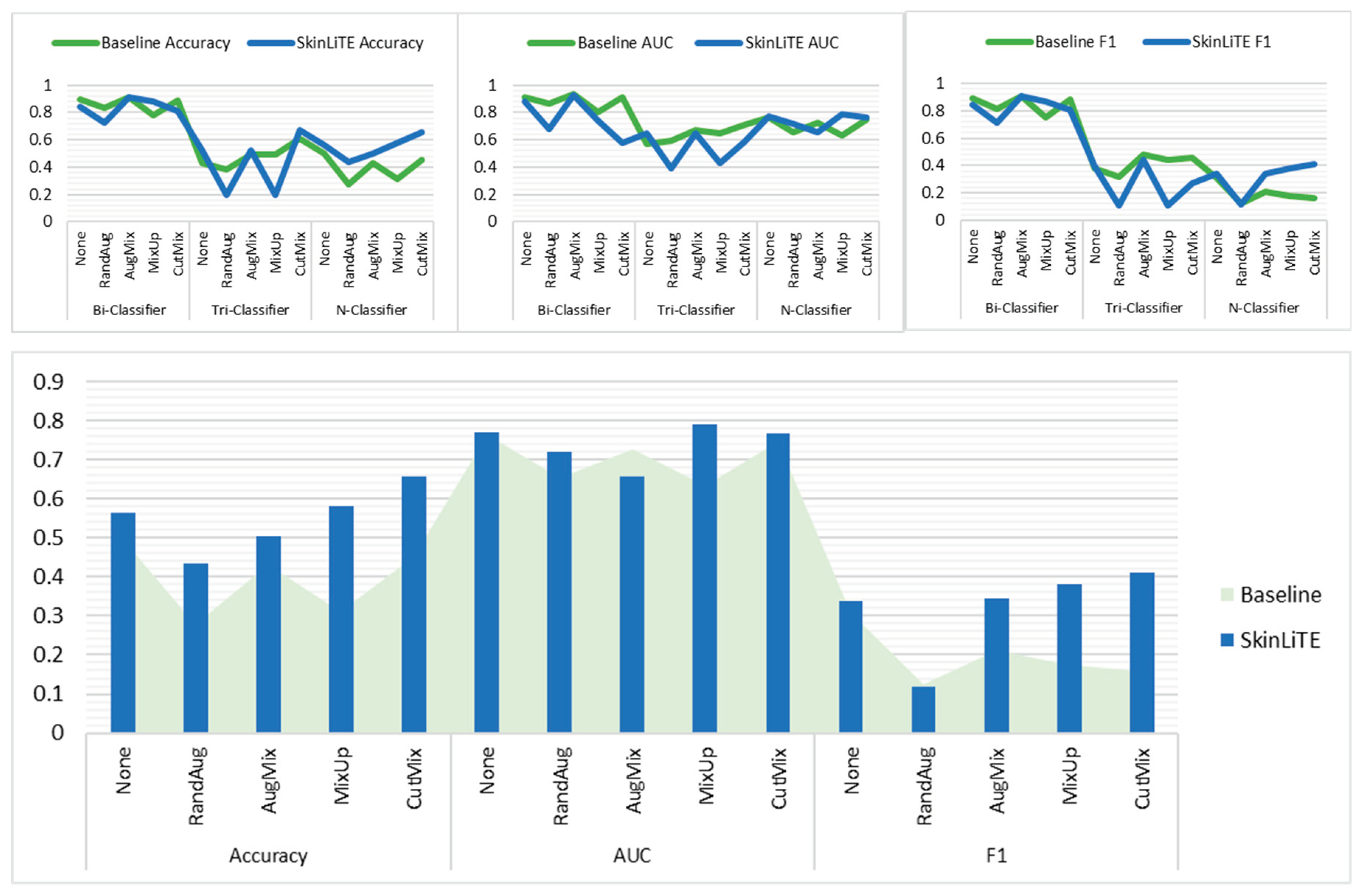
| Bi-Classifier | None | 0.3430 | 0.8956 | 0.9106 | 0.8926 | 2.8242 | 0.4062 | 0.8416 | 0.8808 | 0.8415 |
| RandAug | 0.3805 | 0.8311 | 0.8624 | 0.8164 | 3.2218 | 0.5130 | 0.7231 | 0.6770 | 0.7171 | |
| AugMix | 0.2305 | 0.9096 | 0.9351 | 0.9064 | 3.0800 | 0.4035 | 0.9087 | 0.9264 | 0.9067 | |
| MixUp | 0.5453 | 0.7793 | 0.8057 | 0.7508 | 2.9713 | 0.3774 | 0.8771 | 0.7425 | 0.8706 | |
| CutMix | 0.2972 | 0.8881 | 0.9127 | 0.8830 | 3.2412 | 0.4894 | 0.8091 | 0.5810 | 0.8089 | |
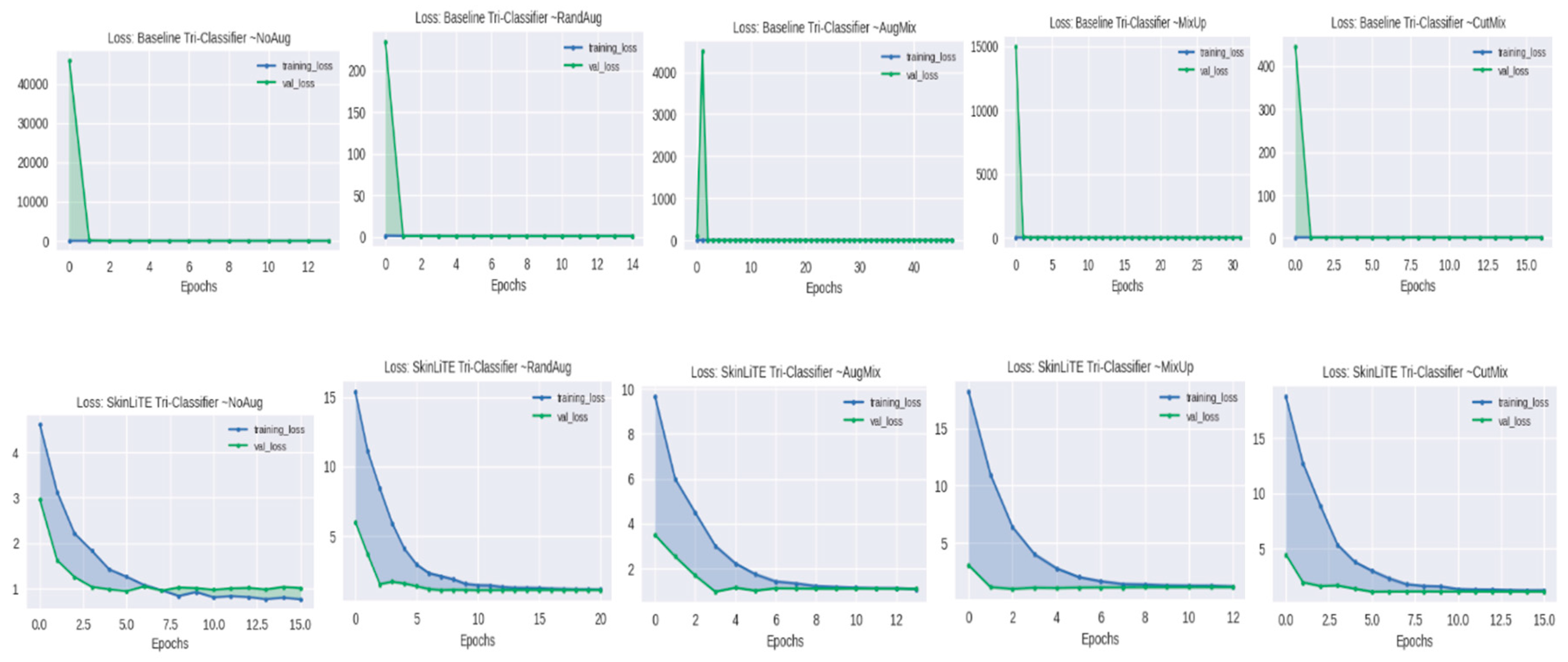
| Tri-Classifier | None | 1.0733 | 0.4291 | 0.5707 | 0.3786 | 2.8309 | 1.0139 | 0.5200 | 0.6497 | 0.3965 |
| RandAug | 1.0937 | 0.3818 | 0.5898 | 0.3210 | 3.4490 | 1.1055 | 0.1945 | 0.3957 | 0.1086 | |
| AugMix | 0.9353 | 0.4891 | 0.6743 | 0.4800 | 3.4023 | 0.9751 | 0.5273 | 0.6469 | 0.4446 | |
| MixUp | 0.9863 | 0.4891 | 0.6467 | 0.4385 | 3.3382 | 1.1188 | 0.1945 | 0.4290 | 0.1086 | |
| CutMix | 0.9240 | 0.6109 | 0.7104 | 0.4550 | 3.4335 | 1.0548 | 0.6691 | 0.5830 | 0.2672 | |
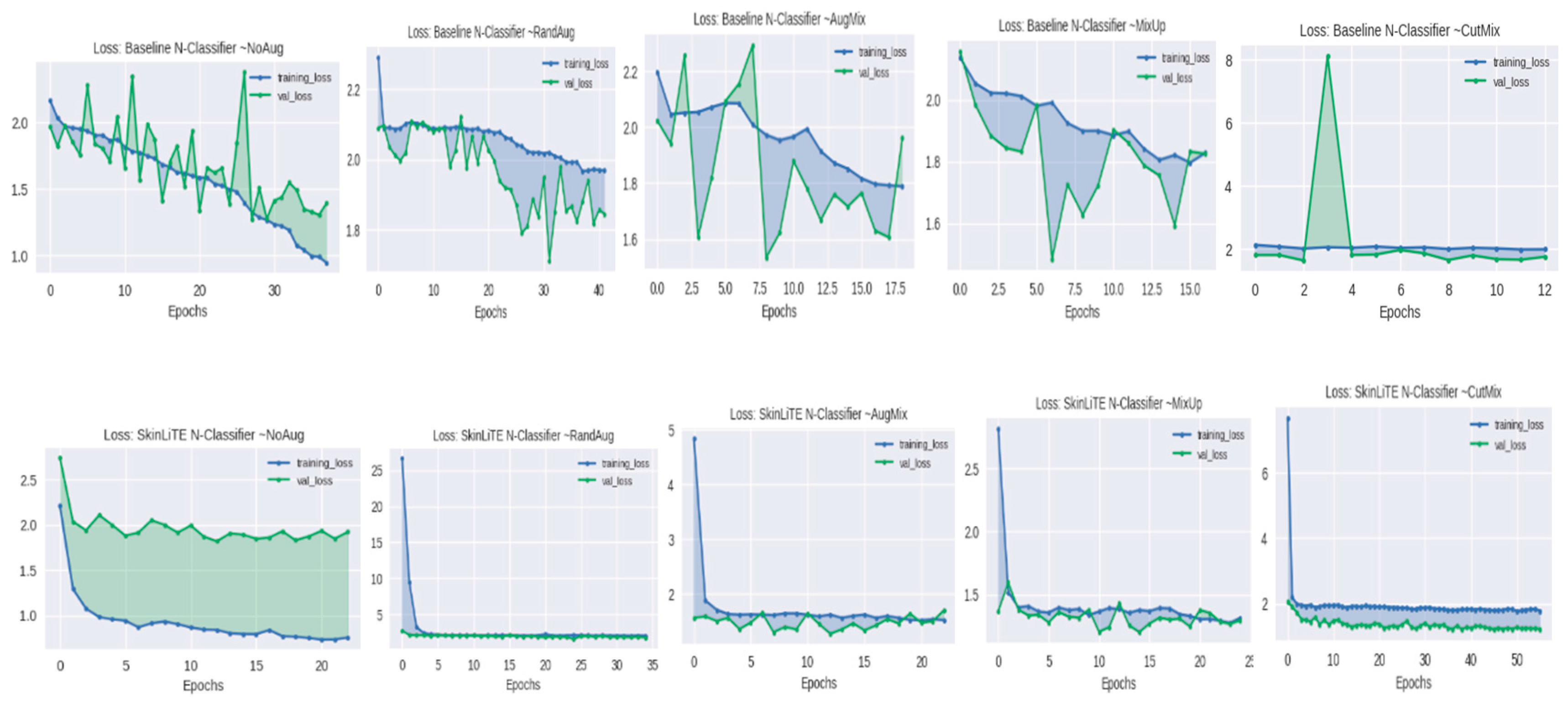
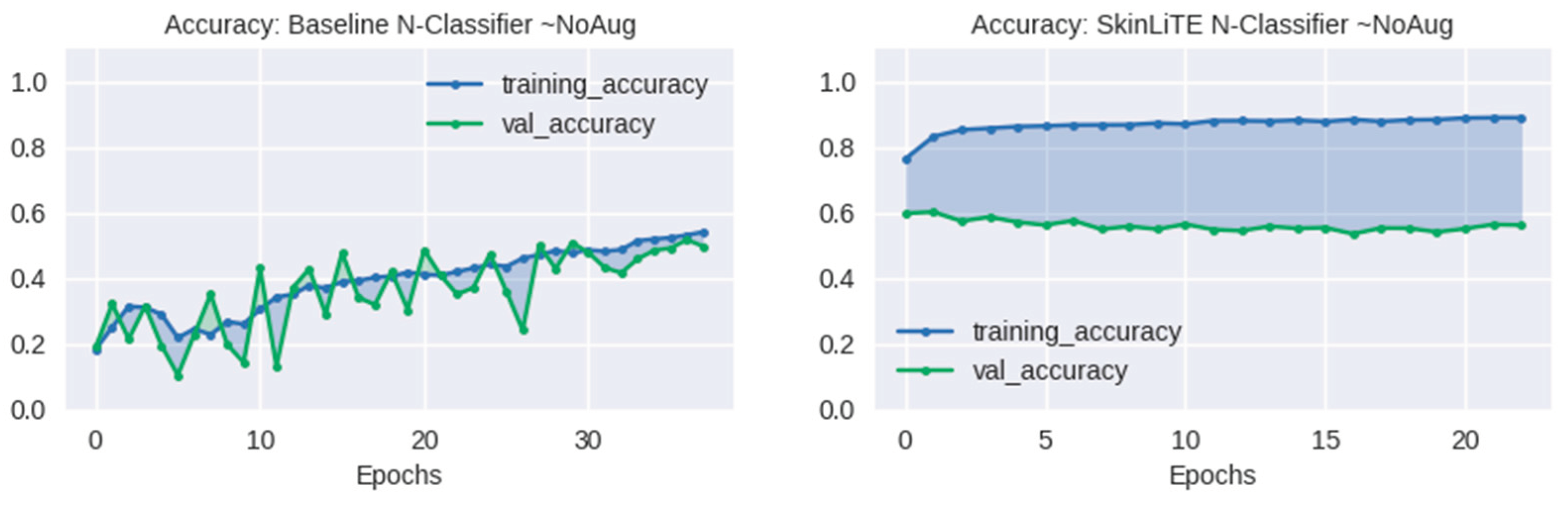
| N-Classifier | None | 1.3930 | 0.4974 | 0.7638 | 0.3063 | 2.1985 | 1.9215 | 0.5626 | 0.7693 | 0.3382 |
| RandAug | 1.8438 | 0.2777 | 0.6546 | 0.1265 | 3.2310 | 1.7804 | 0.4345 | 0.7197 | 0.1181 | |
| AugMix | 1.6061 | 0.4311 | 0.7260 | 0.2101 | 2.8542 | 1.7088 | 0.5034 | 0.6576 | 0.3443 | |
| MixUp | 1.8259 | 0.3121 | 0.6326 | 0.1754 | 2.5672 | 1.2964 | 0.5805 | 0.7900 | 0.3810 | |
| CutMix | 1.7430 | 0.4501 | 0.7477 | 0.1588 | 3.1836 | 1.2298 | 0.6561 | 0.7658 | 0.4126 | |
| Model Name | Manuscript | Use External Data | Top-1% |
| Ensample-24-mcm10 (https://challenge.isic-archive.com/leaderboards/live/) | No | Yes | 0.670 |
| MinJie (https://challenge.isic-archive.com/leaderboards/live/) | No | Yes | 0.662 |
| SkinLiTE (proposed) | Yes | No | 0.656 |
| CASS: Cross Architectural Self-Supervision for Medical Image Analysis [132] | Yes | Yes | 0.652 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





