Submitted:
08 July 2025
Posted:
25 July 2025
You are already at the latest version
Abstract
Keywords:
1. Introduction
- Early Detection Improvement: To develop an automated system that facilitates the early identification of skin cancer, enabling prompt intervention and thereby increasing patient survival rates.
- Accessibility Enhancement: Create a scalable diagnostic tool deployable in resource-limited settings with minimal dermatologist access.
- Clinical Decision Support:To assist medical professionals by improving classification accuracy for diagnostically challenging lesions (especially melanoma) while reducing inter-observer variability.
- System Efficiency:Optimize model performance for integration into mobile health apps and clinical workflows without compromising computational efficiency.
- Hybrid Training Strategy:Introduces a progressive fine-tuning method that combines uncertainty-aware inference, full-network optimization, and transfer learning (TTA + Monte Carlo Dropout). It achieves 92.29% accuracy on HAM10000 dermoscopic image dataset, which is 15% better than baseline frozen-layer transfer learning.
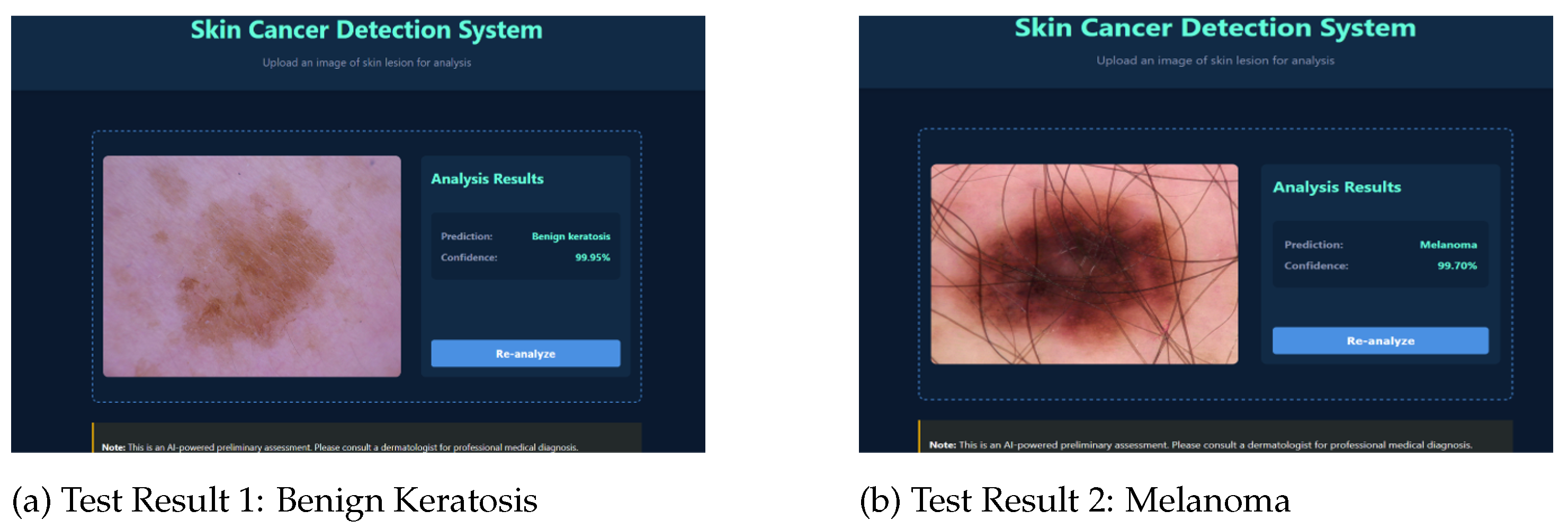
- Web-Based Diagnostic Interface: Web-based platform allows non-experts to upload images and receive instant predictions with confidence scores, addressing accessibility gaps.
- Clinical-Grade Data Handling: Strategic oversampling (akiec/bcc/df/vasc → 1,000 images each) and downsampling (nv → 1,300) improves rare-class recall by 15–20% while maintaining 92.29% overall accuracy.
- Lightweight Architecture: EfficientNet-B0 model implementation ensures high performance suitable for real-time clinical use and edge devices.
2. Materials and Methods


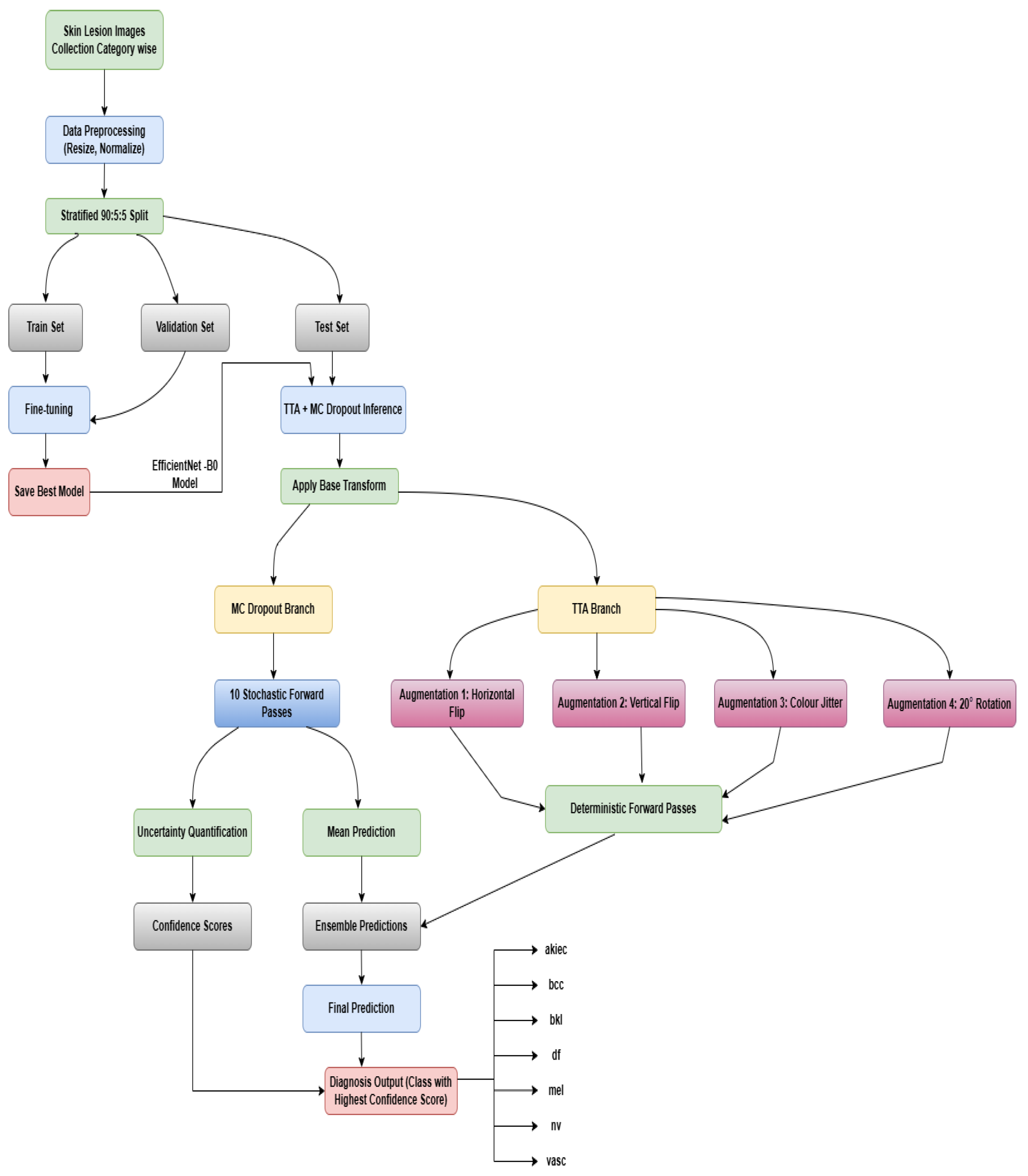
2.1. Data Preparation and Initialization
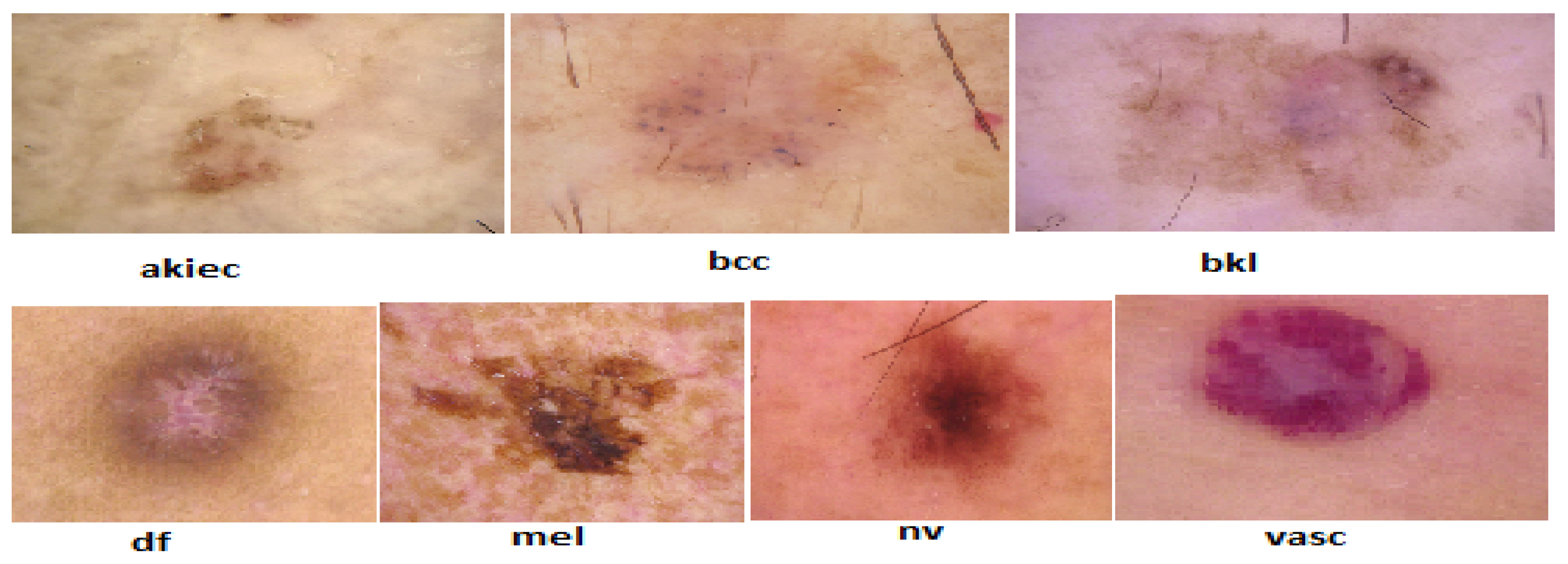
2.1.1. Data Collection and Description
- Actinic keratoses and intraepithelial carcinoma (akiec): Precancerous or early-stage malignant lesions with 327 images.
- Basal cell carcinoma (bcc): A common form of skin cancer represented by 327 images.
- Benign keratosis-like lesions (bkl): Includes benign growths like seborrheic keratoses with 1,099 images.
- Dermatofibroma (df): A rare, benign fibrous skin tumor with 115 images.
- Melanoma (mel): A highly dangerous skin cancer with 1,113 representative images.
- Melanocytic nevi (nv): Benign moles dominating the dataset with 6,705 images.
- Vascular lesions (vasc): Includes blood vessel-related lesions like angiomas, with 142 images.
2.1.2. Data Preprocessing and Balancing
2.1.3. Dataset Splitting
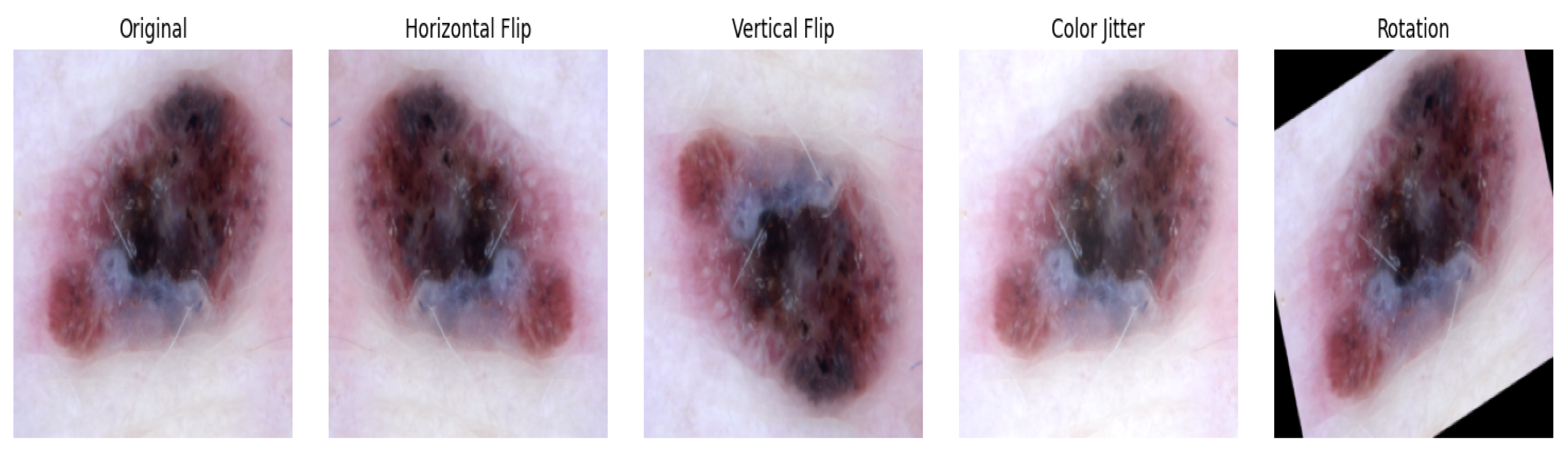
2.1.4. Image Transformation and Loading
2.2. Model Set Up and Implementation
2.2.1. Model Architecture and Transfer Learning
2.2.2. Model Training
2.2.3. Fine-Tuning
2.3. Model Evaluation
3. Results and Discussion
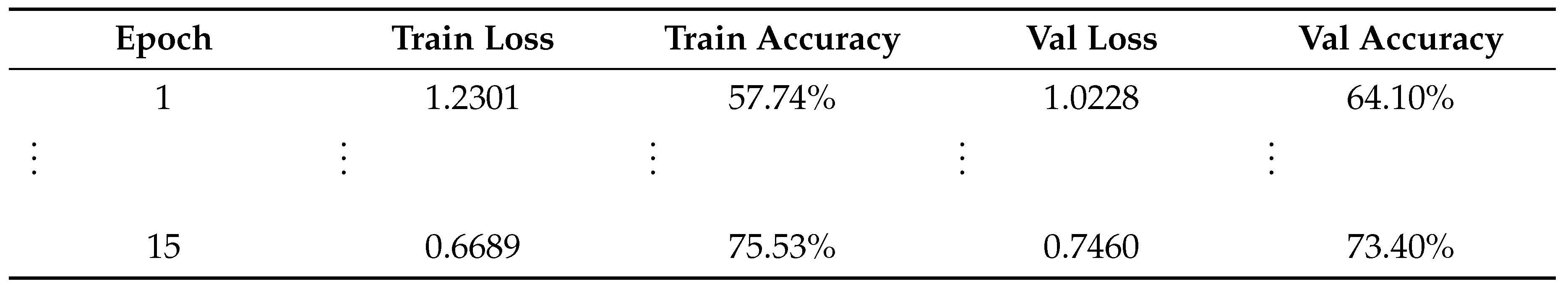
3.1. Training and Validation Performance
| Train:Val:Test Ratio | EfficientNet-B0 | Res Net50 | Dense Net121 | Mobile Net | InceptionV3 |
|---|---|---|---|---|---|
| 60:20:20 | 74.18% | 70.79% | 74.38% | 72.85% | 65.67% |
| 70:15:15 | 74.80% | 70.19% | 74.17% | 73.20% | 65.48% |
| 80:10:10 | 74.60% | 74.20% | 74.20% | 73.80% | 67.42% |
| 90:5:5 | 77.39% | 73.94% | 73.94% | 73.14% | 67.29% |
3.2. Fine-Tuning Performance

3.3. Test Set Evaluation
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| DL | Deep Learning |
| ML | Machine Learning |
| CNN | Convolutional Neural Network |
| HAM | Human Against Machine |
| TTA | Test Time Augmentation |
| AKIEC | Actinic keratoses and intraepithelial carcinoma |
| BCC | Basal Cell Carcinoma, |
| DF | Dermatofibroma |
| VASC | Vascular Lesions |
| BCAT | Brain Computer Aptitude Test |
| BKL | Benign keratosis |
| MEL | Melanoma |
References
- Ashfaq, N., Suhail, Z., Khalid, A., et al. 2025. SkinSight: advancing deep learning for skin cancer diagnosis and classification. Discovery Computing 28: 63. [CrossRef]
- Kavitha, C., Priyanka, S., Praveen Kumar, M., Kusuma, V. 2024. Skin Cancer Detection and Classification using Deep Learning Techniques. Procedia Computer Science 235: 2793–2802. [CrossRef]
- Naeem, A., Farooq, M. S., Khelifi, A., & Abid, A. 2020. Malignant Melanoma Classification Using Deep Learning: Datasets, Performance Measurements, Challenges and Opportunities. IEEE Access 8: 110575–110597. [CrossRef]
- Balaha, H. M., & Hassan, A. E. S. 2023. Skin cancer diagnosis based on deep transfer learning and sparrow search algorithm. Neural Computing & Applications 35: 815–853. [CrossRef]
- Alotaibi, A., & AlSaeed, D. 2025. Skin Cancer Detection Using Transfer Learning and Deep Attention Mechanisms. Diagnostics 15: 99. [CrossRef]
- Djaroudib, K., Lorenz, P., Belkacem Bouzida, R., & Merzougui, H. 2024. Skin Cancer Diagnosis Using VGG16 and Transfer Learning: Analyzing the Effects of Data Quality over Quantity on Model Efficiency. Applied Sciences 14: 7447. [CrossRef]
- Nazari, S., & Garcia, R. 2023. Automatic Skin Cancer Detection Using Clinical Images: A Comprehensive Review. Life 13(11): 2123. [CrossRef]
- Naqvi, M., Gilani, S. Q., Syed, T., Marques, O., & Kim, H.-C. 2023. Skin Cancer Detection Using Deep Learning—A Review. Diagnostics 13: 1911. [CrossRef]
- Naseri, H., & Safaei, A. A. 2025. Diagnosis and prognosis of melanoma from dermoscopy images using machine learning and deep learning: a systematic literature review. BMC Cancer 25: 75. [CrossRef]
- Magalhaes, C., Mendes, J., & Vardasca, R. 2024. Systematic Review of Deep Learning Techniques in Skin Cancer Detection. BioMedInformatics 4: 2251–2270. [CrossRef]
- Imran, A., Nasir, A., Bilal, M., Sun, G., Alzahrani, A., & Almuhaimeed, A. 2022. Skin Cancer Detection Using Combined Decision of Deep Learners. IEEE Access 10: 118198–118212. [CrossRef]
- Moturi, D., Surapaneni, R. K., & Avanigadda, V. S. G. 2024. Developing an efficient method for melanoma detection using CNN techniques. Journal of the Egyptian National Cancer Institute 36: 6. [CrossRef]
- Kreouzi, M., Theodorakis, N., Feretzakis, G., Paxinou, E., Sakagianni, A., Kalles, D., Anastasiou, A., Verykios, V. S., & Nikolaou, M. 2025. Deep Learning for Melanoma Detection: A Deep Learning Approach to Differentiating Malignant Melanoma from Benign Melanocytic Nevi. Cancers 17: 28. [CrossRef]
- Tahir, M., Naeem, A., Malik, H., Tanveer, J., Naqvi, R. A., & Lee, S.-W. 2023. DSCC_Net: Multi-Classification Deep Learning Models for Diagnosing of Skin Cancer Using Dermoscopic Images. Cancers 15: 2179. [CrossRef]
- Naeem, A., Anees, T., Khalil, M., Zahra, K., Naqvi, R. A., & Lee, S.-W. 2024. SNC_Net: Skin Cancer Detection by Integrating Handcrafted and Deep Learning-Based Features Using Dermoscopy Images. Mathematics 12: 1030. [CrossRef]
- Zia Ur Rehman, M., Ahmed, F., Alsuhibany, S. A., Jamal, S. S., Zulfiqar Ali, M., & Ahmad, J. 2022. Classification of Skin Cancer Lesions Using Explainable Deep Learning. Sensors 22: 6915. [CrossRef]
- Karki, R., G C, S., Rezazadeh, J., & Khan, A. 2025. Deep Learning for Early Skin Cancer Detection: Combining Segmentation, Augmentation, and Transfer Learning. Big Data Cogn. Comput. 9: 97. [CrossRef]
- Gouda, W., Sama, N. U., Al-Waakid, G., Humayun, M., & Jhanjhi, N. Z. 2022. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 10: 1183. [CrossRef]
- Natha, P., & Rajeswari, P. R. 2023. Skin Cancer Detection using Machine Learning Classification Models. International Journal of Intelligent Systems and Applications in Engineering 12(6s): 139–145. https://ijisae.org/index.php/IJISAE/article/view/3966.
- Das, S., Kumar, V., & Cicceri, G. 2024. Chatbot Enable Brain Cancer Prediction Using Convolutional Neural Network for Smart Healthcare. In Healthcare-Driven Intelligent Computing Paradigms to Secure Futuristic Smart Cities (pp. 268–279). Chapman and Hall/CRC.
- Ashafuddula, N. I. M., & Islam, R. 2023. Melanoma skin cancer and nevus mole classification using intensity value estimation with convolutional neural network. Computer Science 24(3). [CrossRef]
- Rashad, N. M., Abdelnapi, N. M., Seddik, A. F., et al. 2025. Automating skin cancer screening: a deep learning. J. Eng. Appl. Sci. 72: 6. [CrossRef]

| Class | Precision | Recall | F1-score | Support |
|---|---|---|---|---|
| akiec | 1.00 | 0.96 | 0.98 | 50 |
| bcc | 0.92 | 0.98 | 0.95 | 50 |
| bkl | 0.92 | 0.85 | 0.89 | 55 |
| df | 1.00 | 1.00 | 1.00 | 50 |
| mel | 0.78 | 0.77 | 0.77 | 56 |
| nv | 0.87 | 0.92 | 0.90 | 65 |
| vasc | 1.00 | 1.00 | 1.00 | 50 |
| Accuracy | 0.92 | 376 | ||
| Macro avg | 0.93 | 0.93 | 0.93 | 376 |
| Weighted avg | 0.92 | 0.92 | 0.92 | 376 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).