Submitted:
27 July 2024
Posted:
30 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
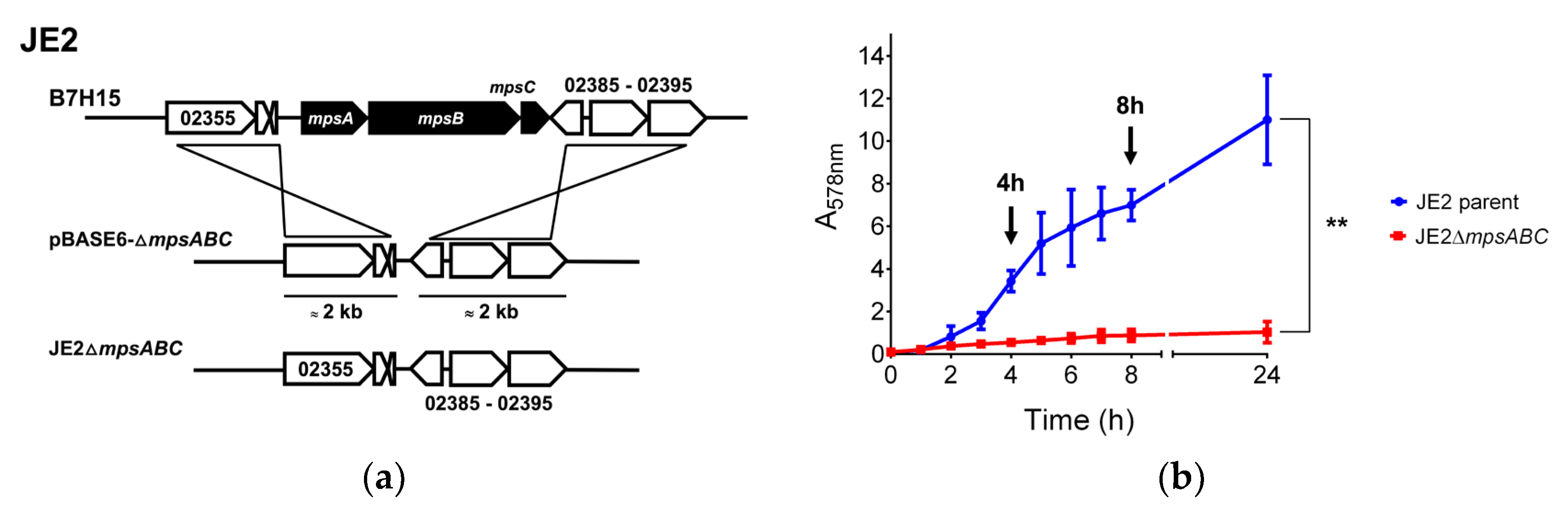
2.1. The mpsABC Deletion Mutant Has a Thicker Cell Wall
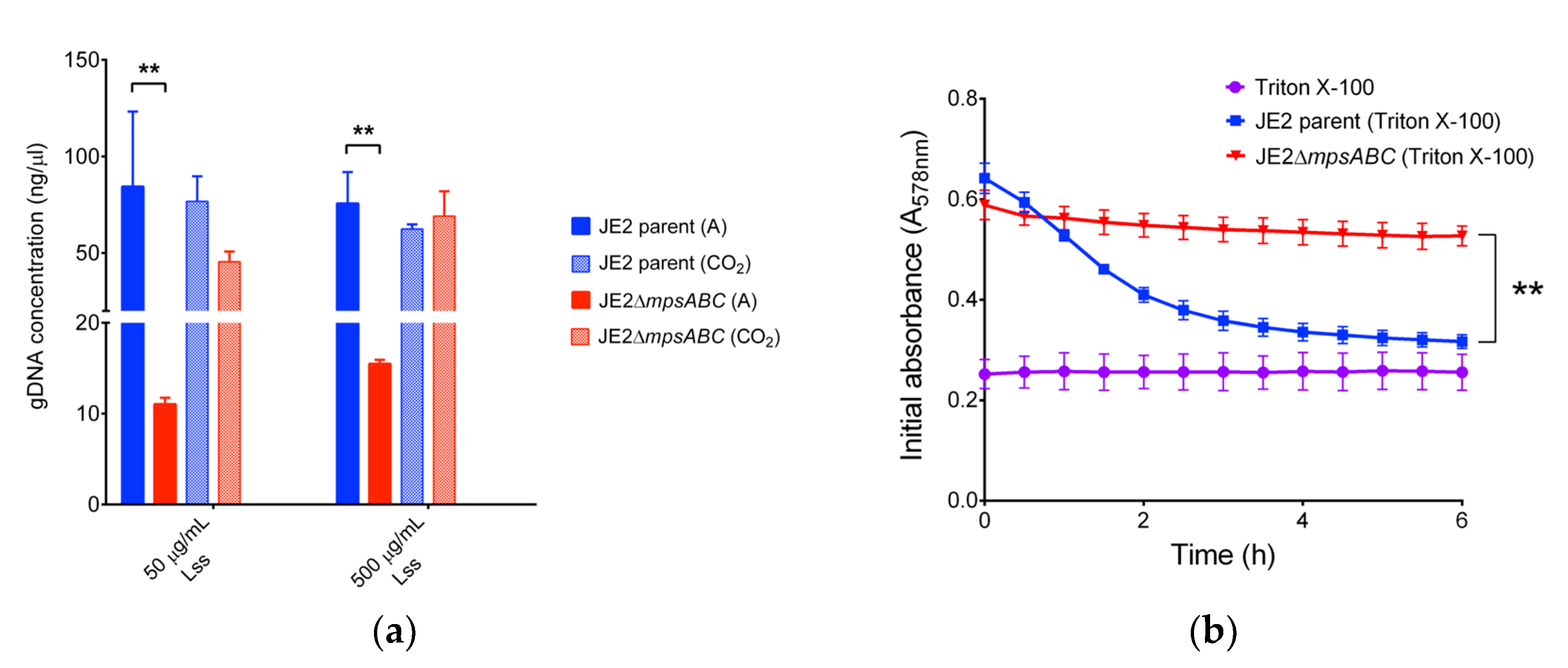
2.2. The ΔmpsABC Mutant Is Largely Inert to Cell Lysis Induced by Lysostaphin and Triton X-100
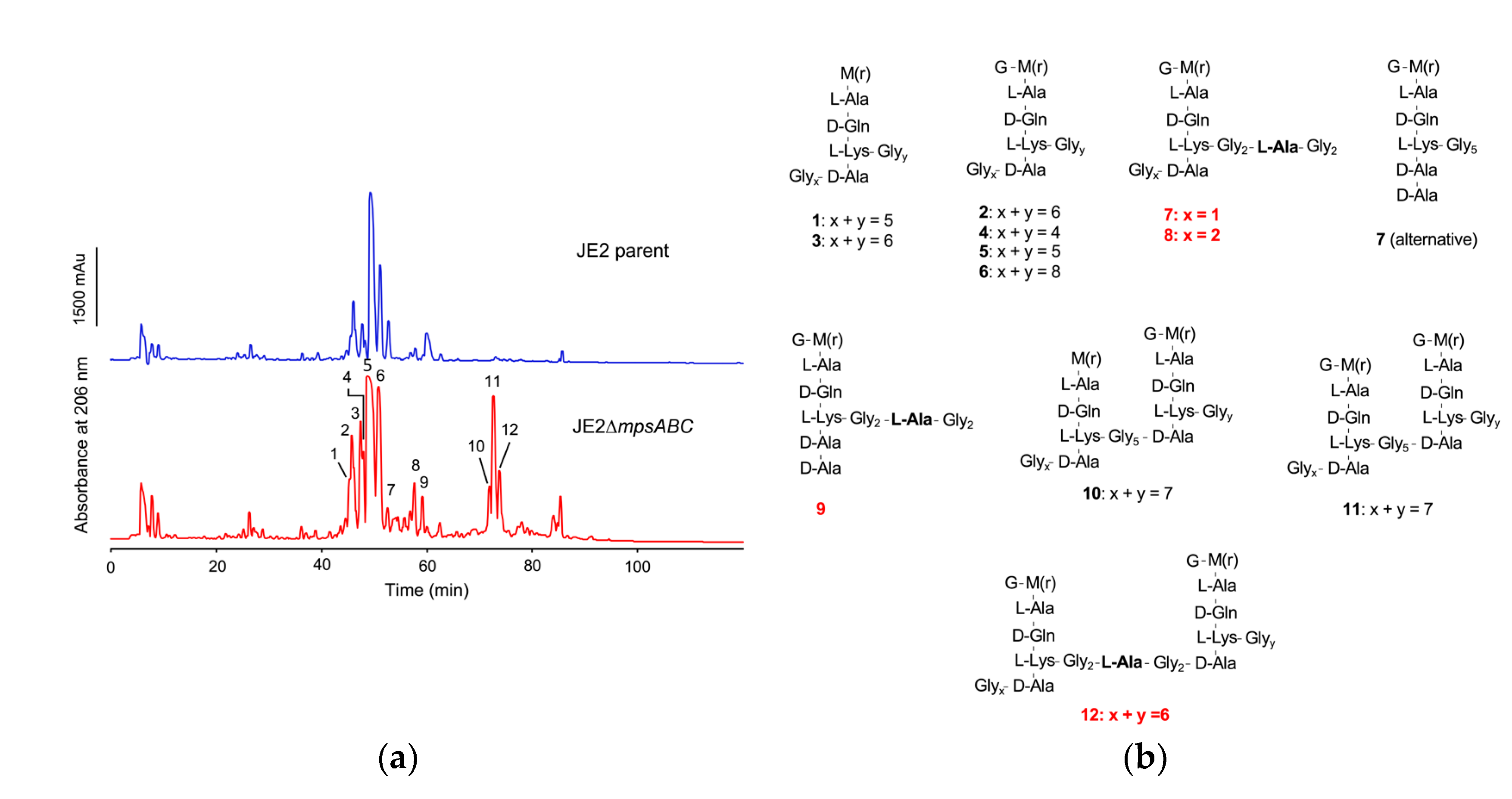
2.3. The Peptidoglycan (PG) of the Mutant Contains Alanine in the Gly5-Bridge
2.4. Comparative Transcriptome Analysis Revealed Differentially Regulated Genes in ΔmpsABC under Ambient Air Condition
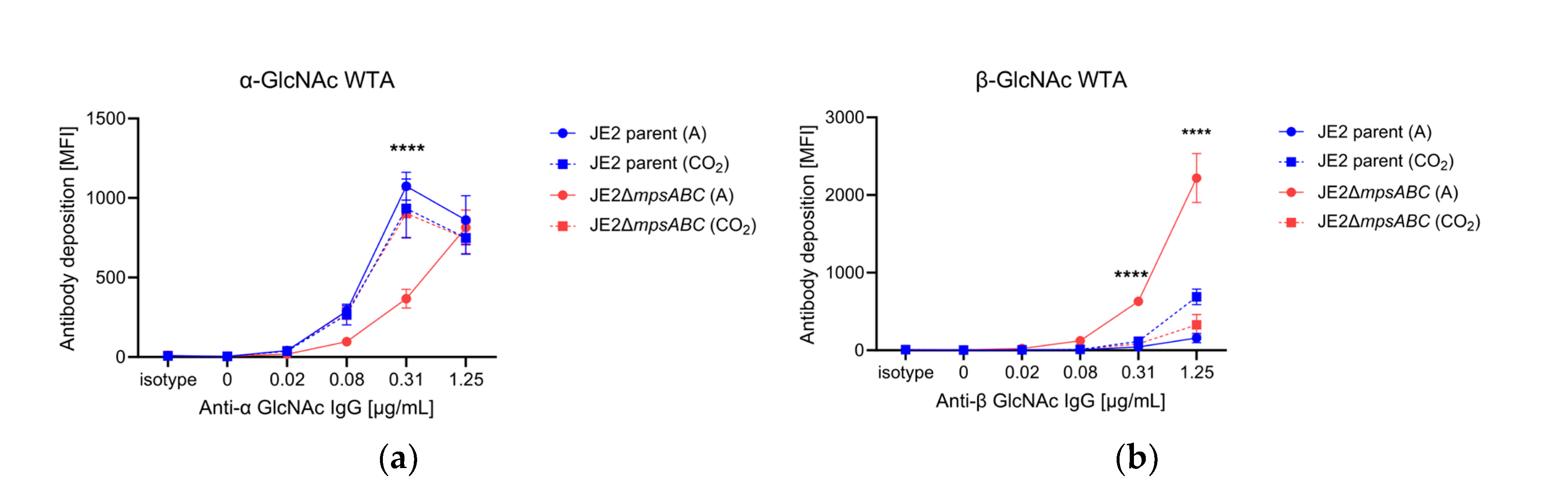
2.5. The ΔmpsABC Mutant’s Wall Teichoic Acids (WTA) Showed a Lower α- and Higher ß-Glycosylation
3. Discussion
4. Materials and Methods
4.1. Bacterial strains and Growth Conditions
4.2. Growth Studies
4.3. RNA Isolation, Library Construction, Sequencing, and Analysis
4.4. Triton X-100-Induced Autolysis Assay
4.5. Lysostaphin Induced Lysis Assay
4.6. Transmission Electron Microscopy
4.7. Muropeptide Analysis
4.8. Mass Spectrometry Analysis of Monomeric Muropeptides
4.9. Flow Cytometry Analysis with WTA Detecting Antibodies
4.10. Statistical Analyses and Quantification of CW Thickness
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fujita Y, Matsuoka H, Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol. 2007, 66, 829–839. [Google Scholar] [CrossRef]
- Tong, L. Structure and function of biotin-dependent carboxylases. Cell Mol Life Sci. 2013, 70, 863–891. [Google Scholar] [CrossRef]
- Merlin C, Masters M, McAteer S, Coulson A. Why is carbonic anhydrase essential to Escherichia coli? J Bacteriol. 2003, 185, 6415–6424. [Google Scholar] [CrossRef]
- Langereis JD, Zomer A, Stunnenberg HG, Burghout P, Hermans PWM. Nontypeable Haemophilus influenzae Carbonic Anhydrase Is Important for Environmental and Intracellular Survival. J Bacteriol. 2013, 195, 2737–2746. [Google Scholar] [CrossRef]
- Casey, JR. Why bicarbonate? Biochem Cell Biol. 2006, 84, 930–939. [Google Scholar] [CrossRef]
- Price GD, Badger MR, Woodger FJ, Long BM. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot. 2008, 59, 1441–1461. [Google Scholar] [CrossRef]
- Price, GD. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth Res. 2011, 109, 47–57. [Google Scholar] [CrossRef]
- Mangiapia M, Usf M, Brown TW, Chaput D, Haller E, Harmer TL, et al. Proteomic and Mutant Analysis of the CO2 Concentrating Mechanism of Hydrothermal Vent Chemolithoautotroph Thiomicrospira crunogena. J Bacteriol. [CrossRef]
- Fan SH, Ebner P, Reichert S, Hertlein T, Zabel S, Lankapalli AK, et al. MpsAB is important for Staphylococcus aureus virulence and growth at atmospheric CO2 levels. Nat Commun. 2019, 10, 3627. [Google Scholar] [CrossRef]
- Mayer S, Steffen W, Steuber J, Götz F. The Staphylococcus aureus NuoL-Like Protein MpsA Contributes to the Generation of Membrane Potential. J Bacteriol. 2015, 197, 794–806. [Google Scholar] [CrossRef]
- Fan SH, Liberini E, Götz F. Staphylococcus aureus Genomes Harbor Only MpsAB-Like Bicarbonate Transporter but Not Carbonic Anhydrase as Dissolved Inorganic Carbon Supply System. Microbiol Spectr. [CrossRef]
- Fan SH, Matsuo M, Huang L, Tribelli PM, Götz F. The MpsAB Bicarbonate Transporter Is Superior to Carbonic Anhydrase in Biofilm-Forming Bacteria with Limited CO2 Diffusion. Microbiol Spectr. [CrossRef]
- Ersoy SC, Manna AC, Proctor RA, Chambers HF, Harrison EM, Bayer AS, et al. The NaHCO(3)-Responsive Phenotype in Methicillin-Resistant Staphylococcus aureus (MRSA) Is Influenced by mecA Genotype. Antimicrob Agents Chemother. 2022, 66, e0025222. [Google Scholar] [CrossRef]
- Fan S-H, Proctor RA, Ersoy SC, Manna AC, Cheung AL, Götz F, et al. Role of the NaHCO(3) Transporter MpsABC in the NaHCO(3). 2013; ß-Lactam-Responsive Phenotype in Methicillin-Resistant Staphylococcus aureus. Microbiology Spectrum. 2023, 11, e00141–23. [Google Scholar] [CrossRef]
- Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio. 2013, 4, e00537–12. [Google Scholar] [CrossRef]
- Maya-Martinez R, Alexander JAN, Otten CF, Ayala I, Vollmer D, Gray J, et al. Recognition of Peptidoglycan Fragments by the Transpeptidase PBP4 From Staphylococcus aureus. Front Microbiol. 2018, 9, 3223. [Google Scholar] [CrossRef]
- Brown S, Xia G, Luhachack LG, Campbell J, Meredith TC, Chen C, et al. Methicillin resistance in Staphylococcus aureus requires glycosylated wall teichoic acids. Proceedings of the National Academy of Sciences. 2012, 109, 18909–18914. [Google Scholar] [CrossRef]
- Xia G, Maier L, Sanchez-Carballo P, Li M, Otto M, Holst O, et al. Glycosylation of wall teichoic acid in Staphylococcus aureus by TarM. J Biol Chem. 2010, 285, 13405–13415. [Google Scholar] [CrossRef]
- Hendriks A, van Dalen R, Ali S, Gerlach D, van der Marel GA, Fuchsberger FF, et al. Impact of Glycan Linkage to Staphylococcus aureus Wall Teichoic Acid on Langerin Recognition and Langerhans Cell Activation. ACS Infect Dis. 2021, 7, 624–635. [Google Scholar] [CrossRef]
- Beveridge, TJ. Ultrastructure, chemistry, and function of the bacterial wall. Int Rev Cytol. 1981, 72, 229–317. [Google Scholar] [CrossRef]
- Giesbrecht P, Kersten T, Maidhof H, Wecke J. Staphylococcal cell wall: morphogenesis and fatal variations in the presence of penicillin. Microbiol Mol Biol Rev. 1998, 62, 1371–1414. [Google Scholar] [CrossRef]
- Hancock R, Park JT. Cell-wall synthesis by Staphylococcus aureus in the presence of chloramphenicol. Nature. 1958, 181, 1050–1052. [Google Scholar] [CrossRef]
- Kim SJ, Chang J, Singh M. Peptidoglycan architecture of Gram-positive bacteria by solid-state NMR. Biochim Biophys Acta. [CrossRef]
- Berscheid A, François P, Strittmatter A, Gottschalk G, Schrenzel J, Sass P, et al. Generation of a vancomycin-intermediate Staphylococcus aureus (VISA) strain by two amino acid exchanges in VraS. J Antimicrob Chemother. 2014, 69, 3190–3198. [Google Scholar] [CrossRef]
- Sass P, Berscheid A, Jansen A, Oedenkoven M, Szekat C, Strittmatter A, et al. Genome sequence of Staphylococcus aureus VC40, a vancomycin- and daptomycin-resistant strain, to study the genetics of development of resistance to currently applied last-resort antibiotics. J Bacteriol. 2012, 194, 2107–2108. [Google Scholar] [CrossRef]
- Stapleton MR, Horsburgh MJ, Hayhurst EJ, Wright L, Jonsson IM, Tarkowski A, et al. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol. 2007, 189, 7316–7325. [Google Scholar] [CrossRef]
- Buist G, Steen A, Kok J, Kuipers OR. LysM, a widely distributed protein motif for binding to (peptido)glycans. Molecular Microbiology. 2008, 68, 838–847. [Google Scholar] [CrossRef]
- Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun. 2014, 5, 4269. [Google Scholar] [CrossRef]
- Gally D, Archibald AR. Cell wall assembly in Staphylococcus aureus: proposed absence of secondary crosslinking reactions. J Gen Microbiol. 1993, 139, 1907–1913. [Google Scholar] [CrossRef]
- Snowden MA, Perkins HR. Peptidoglycan cross-linking in Staphylococcus aureus. An apparent random polymerisation process. Eur J Biochem. 1990, 191, 373–377. [Google Scholar] [CrossRef]
- Vollmer W, Seligman SJ. Architecture of peptidoglycan: more data and more models. Trends Microbiol. 2010, 18, 59–66. [Google Scholar] [CrossRef]
- DeHart HP, Heath HE, Heath LS, LeBlanc PA, Sloan GL. The lysostaphin endopeptidase resistance gene (epr) specifies modification of peptidoglycan cross bridges in Staphylococcus simulans and Staphylococcus aureus. Appl Environ Microbiol. 1995, 61, 1475–1479. [Google Scholar] [CrossRef]
- Schindler CA, Schuhardt VT. LYSOSTAPHIN: A NEW BACTERIOLYTIC AGENT FOR THE STAPHYLOCOCCUS. Proc Natl Acad Sci U S A. 1964, 51, 414–421. [Google Scholar] [CrossRef]
- Monteiro JM, Covas G, Rausch D, Filipe SR, Schneider T, Sahl HG, et al. The pentaglycine bridges of Staphylococcus aureus peptidoglycan are essential for cell integrity. Sci Rep. 2019, 9, 5010. [Google Scholar] [CrossRef]
- Willing S, Dyer E, Schneewind O, Missiakas D. FmhA and FmhC of Staphylococcus aureus incorporate serine residues into peptidoglycan cross-bridges. J Biol Chem. 2020, 295, 13664–13676. [Google Scholar] [CrossRef]
- Strauss A, Thumm G, Götz F. Influence of Lif, the lysostaphin immunity factor, on acceptors of surface proteins and cell wall sorting efficiency in Staphylococcus carnosus. J Bacteriol. 1998, 180, 4960–4962. [Google Scholar] [CrossRef]
- Thumm G, Götz F. Studies on prolysostaphin processing and characterization of the lysostaphin immunity factor (Lif) of Staphylococcus simulans biovar staphylolyticus. Mol Microbiol. 1997, 23, 1251–1265. [Google Scholar] [CrossRef] [PubMed]
- Rohrer S, Berger-Bachi B. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci. Antimicrob Agents Chemother. 2003, 47, 837–846. [Google Scholar] [CrossRef] [PubMed]
- van Dalen R, Peschel A, van Sorge NM. Wall Teichoic Acid in Staphylococcus aureus Host Interaction. Trends in Microbiology. 2020, 28, 985–998. [Google Scholar] [CrossRef]
- Baddiley J, Buchanan JG, Martin RO, Rajbhandary UL. Teichoic acid from the walls of Staphylococcus aureus H. 2. Location of phosphate and alanine residues. Biochem J. 1962, 85, 49–56. [Google Scholar] [CrossRef]
- Fischer, W. Physiology of lipoteichoic acids in bacteria. Adv Microb Physiol. [CrossRef]
- Peschel A, Otto M. Phenol-soluble modulins and staphylococcal infection. Nat Rev Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef]
- Hort M, Bertsche U, Nozinovic S, Dietrich A, Schrötter AS, Mildenberger L, et al. The Role of β-Glycosylated Wall Teichoic Acids in the Reduction of Vancomycin Susceptibility in Vancomycin-Intermediate Staphylococcus aureus. Microbiol Spectr. 2021, 9, e0052821. [Google Scholar] [CrossRef]
- Alam A, Broms JE, Kumar R, Sjostedt A. The Role of ClpB in Bacterial Stress Responses and Virulence. Front Mol Biosci. 6689. [CrossRef]
- Scovill WH, Schreier HJ, Bayles KW. Identification and characterization of the pckA gene from Staphylococcus aureus. J Bacteriol. 1996, 178, 3362–3364. [Google Scholar] [CrossRef]
- Schlag M, Biswas R, Krismer B, Köhler T, Zoll S, Yu W, et al. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol. 2010, 75, 864–873. [Google Scholar] [CrossRef]
- 10.1111/j.1365-2958.2009.07007.x.
- Bera A, Herbert S, Jakob A, Vollmer W, Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005, 55, 778–787. [Google Scholar] [CrossRef] [PubMed]
- de Jonge BL, Chang YS, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem. 1992, 267, 11248–11254. [Google Scholar] [CrossRef]
- Figueiredo TA, Sobral RG, Ludovice AM, Almeida JM, Bui NK, Vollmer W, et al. Identification of genetic determinants and enzymes involved with the amidation of glutamic acid residues in the peptidoglycan of Staphylococcus aureus. PLoS Pathog. 2012, 8, e1002508. [Google Scholar] [CrossRef]
- Bui NK, Gray J, Schwarz H, Schumann P, Blanot D, Vollmer W. The peptidoglycan sacculus of Myxococcus xanthus has unusual structural features and is degraded during glycerol-induced myxospore development. J Bacteriol. 2009, 191, 494–505. [Google Scholar] [CrossRef] [PubMed]
- Lehar SM, Pillow T, Xu M, Staben L, Kajihara KK, Vandlen R, et al. Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015, 527, 323–328. [CrossRef]
- van Dalen R, Molendijk MM, Ali S, van Kessel KPM, Aerts P, van Strijp JAG, et al. Do not discardsStaphylococcus aureus WTA as a vaccine antigen. Nature. 2019, 572, E1–E2. [Google Scholar] [CrossRef]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012, 9, 676–682. [Google Scholar] [CrossRef]
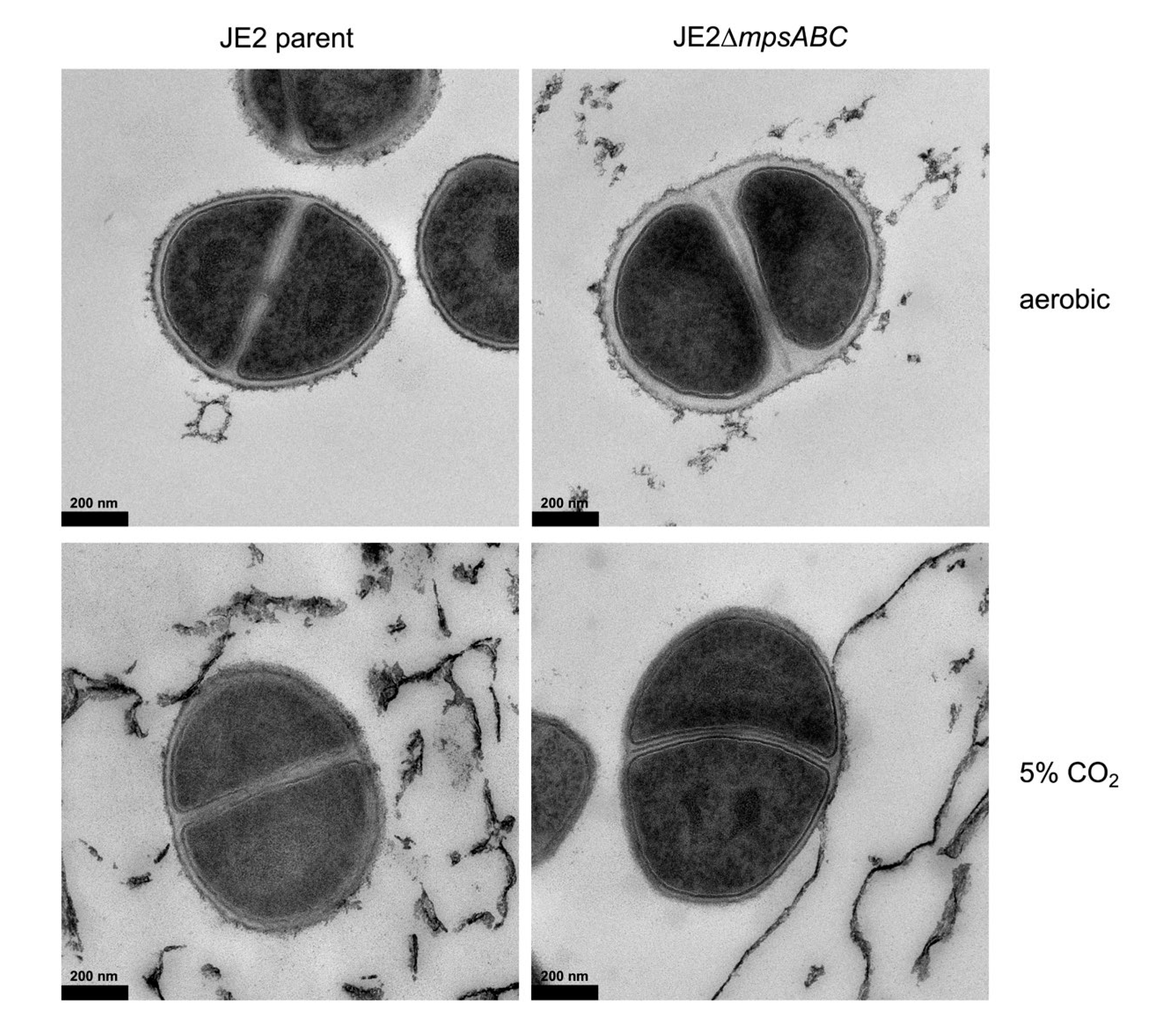
| Cell wall thickness in nm (mean ± SD) | ||
| Strain | Ambient air | 5% CO2 |
| JE2 parent | 15.37 ± 2.64 | 12.05 ± 2.44 |
| JE2ΔmpsABC | 33.43 ± 7.71 | 15.00 ± 2.78 |
| Cell wall thickness in nm (mean ± SD) | ||
| Strain | Ambient air | 5% CO2 |
| JE2 parent | 15.37 ± 2.64 | 12.05 ± 2.44 |
| JE2ΔmpsABC | 33.43 ± 7.71 | 15.00 ± 2.78 |
| Protein Number | Function | Fold change Mutant vs. Parent |
||
|---|---|---|---|---|
| 4h | 8h | |||
| Cell wall (CW) lytic enzymes | ||||
| B7H15_11645 | SceD, lytic transglycosylase | 592.3 | 517.1 | |
| B7H15_14290 | IsaA, lytic transglycosylase | 4.3 | 2.1 | |
| B7H15_12775 | SsaA, secretory antigen | 20 | 5.2 | |
| B7H15_10875 | Amidase, 251 aa, prophage-encoded | 16.8 | 9.9 | |
| B7H15_01500 | LytM, glycine-glycine endopeptidase | 14.8 | 8.4 | |
| B7H15_03720 | LysM, peptidoglycan-binding domain-containing protein, probably autolysin | 8 | 2 | |
| Wall teichoic acid (WTA) biosynthesis | ||||
| B7H15_05355 | TarM, poly(ribitol-phosphate) α-N-acetylglucosaminyltransferase | - 43.3 | - 61.5 | |
| CW anchored proteins | ||||
| B7H15_00750 | SasD, cell-wall-anchored protein | - 27.3 | - 3.2 | |
| B7H15_14775 | SasA, serine-rich repeat glycoprotein adhesin | - 23.4 | - 15 | |
| B7H15_13880 | SIRK signal domain/LPXTG anchor domain surface protein | - 22.2 | - 3.3 | |
| B7H15_00135 | AdsA, LPXTG-anchored adenosine synthase | - 11.5 | - 4.1 | |
| B7H15_00620 | Spa, staphylococcal protein A | - 6.4 | - 1.6 | |
| Secreted enzymes | ||||
| B7H15_01535 | Peptidase C51 domain-containing protein | - 64.6 | - 31 | |
| B7H15_14690 | Aureolysin, zinc metalloproteinase | - 44.6 | - 1.2 | |
| B7H15_00545 | Phospholipase C, phosphatidylinositol | - 23.1 | - 1.7 | |
| B7H15_01765 | Lip2(geh), triacylglycerol lipase | - 20.3 | - 200 | |
| Transporters | ||||
| B7H15_01540 | Type VII secretion effector EsxA (and all other type VII secretion protein genes) | - 56.5 | - 102 | |
| B7H15_01675 | Formate/nitrite transporter family protein | - 33.7 | - 1.7 | |
| B7H15_01120 | Peptide ABC transporter substrate-binding protein | - 15.3 | - 4.7 | |
| B7H15_01110 | ABC transporter permease | - 14.9 | - 6 | |
| B7H15_00420 | YdhK family protein, lipoprotein, putative | - 13.5 | - 29 | |
| B7H15_00415 | Copper-translocating P-type ATPase | - 11.2 | - 21.6 | |
| B7H15_03550 | Metal ABC transporter permease | - 11.2 | - 176 | |
| Toxins | ||||
| B7H15_02365 | PSM-α3, phenol-soluble modulin | - 479 | - 6360 | |
| B7H15_11280 | δ-lysin, phenol-soluble modulin | - 336 | - 11215 | |
| B7H15_06120 | PSM-ß, phenol-soluble modulin | - 235 | - 5200 | |
| B7H15_06050 | Hla, α-hemolysin | - 61 | - 2408 | |
| Regulators | ||||
| B7H15_14830 | IcaR, ica operon transcriptional repressor | - 20.7 | - 15.6 | |
| B7H15_11300 | AgrA, accessory gene regulator protein A | - 25 | - 91 | |
| B7H15_11285 | AgrB, accessory gene regulator protein B | - 24 | - 89 | |
| B7H15_11290 | AgrD, accessory gene regulator protein D | - 24 | - 69 | |
| B7H15_11295 | AgrC, accessory gene regulator protein C | - 28 | - 90 | |
| B7H15_03430 | SarA, staphylococcal accessory regulator | - 1.7 | - 3 | |
| B7H15_06535 | CodY, transcriptional repressor | - 1.2 | - 1.6 | |
| Prophage genes | ||||
| B7H15_10915 | Phage tail family protein (and most phage related genes) | 12.5 | nd | |
| B7H15_11045 | PVL, phi PVL orf 51-like protein | 16.5 | 13 | |
| Cross bridge synthesis | ||||
| B7H15_13360 | FmhA, fem-like factor | 1.02 | 1.7 | |
| B7H15_06500 | FmhC, fem-like factor | 0.98 | - 2.3 | |
| Fatty acid biosynthesis | ||||
| B7H15_06405 | FabD, ACP S-malonyltransferase | 1.2 | 1.5 | |
| B7H15_05075 | FabF, ß-ketoacyl-ACP synthase II | - 1.5 | - 1.2 | |
| B7H15_06410 | FabG, 3-oxoacyl-acyl-carrier-protein reductase | - 1.2 | - 1.1 | |
| B7H15_05210 | FabI, enoyl-ACP reducatse | 1.5 | 1.7 | |
| B7H15_11660 | FabZ, 3-hydroxyacyl-ACP dehydratase | 2.2 | 1.6 | |
| B7H15_07130 | PlsY, glycerol-3-phosphate acyltransferase | - 1.2 | - 1.5 | |
| B7H15_06400 | PlsX, phosphate acyltransferase | - 1.5 | 1 | |
| B7H15_06385 | FakA, fatty acid kinase | 1.1 | 1 | |
| B7H15_04195 | FakB1, fatty acid binding protein 1 | - 1.2 | 1 | |
| B7H15_07520 | FakB2, fatty acid binding protein 2 | 1 | 1 | |
| Carboxylating Enzymes | ||||
| B7H15_09750 | PckA, PEP carboxylase | - 5 | - 100 | |
| B7H15_08395 | AccC, acetyl-coA carboxylase | 1.2 | 2 | |
| Cell wall thickness in nm (mean ± SD) | ||
| Strain | Ambient air | 5% CO2 |
| JE2 parent | 15.37 ± 2.64 | 12.05 ± 2.44 |
| JE2ΔmpsABC | 33.43 ± 7.71 | 15.00 ± 2.78 |
| Cell wall thickness in nm (mean ± SD) | ||
| Strain | Ambient air | 5% CO2 |
| JE2 parent | 15.37 ± 2.64 | 12.05 ± 2.44 |
| JE2ΔmpsABC | 33.43 ± 7.71 | 15.00 ± 2.78 |
| Protein Number | Function | Fold change Mutant vs. Parent |
||
|---|---|---|---|---|
| 4h | 8h | |||
| Cell wall (CW) lytic enzymes | ||||
| B7H15_11645 | SceD, lytic transglycosylase | 592.3 | 517.1 | |
| B7H15_14290 | IsaA, lytic transglycosylase | 4.3 | 2.1 | |
| B7H15_12775 | SsaA, secretory antigen | 20 | 5.2 | |
| B7H15_10875 | Amidase, 251 aa, prophage-encoded | 16.8 | 9.9 | |
| B7H15_01500 | LytM, glycine-glycine endopeptidase | 14.8 | 8.4 | |
| B7H15_03720 | LysM, peptidoglycan-binding domain-containing protein, probably autolysin | 8 | 2 | |
| Wall teichoic acid (WTA) biosynthesis | ||||
| B7H15_05355 | TarM, poly(ribitol-phosphate) α-N-acetylglucosaminyltransferase | - 43.3 | - 61.5 | |
| CW anchored proteins | ||||
| B7H15_00750 | SasD, cell-wall-anchored protein | - 27.3 | - 3.2 | |
| B7H15_14775 | SasA, serine-rich repeat glycoprotein adhesin | - 23.4 | - 15 | |
| B7H15_13880 | SIRK signal domain/LPXTG anchor domain surface protein | - 22.2 | - 3.3 | |
| B7H15_00135 | AdsA, LPXTG-anchored adenosine synthase | - 11.5 | - 4.1 | |
| B7H15_00620 | Spa, staphylococcal protein A | - 6.4 | - 1.6 | |
| Secreted enzymes | ||||
| B7H15_01535 | Peptidase C51 domain-containing protein | - 64.6 | - 31 | |
| B7H15_14690 | Aureolysin, zinc metalloproteinase | - 44.6 | - 1.2 | |
| B7H15_00545 | Phospholipase C, phosphatidylinositol | - 23.1 | - 1.7 | |
| B7H15_01765 | Lip2(geh), triacylglycerol lipase | - 20.3 | - 200 | |
| Transporters | ||||
| B7H15_01540 | Type VII secretion effector EsxA (and all other type VII secretion protein genes) | - 56.5 | - 102 | |
| B7H15_01675 | Formate/nitrite transporter family protein | - 33.7 | - 1.7 | |
| B7H15_01120 | Peptide ABC transporter substrate-binding protein | - 15.3 | - 4.7 | |
| B7H15_01110 | ABC transporter permease | - 14.9 | - 6 | |
| B7H15_00420 | YdhK family protein, lipoprotein, putative | - 13.5 | - 29 | |
| B7H15_00415 | Copper-translocating P-type ATPase | - 11.2 | - 21.6 | |
| B7H15_03550 | Metal ABC transporter permease | - 11.2 | - 176 | |
| Toxins | ||||
| B7H15_02365 | PSM-α3, phenol-soluble modulin | - 479 | - 6360 | |
| B7H15_11280 | δ-lysin, phenol-soluble modulin | - 336 | - 11215 | |
| B7H15_06120 | PSM-ß, phenol-soluble modulin | - 235 | - 5200 | |
| B7H15_06050 | Hla, α-hemolysin | - 61 | - 2408 | |
| Regulators | ||||
| B7H15_14830 | IcaR, ica operon transcriptional repressor | - 20.7 | - 15.6 | |
| B7H15_11300 | AgrA, accessory gene regulator protein A | - 25 | - 91 | |
| B7H15_11285 | AgrB, accessory gene regulator protein B | - 24 | - 89 | |
| B7H15_11290 | AgrD, accessory gene regulator protein D | - 24 | - 69 | |
| B7H15_11295 | AgrC, accessory gene regulator protein C | - 28 | - 90 | |
| B7H15_03430 | SarA, staphylococcal accessory regulator | - 1.7 | - 3 | |
| B7H15_06535 | CodY, transcriptional repressor | - 1.2 | - 1.6 | |
| Prophage genes | ||||
| B7H15_10915 | Phage tail family protein (and most phage related genes) | 12.5 | nd | |
| B7H15_11045 | PVL, phi PVL orf 51-like protein | 16.5 | 13 | |
| Cross bridge synthesis | ||||
| B7H15_13360 | FmhA, fem-like factor | 1.02 | 1.7 | |
| B7H15_06500 | FmhC, fem-like factor | 0.98 | - 2.3 | |
| Fatty acid biosynthesis | ||||
| B7H15_06405 | FabD, ACP S-malonyltransferase | 1.2 | 1.5 | |
| B7H15_05075 | FabF, ß-ketoacyl-ACP synthase II | - 1.5 | - 1.2 | |
| B7H15_06410 | FabG, 3-oxoacyl-acyl-carrier-protein reductase | - 1.2 | - 1.1 | |
| B7H15_05210 | FabI, enoyl-ACP reducatse | 1.5 | 1.7 | |
| B7H15_11660 | FabZ, 3-hydroxyacyl-ACP dehydratase | 2.2 | 1.6 | |
| B7H15_07130 | PlsY, glycerol-3-phosphate acyltransferase | - 1.2 | - 1.5 | |
| B7H15_06400 | PlsX, phosphate acyltransferase | - 1.5 | 1 | |
| B7H15_06385 | FakA, fatty acid kinase | 1.1 | 1 | |
| B7H15_04195 | FakB1, fatty acid binding protein 1 | - 1.2 | 1 | |
| B7H15_07520 | FakB2, fatty acid binding protein 2 | 1 | 1 | |
| Carboxylating Enzymes | ||||
| B7H15_09750 | PckA, PEP carboxylase | - 5 | - 100 | |
| B7H15_08395 | AccC, acetyl-coA carboxylase | 1.2 | 2 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





