Submitted:
24 July 2024
Posted:
25 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Identification of Reported Minor CMPs Genes
1.2. Gathering Sources
1.3. Literature Review
2. Genes Classification
2.1. Definitive Genes Associated with ARVC, DCM and HCM2.2. Minor genes in ARVC, DCM and HCM
3. Gene-Disease Validity and Clinical Evidence
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Wilcox JE, Hershberger RE. Genetic cardiomyopathies. Curr Opin Cardiol. 2018;33: 354–362. [CrossRef]
- Walsh, R.; Mazzarotto, F.; Whiffin, N.; Buchan, R.; Midwinter, W.; Wilk, A.; Li, N.; Felkin, L.; Ingold, N.; Govind, R.; et al. Quantitative approaches to variant classification increase the yield and precision of genetic testing in Mendelian diseases: the case of hypertrophic cardiomyopathy. Genome Med. 2019, 11, 1–18. [CrossRef]
- Aiyer, S.; Kalutskaya, E.; Agdamag, A.C.; Tang, W.H.W. Genetic Evaluation and Screening in Cardiomyopathies: Opportunities and Challenges for Personalized Medicine. J. Pers. Med. 2023, 13, 887. [CrossRef]
- Ingles, J.; Goldstein, J.; Thaxton, C.; Caleshu, C.; Corty, E.W.; Crowley, S.B.; Dougherty, K.; Harrison, S.M.; McGlaughon, J.; Milko, L.V.; et al. Evaluating the Clinical Validity of Hypertrophic Cardiomyopathy Genes. Circ. Genom. Precis. Med. 2019, 12, e002460–e002460. [CrossRef]
- Jordan, E.; Peterson, L.; Ai, T.; Asatryan, B.; Bronicki, L.; Brown, E.; Celeghin, R.; Edwards, M.; Fan, J.; Ingles, J.; et al. Evidence-Based Assessment of Genes in Dilated Cardiomyopathy. Circulation 2021, 144, 7–19. [CrossRef]
- Garcia, J.; Tahiliani, J.; Johnson, N.M.; Aguilar, S.; Beltran, D.; Daly, A.; Decker, E.; Haverfield, E.; Herrera, B.; Murillo, L.; et al. Clinical Genetic Testing for the Cardiomyopathies and Arrhythmias: A Systematic Framework for Establishing Clinical Validity and Addressing Genotypic and Phenotypic Heterogeneity. Front. Cardiovasc. Med. 2016, 3, 20. [CrossRef]
- de Oca, M.M.; Varela, M.V.L.; Jardim, J.; Stirvulov, R.; Surmont, F. Bronchodilator treatment for COPD in primary care of four Latin America countries: The multinational, cross-sectional, non-interventional PUMA study. Pulm. Pharmacol. Ther. 2016, 38, 10–16. [CrossRef]
- Vimalanathan, A.K.; Ehler, E.; Gehmlich, K. Genetics of and pathogenic mechanisms in arrhythmogenic right ventricular cardiomyopathy. Biophys. Rev. 2018, 10, 973–982. [CrossRef]
- Mahmaljy H, Yelamanchili VS, Singhal M. Dilated Cardiomyopathy. StatPearls. Treasure Island (FL): StatPearls Publishing; 2023.
- McNally EM, Mestroni L. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ Res. 2017;121: 731–748. [CrossRef]
- Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381: 242–255.
- Abbas, M.T.; Ali, N.B.; Farina, J.M.; Mahmoud, A.K.; Pereyra, M.; Scalia, I.G.; Kamel, M.A.; Barry, T.; Lester, S.J.; Cannan, C.R.; et al. Role of Genetics in Diagnosis and Management of Hypertrophic Cardiomyopathy: A Glimpse into the Future. Biomedicines 2024, 12, 682. [CrossRef]
- Salazar-Mendiguchía, J.; Ochoa, J.P.; Palomino-Doza, J.; Domínguez, F.; Díez-López, C.; Akhtar, M.; Ramiro-León, S.; Clemente, M.M.; Pérez-Cejas, A.; Robledo, M.; et al. Mutations in cause an autosomal-recessive form of hypertrophic cardiomyopathy. Heart 2020, 106, 1342–1348. [CrossRef]
- Piqueras-Flores J, Villacorta-Argüelles E, Galvin J, Climent-Payá V, Escobar-López LE, Amor-Salamanca A, et al. Intermediate-effect size p.Arg637Gln in increases risk of HCM and is associated with an aggressive phenotype in homozygous carriers. J Med Genet. 2024;61: 423–427. [CrossRef]
- Hayesmoore, J.B.; Bhuiyan, Z.A.; Coviello, D.A.; du Sart, D.; Edwards, M.; Iascone, M.; Morris-Rosendahl, D.J.; Sheils, K.; van Slegtenhorst, M.; Thomson, K.L. EMQN: Recommendations for genetic testing in inherited cardiomyopathies and arrhythmias. Eur. J. Hum. Genet. 2023, 31, 1003–1009. [CrossRef]
- Homepage. In: EMQN [Internet]. 29 Jan 2024 [cited 17 Jul 2024]. Available: https://www.emqn.org/.
- Clinical Genome Resource. Welcome to ClinGen. [cited 17 Jul 2024]. Available: https://clinicalgenome.org/.
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.F.; McBride, K.L.; Morales, A.; Taylor, M.R.G.; Vatta, M.; Ware, S.M.; et al. Genetic evaluation of cardiomyopathy: A clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet. Med. 2018, 20, 899–909. [CrossRef]
- ClinVar. Available online: https://www.ncbi.nlm.nih.gov/clinvar/ (accessed on 3 March 2020).
- National Center for Biotechnology Information. [cited 17 Jul 2024]. Available: https://www.ncbi.nlm.nih.gov/.
- National Library of Medicine - National Institutes of Health. 1993 [cited 17 Jul 2024]. Available: https://www.nlm.nih.gov/.
- National Institutes of Health (NIH). In: National Institutes of Health (NIH) [Internet]. [cited 17 Jul 2024]. Available: https://www.nih.gov/.
- Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, et al. GeneReviews®. 2024 [cited 17 Jul 2024]. Available: https://www.ncbi.nlm.nih.gov/books/NBK1116/.
- Home - OMIM. [cited 17 Jul 2024]. Available: https://www.omim.org/.
- Wilde AAM, Semsarian C, Márquez MF, Sepehri Shamloo A, Ackerman MJ, Ashley EA, et al. European Heart Rhythm Association (EHRA)/Heart Rhythm Society (HRS)/Asia Pacific Heart Rhythm Society (APHRS)/Latin American Heart Rhythm Society (LAHRS) Expert Consensus Statement on the State of Genetic Testing for Cardiac Diseases. Heart Rhythm. 2022;19: e1–e60.
- Kane, G.C.; Liu, X.-K.; Yamada, S.; Olson, T.M.; Terzic, A. Cardiac KATP channels in health and disease. J. Mol. Cell. Cardiol. 2005, 38, 937–943. [CrossRef]
- Solbach, T.F.; König, J.; Fromm, M.F.; Zolk, O. ATP-Binding Cassette Transporters in the Heart. Trends Cardiovasc. Med. 2005, 16, 7–15. [CrossRef]
- Bienengraeber, M.; Olson, T.M.; A Selivanov, V.; Kathmann, E.C.; O’Cochlain, F.; Gao, F.; Karger, A.B.; Ballew, J.D.; Hodgson, D.M.; Zingman, L.V.; et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat. Genet. 2004, 36, 382–387. [CrossRef]
- Carnevale, A.; Rosas-Madrigal, S.; Rosendo-Gutiérrez, R.; López-Mora, E.; Romero-Hidalgo, S.; Avila-Vazzini, N.; Jacobo-Albavera, L.; Domínguez-Pérez, M.; Vargas-Alarcón, G.; Pérez-Villatoro, F.; et al. Genomic study of dilated cardiomyopathy in a group of Mexican patients using site-directed next generation sequencing. Mol. Genet. Genom. Med. 2020, 8. [CrossRef]
- Shen, C.; Xu, L.; Sun, X.; Sun, A.; Ge, J. Genetic variants in Chinese patients with sporadic dilated cardiomyopathy: a cross-sectional study. Ann. Transl. Med. 2022, 10, 129–129. [CrossRef]
- Fahrenbach, J.P.; Stoller, D.; Kim, G.; Aggarwal, N.; Yerokun, B.; Earley, J.U.; Hadhazy, M.; Makielski, J.C.; McNally, E.M.; Shi, N.-Q. Abcc9is required for the transition to oxidative metabolism in the newborn heart. FASEB J. 2014, 28, 2804–2815. [CrossRef]
- Zaytseva, A.; Tulintseva, T.; Fomicheva, Y.; Mikhailova, V.; Treshkur, T.; Kostareva, A. Case Report: Loss-of-Function ABCC9 Genetic Variant Associated With Ventricular Fibrillation. Front. Genet. 2022, 13, 718853. [CrossRef]
- Lornage, X.; Romero, N.B.; Grosgogeat, C.A.; Malfatti, E.; Donkervoort, S.; Marchetti, M.M.; Neuhaus, S.B.; Foley, A.R.; Labasse, C.; Schneider, R.; et al. ACTN2 mutations cause "Multiple structured Core Disease" (MsCD). Acta Neuropathol. 2019, 137, 501–519. [CrossRef]
- Prondzynski, M.; Lemoine, M.D.; Zech, A.T.; Horváth, A.; Di Mauro, V.; Koivumäki, J.T.; Kresin, N.; Busch, J.; Krause, T.; Krämer, E.; et al. Disease modeling of a mutation in α-actinin 2 guides clinical therapy in hypertrophic cardiomyopathy. EMBO Mol. Med. 2019, 11, e11115. [CrossRef]
- Chiu C, Bagnall RD, Ingles J, Yeates L, Kennerson M, Donald JA, et al. Mutations in alpha-actinin-2 cause hypertrophic cardiomyopathy: a genome-wide analysis. J Am Coll Cardiol. 2010;55: 1127–1135.
- Mohapatra, B.; Jimenez, S.; Lin, J.H.; Bowles, K.R.; Coveler, K.J.; Marx, J.G.; A Chrisco, M.; Murphy, R.T.; Lurie, P.R.; Schwartz, R.J.; et al. Mutations in the muscle LIM protein and α-actinin-2 genes in dilated cardiomyopathy and endocardial fibroelastosis. Mol. Genet. Metab. 2003, 80, 207–215. [CrossRef]
- Fan, L.-L.; Huang, H.; Jin, J.-Y.; Li, J.-J.; Chen, Y.-Q.; Xiang, R. Whole-Exome Sequencing Identifies a Novel Mutation (p.L320R) of Alpha-Actinin 2 in a Chinese Family with Dilated Cardiomyopathy and Ventricular Tachycardia. Cytogenet. Genome Res. 2019, 157, 148–152. [CrossRef]
- Atang, A.E.; Rebbeck, R.T.; Thomas, D.D.; Avery, A.W. Cardiomyopathy-associated variants alter the structure and function of the α-actinin-2 actin-binding domain. Biochem. Biophys. Res. Commun. 2023, 670, 12–18. [CrossRef]
- Hou, C.R.; Cortez, D. Novel ACTN2 missense variant is associated with idiopathic ventricular fibrillation: a case report. Eur. Hear. J. - Case Rep. 2022, 6, ytac229. [CrossRef]
- Zhang, N.; Xie, X.-J.; Wang, J.-A. Multifunctional protein: cardiac ankyrin repeat protein. J Zhejiang Univ Sci 2016, 17, 333–341. [CrossRef]
- Arimura, T.; Bos, J.M.; Sato, A.; Kubo, T.; Okamoto, H.; Nishi, H.; Harada, H.; Koga, Y.; Moulik, M.; Doi, Y.L.; et al. Cardiac Ankyrin Repeat Protein Gene (ANKRD1) Mutations in Hypertrophic Cardiomyopathy. Circ. 2009, 54, 334–342. [CrossRef]
- Crocini, C.; Arimura, T.; Reischmann, S.; Eder, A.; Braren, I.; Hansen, A.; Eschenhagen, T.; Kimura, A.; Carrier, L. Impact of ANKRD1 mutations associated with hypertrophic cardiomyopathy on contraction parameters of engineered heart tissue. Basic Res. Cardiol. 2013, 108, 349. [CrossRef]
- Duboscq-Bidot, L.; Charron, P.; Ruppert, V.; Fauchier, L.; Richter, A.; Tavazzi, L.; Arbustini, E.; Wichter, T.; Maisch, B.; Komajda, M.; et al. Mutations in the ANKRD1 gene encoding CARP are responsible for human dilated cardiomyopathy. Eur. Hear. J. 2009, 30, 2128–2136. [CrossRef]
- Moulik, M.; Vatta, M.; Witt, S.H.; Arola, A.M.; Murphy, R.T.; McKenna, W.J.; Boriek, A.M.; Oka, K.; Labeit, S.; Bowles, N.E.; et al. ANKRD1, the Gene Encoding Cardiac Ankyrin Repeat Protein, Is a Novel Dilated Cardiomyopathy Gene. Circ. 2009, 54, 325–333. [CrossRef]
- Chiu, C.; Tebo, M.; Ingles, J.; Yeates, L.; Arthur, J.W.; Lind, J.M.; Semsarian, C. Genetic screening of calcium regulation genes in familial hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 2007, 43, 337–343. [CrossRef]
- Verhagen, J.M.A.; Veldman, J.H.; van der Zwaag, P.A.; von der Thüsen, J.H.; Brosens, E.; Christiaans, I.; Dooijes, D.; Enden, A.T.J.M.H.-V.D.; Deprez, R.H.L.; Michels, M.; et al. Lack of evidence for a causal role of CALR3 in monogenic cardiomyopathy. Eur. J. Hum. Genet. 2018, 26, 1603–1610. [CrossRef]
- Ikawa, M.; Tokuhiro, K.; Yamaguchi, R.; Benham, A.M.; Tamura, T.; Wada, I.; Satouh, Y.; Inoue, N.; Okabe, M. Calsperin Is a Testis-specific Chaperone Required for Sperm Fertility. J. Biol. Chem. 2011, 286, 5639–5646. [CrossRef]
- Hung, I.-C.; Cherng, B.-W.; Hsu, W.-M.; Lee, S.-J. Calnexin is required for zebrafish posterior lateral line development. Int. J. Dev. Biol. 2013, 57, 427–438. [CrossRef]
- Mayosi, B.M.; Fish, M.; Shaboodien, G.; Mastantuono, E.; Kraus, S.; Wieland, T.; Kotta, M.-C.; Chin, A.; Laing, N.; Ntusi, N.B.; et al. Identification of Cadherin 2 ( CDH2 ) Mutations in Arrhythmogenic Right Ventricular Cardiomyopathy. Circ. Cardiovasc. Genet. 2017, 10. [CrossRef]
- Tiwari P, Mrigwani A, Kaur H, Kaila P, Kumar R, Guptasarma P. Structural-Mechanical and Biochemical Functions of Classical Cadherins at Cellular Junctions: A Review and Some Hypotheses. Adv Exp Med Biol. 2018;1112: 107–138. [CrossRef]
- Kwon, Y.-S.; Park, T.I.; Cho, Y.; Bae, M.H.; Kim, S. Clinical usefulness of immunohistochemistry for plakoglobin, N-cadherin, and connexin-43 in the diagnosis of arrhythmogenic right ventricular cardiomyopathy. Int J Clin Exp Pathol. 2013, 6, 2928–35.
- Vafiadaki, E.; Arvanitis, D.A.; Sanoudou, D. Muscle LIM Protein: Master regulator of cardiac and skeletal muscle functions. Gene 2015, 566, 1–7. [CrossRef]
- Liang, S.; Zhou, Y.; Chang, Y.; Li, J.; Zhang, M.; Gao, P.; Li, Q.; Yu, H.; Kawakami, K.; Ma, J.; et al. A novel gene-trap line reveals the dynamic patterns and essential roles of cysteine and glycine-rich protein 3 in zebrafish heart development and regeneration. Cell. Mol. Life Sci. 2024, 81, 1–14. [CrossRef]
- Geier, C.; Gehmlich, K.; Ehler, E.; Hassfeld, S.; Perrot, A.; Hayess, K.; Cardim, N.; Wenzel, K.; Erdmann, B.; Krackhardt, F.; et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum. Mol. Genet. 2008, 17, 2753–2765. [CrossRef]
- Walsh, R.; Buchan, R.; Wilk, A.; John, S.; Felkin, L.E.; Thomson, K.L.; Chiaw, T.H.; Loong, C.C.W.; Pua, C.J.; Raphael, C.; et al. Defining the genetic architecture of hypertrophic cardiomyopathy: re-evaluating the role of non-sarcomeric genes. Eur. Hear. J. 2017, 38, 3461–3468. [CrossRef]
- Salazar-Mendiguchía, J.; Barriales-Villa, R.; Lopes, L.R.; Ochoa, J.P.; Rodríguez-Vilela, A.; Palomino-Doza, J.; Larrañaga-Moreira, J.M.; Cicerchia, M.; Cárdenas-Reyes, I.; García-Giustiniani, D.; et al. The p.(Cys150Tyr) variant in CSRP3 is associated with late-onset hypertrophic cardiomyopathy in heterozygous individuals. Eur. J. Med Genet. 2020, 63, 104079. [CrossRef]
- Arber, S.; Hunter, J.J.; Ross, J., Jr.; Hongo, M.; Sansig, G.; Borg, J.; Perriard, J.-C.; Chien, K.R.; Caroni, P. MLP-Deficient Mice Exhibit a Disruption of Cardiac Cytoarchitectural Organization, Dilated Cardiomyopathy, and Heart Failure. Cell 1997, 88, 393–403. [CrossRef]
- Hershberger RE, Parks SB, Kushner JD, Li D, Ludwigsen S, Jakobs P, et al. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Transl Sci. 2008;1: 21–26. [CrossRef]
- Zimmerman, R.S.; Cox, S.; Lakdawala, N.K.; Cirino, A.; Mancini-Dinardo, D.; Clark, E.; Leon, A.; Duffy, E.; White, E.; Baxter, S.; et al. A novel custom resequencing array for dilated cardiomyopathy. Genet Med. 2010, 12, 268–278. [CrossRef]
- Giri, P.; Jain, D.; Kumar, A.; Mohapatra, B. Identification and in silico characterization of CSRP3 synonymous variants in dilated cardiomyopathy. Mol. Biol. Rep. 2023, 50, 4105–4117. [CrossRef]
- Erdmann, J.; Hassfeld, S.; Kallisch, H.; Fleck, E.; Regitz-Zagrose, V. Genetic variants in the promoter (g983G>T) and coding region (A92T) of the human cardiotrophin-1 gene (CTF1) in patients with dilated cardiomyopathy. Hum. Mutat. 2000, 16, 448. [CrossRef]
- Pugh, T.J.; Kelly, M.A.; Gowrisankar, S.; Hynes, E.; Seidman, M.A.; Baxter, S.M.; Bowser, M.; Harrison, B.; Aaron, D.; Mahanta, L.M.; et al. The landscape of genetic variation in dilated cardiomyopathy as surveyed by clinical DNA sequencing. Genet Med. 2014, 16, 601–608. [CrossRef]
- Akinrinade, O.; Ollila, L.; Vattulainen, S.; Tallila, J.; Gentile, M.; Salmenperä, P.; Koillinen, H.; Kaartinen, M.; Nieminen, M.S.; Myllykangas, S.; et al. Genetics and genotype–phenotype correlations in Finnish patients with dilated cardiomyopathy. Eur. Hear. J. 2015, 36, 2327–2337. [CrossRef]
- Zolk, O.; Ng, L.L.; O’brien, R.J.; Weyand, M.; Eschenhagen, T. Augmented Expression of Cardiotrophin-1 in Failing Human Hearts Is Accompanied by Diminished Glycoprotein 130 Receptor Protein Abundance. Circulation 2002, 106, 1442–1446. [CrossRef]
- Goossens S, Janssens B, Bonné S, De Rycke R, Braet F, van Hengel J, et al. A unique and specific interaction between alphaT-catenin and plakophilin-2 in the area composita, the mixed-type junctional structure of cardiac intercalated discs. J Cell Sci. 2007;120: 2126–2136. [CrossRef]
- van Tintelen, J.; Entius, M.M.; Bhuiyan, Z.A.; Jongbloed, R.; Wiesfeld, A.C.; Wilde, A.A.; van der Smagt, J.; Boven, L.G.; Mannens, M.M.; van Langen, I.M.; et al. Plakophilin-2 Mutations Are the Major Determinant of Familial Arrhythmogenic Right Ventricular Dysplasia/Cardiomyopathy. Circulation 2006, 113, 1650–1658. [CrossRef]
- Hung, P.-F.; Chung, F.-P.; Hung, C.-L.; Lin, Y.-J.; Kuo, T.-T.; Liao, J.-N.; Chen, Y.-Y.; Pan, C.-H.; Shaw, K.-P.; Chen, S.-A. Decreased Expression of Plakophilin-2 and αT-Catenin in Arrhythmogenic Right Ventricular Cardiomyopathy: Potential Markers for Diagnosis. Int. J. Mol. Sci. 2022, 23, 5529. [CrossRef]
- Christensen, A.H.; Benn, M.; Tybjærg-Hansen, A.; Haunso, S.; Svendsen, J.H. Screening of Three Novel Candidate Genes in Arrhythmogenic Right Ventricular Cardiomyopathy. Genet. Test. Mol. Biomarkers 2011, 15, 267–271. [CrossRef]
- van Hengel J, Calore M, Bauce B, Dazzo E, Mazzotti E, De Bortoli M, et al. Mutations in the area composita protein αT-catenin are associated with arrhythmogenic right ventricular cardiomyopathy. Eur Heart J. 2013;34: 201–210. [CrossRef]
- Li J, Goossens S, van Hengel J, Gao E, Cheng L, Tyberghein K, et al. Loss of αT-catenin alters the hybrid adhering junctions in the heart and leads to dilated cardiomyopathy and ventricular arrhythmia following acute ischemia. J Cell Sci. 2012;125: 1058–1067. [CrossRef]
- Cao, Q.; Shen, Y.; Liu, X.; Yu, X.; Yuan, P.; Wan, R.; Liu, X.; Peng, X.; He, W.; Pu, J.; et al. Phenotype and Functional Analyses in a Transgenic Mouse Model of Left Ventricular Noncompaction Caused by a DTNA Mutation. Int. Hear. J. 2017, 58, 939–947. [CrossRef]
- Walsh, R.; Thomson, K.L.; Ware, J.S.; Funke, B.H.; Woodley, J.; McGuire, K.J.; Mazzarotto, F.; Blair, E.; Seller, A.; Taylor, J.C.; et al. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genet. Med. 2017, 19, 192–203. [CrossRef]
- Abe, S.; Takeda, H.; Nishio, S.-Y.; Usami, S.-I. Sensorineural hearing loss and mild cardiac phenotype caused by an EYA4 mutation. Hum. Genome Var. 2018, 5, 23. [CrossRef]
- Schönberger, J.; Wang, L.; Shin, J.T.; Kim, S.D.; Depreux, F.F.S.; Zhu, H.; Zon, L.; Pizard, A.; Kim, J.B.; A MacRae, C.; et al. Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss. Nat. Genet. 2005, 37, 418–422. [CrossRef]
- Williams, T.; Hundertmark, M.; Nordbeck, P.; Voll, S.; Muehlfelder, M.; Schraut, S.; Elsner, I.; Schoenberger, J.; Ritter, O. Abstract 080: Eya4 Induces Hypertrophy Via Regulation Of p27kip1. Circ Cardiovasc Genet. 2013, 113. [CrossRef]
- Ushijima, T.; Fujimoto, N.; Matsuyama, S.; Kan-O, M.; Kiyonari, H.; Shioi, G.; Kage, Y.; Yamasaki, S.; Takeya, R.; Sumimoto, H. The actin-organizing formin protein Fhod3 is required for postnatal development and functional maintenance of the adult heart in mice. J. Biol. Chem. 2018, 293, 148–162. [CrossRef]
- Ochoa, J.P.; Sabater-Molina, M.; García-Pinilla, J.M.; Mogensen, J.; Restrepo-Córdoba, A.; Palomino-Doza, J.; Villacorta, E.; Martinez-Moreno, M.; Ramos-Maqueda, J.; Zorio, E.; et al. Formin Homology 2 Domain Containing 3 (FHOD3) Is a Genetic Basis for Hypertrophic Cardiomyopathy. J Am Coll Cardiol. 2018, 72, 2457–2467. [CrossRef]
- Vodnjov, N.; Toplišek, J.; Maver, A.; Čuturilo, G.; Jaklič, H.; Teran, N.; Višnjar, T.; Pušenjak, M..; Hodžić, A.; Miljanović, O.; et al. A novel splice-site FHOD3 founder variant is a common cause of hypertrophic cardiomyopathy in the population of the Balkans–A cohort study. PLOS ONE 2023, 18, e0294969. [CrossRef]
- Wu, G.; Ruan, J.; Liu, J.; Zhang, C.; Kang, L.; Wang, J.; Zou, Y.; Song, L. Variant Spectrum of Formin Homology 2 Domain-Containing 3 Gene in Chinese Patients With Hypertrophic Cardiomyopathy. J. Am. Hear. Assoc. 2021, 10, e018236. [CrossRef]
- Ochoa, J.P.; Lopes, L.R.; Perez-Barbeito, M.; Cazón-Varela, L.; de la Torre-Carpente, M.M.; Sonicheva-Paterson, N.; De Uña-Iglesias, D.; Quinn, E.; Kuzmina-Krutetskaya, S.S.; Garrote, J.A.; et al. Deletions of specific exons of FHOD3 detected by next-generation sequencing are associated with hypertrophic cardiomyopathy. Clin. Genet. 2020, 98, 86–90. [CrossRef]
- Semsarian, C.; Ingles, J.; Bagnall, R.D. Revisiting Genome Sequencing Data in Light of Novel Disease Gene Associations. Journal of the American College of Cardiology 2019, 73, 1365–1366. [CrossRef]
- Lesurf, R.; Said, A.; Akinrinade, O.; Breckpot, J.; Delfosse, K.; Liu, T.; Yao, R.; Persad, G.; McKenna, F.; Noche, R.R.; et al. Whole genome sequencing delineates regulatory, copy number, and cryptic splice variants in early onset cardiomyopathy. npj Genom. Med. 2022, 7, 1–17. [CrossRef]
- Matsuyama, S.; Kage, Y.; Fujimoto, N.; Ushijima, T.; Tsuruda, T.; Kitamura, K.; Shiose, A.; Asada, Y.; Sumimoto, H.; Takeya, R. Interaction between cardiac myosin-binding protein C and formin Fhod3. Proc. Natl. Acad. Sci. 2018, 115, E4386–E4395. [CrossRef]
- Vermeulen, M.; Eberl, H.C.; Matarese, F.; Marks, H.; Denissov, S.; Butter, F.; Lee, K.K.; Olsen, J.V.; Hyman, A.A.; Stunnenberg, H.G.; et al. Quantitative Interaction Proteomics and Genome-wide Profiling of Epigenetic Histone Marks and Their Readers. Cell 2010, 142, 967–980. [CrossRef]
- Kaneda, R.; Takada, S.; Yamashita, Y.; Choi, Y.L.; Nonaka-Sarukawa, M.; Soda, M.; Misawa, Y.; Isomura, T.; Shimada, K.; Mano, H. Genome-wide histone methylation profile for heart failure. Genes Cells 2008, 14, 69–77. [CrossRef]
- Theis, J.L.; Sharpe, K.M.; Matsumoto, M.E.; Chai, H.S.; Nair, A.A.; Theis, J.D.; de Andrade, M.; Wieben, E.D.; Michels, V.V.; Olson, T.M. Homozygosity Mapping and Exome Sequencing Reveal GATAD1 Mutation in Autosomal Recessive Dilated Cardiomyopathy. Circ. Cardiovasc. Genet. 2011, 4, 585–594. [CrossRef]
- Yang, J.; Shah, S.; Olson, T.M.; Xu, X. Modeling GATAD1-Associated Dilated Cardiomyopathy in Adult Zebrafish. J. Cardiovasc. Dev. Dis. 2016, 3, 6. [CrossRef]
- Bendig, G.; Grimmler, M.; Huttner, I.G.; Wessels, G.; Dahme, T.; Just, S.; Trano, N.; Katus, H.A.; Fishman, M.C.; Rottbauer, W. Integrin-linked kinase, a novel component of the cardiac mechanical stretch sensor, controls contractility in the zebrafish heart. Genes Dev. 2006, 20, 2361–2372. [CrossRef]
- Knöll, R.; Postel, R.; Wang, J.; Krätzner, R.; Hennecke, G.; Vacaru, A.M.; Vakeel, P.; Schubert, C.; Murthy, K.; Rana, B.K.; et al. Laminin-α4 and Integrin-Linked Kinase Mutations Cause Human Cardiomyopathy Via Simultaneous Defects in Cardiomyocytes and Endothelial Cells. Circulation 2007, 116, 515–525. [CrossRef]
- Gu, R.; Bai, J.; Ling, L.; Ding, L.; Zhang, N.; Ye, J.; Ferro, A.; Xu, B. Increased Expression of Integrin-Linked Kinase Improves Cardiac Function and Decreases Mortality in Dilated Cardiomyopathy Model of Rats. PLOS ONE 2012, 7, e31279. [CrossRef]
- Sopko, N.; Qin, Y.; Finan, A.; Dadabayev, A.; Chigurupati, S.; Qin, J.; Penn, M.S.; Gupta, S. Significance of Thymosin β4 and Implication of PINCH-1-ILK-α-Parvin (PIP) Complex in Human Dilated Cardiomyopathy. PLOS ONE 2011, 6, e20184. [CrossRef]
- Roura, S.; Gálvez-Montón, C.; Pujal, J.M.; Casani, L.; Fernández, M.A.; Astier, L.; Gastelurrutia, P.; Domingo, M.; Prat-Vidal, C.; Soler-Botija, C.; et al. New insights into lipid raft function regulating myocardial vascularization competency in human idiopathic dilated cardiomyopathy. Atherosclerosis 2013, 230, 354–364. [CrossRef]
- Lehnart, S.E.; Wehrens, X.H.T. The role of junctophilin proteins in cellular function. Physiol. Rev. 2022, 102, 1211–1261. [CrossRef]
- Valtonen, J.; Prajapati, C.; Cherian, R.M.; Vanninen, S.; Ojala, M.; Leivo, K.; Heliö, T.; Koskenvuo, J.; Aalto-Setälä, K. The Junctophilin-2 Mutation p.(Thr161Lys) Is Associated with Hypertrophic Cardiomyopathy Using Patient-Specific iPS Cardiomyocytes and Demonstrates Prolonged Action Potential and Increased Arrhythmogenicity. Biomedicines 2023, 11, 1558. [CrossRef]
- Vanninen, S.U.M.; Leivo, K.; Seppälä, E.H.; Aalto-Setälä, K.; Pitkänen, O.; Suursalmi, P.; Annala, A.-P.; Anttila, I.; Alastalo, T.-P.; Myllykangas, S.; et al. Heterozygous junctophilin-2 (JPH2) p.(Thr161Lys) is a monogenic cause for HCM with heart failure. PLOS ONE 2018, 13, e0203422. [CrossRef]
- Landstrom, A.P.; Weisleder, N.; Batalden, K.B.; Bos, J.M.; Tester, D.J.; Ommen, S.R.; Wehrens, X.H.; Claycomb, W.C.; Ko, J.-K.; Hwang, M.; et al. Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J. Mol. Cell. Cardiol. 2007, 42, 1026–1035. [CrossRef]
- Parker, L.E.; Kramer, R.J.; Kaplan, S.; Landstrom, A.P. One gene, two modes of inheritance, four diseases: A systematic review of the cardiac manifestation of pathogenic variants in JPH2-encoded junctophilin-2. Trends Cardiovasc. Med. 2022, 33, 1–10. [CrossRef]
- Lahiri, S.K.; Jin, F.; Zhou, Y.; Quick, A.P.; Kramm, C.F.; Wang, M.C.; Wehrens, X.H. Altered myocardial lipid regulation in junctophilin-2–associated familial cardiomyopathies. Life Sci. Alliance 2024, 7, e202302330. [CrossRef]
- Wu T, Li X, Jia X, Zhu Z, Lu J, Feng H, et al. Krüppel like factor 10 prevents intervertebral disc degeneration via TGF-β signaling pathway both and. J Orthop Translat. 2021;29: 19–29. [CrossRef]
- Bos JM, Subramaniam M, Hawse JR, Christiaans I, Rajamannan NM, Maleszewski JJ, et al. TGFβ-inducible early gene-1 (TIEG1) mutations in hypertrophic cardiomyopathy. J Cell Biochem. 2012;113: 1896–1903. [CrossRef]
- Al-Hassnan, Z.N.; Almesned, A.; Tulbah, S.; Alakhfash, A.; Alhadeq, F.; Alruwaili, N.; Alkorashy, M.; Alhashem, A.; Alrashdan, A.; Faqeih, E.; et al. Categorized Genetic Analysis in Childhood-Onset Cardiomyopathy. Circ. Genom. Precis. Med. 2020, 13, 504–514. [CrossRef]
- Maurer, C.; Boleti, O.; Torbati, P.N.; Norouzi, F.; Fowler, A.N.R.; Minaee, S.; Salih, K.H.; Taherpour, M.; Birjandi, H.; Alizadeh, B.; et al. Genetic Insights from Consanguineous Cardiomyopathy Families. Genes 2023, 14, 182. [CrossRef]
- Lin, Z.; Li, S.; Feng, C.; Yang, S.; Wang, H.; Ma, D.; Zhang, J.; Gou, M.; Bu, D.; Zhang, T.; et al. Stabilizing mutations of KLHL24 ubiquitin ligase cause loss of keratin 14 and human skin fragility. Nat. Genet. 2016, 48, 1508–1516. [CrossRef]
- Schwieger-Briel, A.; Fuentes, I.; Castiglia, D.; Barbato, A.; Greutmann, M.; Leppert, J.; Duchatelet, S.; Hovnanian, A.; Burattini, S.; Yubero, M.J.; et al. Epidermolysis Bullosa Simplex with KLHL24 Mutations Is Associated with Dilated Cardiomyopathy. J. Investig. Dermatol. 2018, 139, 244–249. [CrossRef]
- Walsh, R.; Offerhaus, J.A.; Tadros, R.; Bezzina, C.R. Minor hypertrophic cardiomyopathy genes, major insights into the genetics of cardiomyopathies. Nat. Rev. Cardiol. 2021, 19, 151–167. [CrossRef]
- Wang, J.; Hoshijima, M.; Lam, J.; Zhou, Z.; Jokiel, A.; Dalton, N.D.; Hultenby, K.; Ruiz-Lozano, P.; Ross, J.; Tryggvason, K.; et al. Cardiomyopathy Associated with Microcirculation Dysfunction in Laminin α4 Chain-deficient Mice. J. Biol. Chem. 2006, 281, 213–220. [CrossRef]
- Lopez-Ayala, J.M.; Ortiz-Genga, M.; Gomez-Milanes, I.; Lopez-Cuenca, D.; Ruiz-Espejo, F.; Sanchez-Munoz, J.J.; Oliva-Sandoval, M.J.; Monserrat, L.; Gimeno, J.R. A mutation in the Z-line Cypher/ZASP protein is associated with arrhythmogenic right ventricular cardiomyopathy. Clin. Genet. 2014, 88, 172–176. [CrossRef]
- Arimura, T.; Hayashi, T.; Terada, H.; Lee, S.-Y.; Zhou, Q.; Takahashi, M.; Ueda, K.; Nouchi, T.; Hohda, S.; Shibutani, M.; et al. A Cypher/ZASP Mutation Associated with Dilated Cardiomyopathy Alters the Binding Affinity to Protein Kinase C. J. Biol. Chem. 2004, 279, 6746–6752. [CrossRef]
- Vatta M, Mohapatra B, Jimenez S, Sanchez X, Faulkner G, Perles Z, et al. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J Am Coll Cardiol. 2003;42: 2014–2027.
- Zheng, M.; Cheng, H.; Li, X.; Zhang, J.; Cui, L.; Ouyang, K.; Han, L.; Zhao, T.; Gu, Y.; Dalton, N.D.; et al. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum. Mol. Genet. 2008, 18, 701–713. [CrossRef]
- van der Meer, D.L.; Marques, I.J.; Leito, J.T.; Besser, J.; Bakkers, J.; Schoonheere, E.; Bagowski, C.P. Zebrafish cypher is important for somite formation and heart development. Dev. Biol. 2006, 299, 356–372. [CrossRef]
- Koopmann, T.T.; Jamshidi, Y.; Naghibi-Sistani, M.; van der Klift, H.M.; Birjandi, H.; Al-Hassnan, Z.; Alwadai, A.; Zifarelli, G.; Karimiani, E.G.; Sedighzadeh, S.; et al. Biallelic loss of LDB3 leads to a lethal pediatric dilated cardiomyopathy. Eur. J. Hum. Genet. 2022, 31, 97–104. [CrossRef]
- Xuan, T.; Wang, D.; Lv, J.; Pan, Z.; Fang, J.; Xiang, Y.; Cheng, H.; Wang, X.; Guo, X. Downregulation of Cypher induces apoptosis in cardiomyocytes via Akt/p38 MAPK signaling pathway. Int. J. Med Sci. 2020, 17, 2328–2337. [CrossRef]
- Schiaffino, S.; Reggiani, C. Molecular diversity of myofibrillar proteins: gene regulation and functional significance. Physiol. Rev. 1996, 76, 371–423. [CrossRef]
- Niimura, H.; Patton, K.K.; McKenna, W.J.; Soults, J.; Maron, B.J.; Seidman, J.; Seidman, C.E. Sarcomere Protein Gene Mutations in Hypertrophic Cardiomyopathy of the Elderly. Circulation 2002, 105, 446–451. [CrossRef]
- Reiser, P.J.; Portman, M.A.; Ning, X.-H.; Moravec, C.S. Human cardiac myosin heavy chain isoforms in fetal and failing adult atria and ventricles. Am J Physiol Heart Circ Physiol. 2001, 280, H1814–H1820. [CrossRef]
- Carniel E, Taylor MRG, Sinagra G, Di Lenarda A, Ku L, Fain PR, et al. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation. 2005;112: 54–59. [CrossRef]
- Hershberger RE, Norton N, Morales A, Li D, Siegfried JD, Gonzalez-Quintana J. Coding sequence rare variants identified in MYBPC3, MYH6, TPM1, TNNC1, and TNNI3 from 312 patients with familial or idiopathic dilated cardiomyopathy. Circ Cardiovasc Genet. 2010;3: 155–161. [CrossRef]
- Zhao T, Ma Y, Zhang Z, Xian J, Geng X, Wang F, et al. Young and early-onset dilated cardiomyopathy with malignant ventricular arrhythmia and sudden cardiac death induced by the heterozygous LDB3, MYH6, and SYNE1 missense mutations. Ann Noninvasive Electrocardiol. 2021;26: e12840. [CrossRef]
- Ntelios, D.; Meditskou, S.; Efthimiadis, G.; Pitsis, A.; Zegkos, T.; Parcharidou, D.; Theotokis, P.; Alexouda, S.; Karvounis, H.; Tzimagiorgis, G. α-Myosin heavy chain (MYH6) in hypertrophic cardiomyopathy: Prominent expression in areas with vacuolar degeneration of myocardial cells. Pathol. Int. 2022, 72, 308–310. [CrossRef]
- Klos, M.; Mundada, L.; Banerjee, I.; Morgenstern, S.; Myers, S.; Leone, M.; Kleid, M.; Herron, T.; Devaney, E. Altered myocyte contractility and calcium homeostasis in alpha-myosin heavy chain point mutations linked to familial dilated cardiomyopathy. Arch. Biochem. Biophys. 2017, 615, 53–60. [CrossRef]
- Davis, J.S.; Hassanzadeh, S.; Winitsky, S.; Lin, H.; Satorius, C.; Vemuri, R.; Aletras, A.H.; Wen, H.; Epstein, N.D. The Overall Pattern of Cardiac Contraction Depends on a Spatial Gradient of Myosin Regulatory Light Chain Phosphorylation. Cell 2001, 107, 631–641. [CrossRef]
- Qin, X.; Li, P.; Qu, H.-Q.; Liu, Y.; Xia, Y.; Chen, S.; Yang, Y.; Huang, S.; Wen, P.; Zhou, X.; et al. FLNC and MYLK2 Gene Mutations in a Chinese Family with Different Phenotypes of Cardiomyopathy. Int. Hear. J. 2021, 62, 127–134. [CrossRef]
- Burstein, D.S.; Gaynor, J.W.; Griffis, H.; Ritter, A.; Connor, M.J.O.; Rossano, J.W.; Lin, K.Y.; Ahrens-Nicklas, R.C. Genetic variant burden and adverse outcomes in pediatric cardiomyopathy. Pediatr. Res. 2020, 89, 1470–1476. [CrossRef]
- Lange, S.; Himmel, M.; Auerbach, D.; Agarkova, I.; Hayess, K.; Fürst, D.O.; Perriard, J.-C.; Ehler, E. Dimerisation of Myomesin: Implications for the Structure of the Sarcomeric M-band. J. Mol. Biol. 2004, 345, 289–298. [CrossRef]
- Siegert, R.; Perrot, A.; Keller, S.; Behlke, J.; Michalewska-Włudarczyk, A.; Wycisk, A.; Tendera, M.; Morano, I.; Özcelik, C. A myomesin mutation associated with hypertrophic cardiomyopathy deteriorates dimerisation properties. Biochem. Biophys. Res. Commun. 2011, 405, 473–479. [CrossRef]
- Cecconi, M.; Parodi, M.I.; Formisano, F.; Spirito, P.; Autore, C.; Musumeci, M.B.; Favale, S.; Forleo, C.; Rapezzi, C.; Biagini, E.; et al. Targeted next-generation sequencing helps to decipher the genetic and phenotypic heterogeneity of hypertrophic cardiomyopathy. Int. J. Mol. Med. 2016, 38, 1111–1124. [CrossRef]
- Bottillo, I.; D’Angelantonio, D.; Caputo, V.; Paiardini, A.; Lipari, M.; De Bernardo, C.; Giannarelli, D.; Pizzuti, A.; Majore, S.; Castori, M.; et al. Molecular analysis of sarcomeric and non-sarcomeric genes in patients with hypertrophic cardiomyopathy. Gene 2015, 577, 227–235. [CrossRef]
- Guo, X.; Fan, C.; Tian, L.; Liu, Y.; Wang, H.; Zhao, S.; Duan, F.; Zhang, X.; Zhao, X.; Wang, F.; et al. The clinical features, outcomes and genetic characteristics of hypertrophic cardiomyopathy patients with severe right ventricular hypertrophy. PLOS ONE 2017, 12, e0174118. [CrossRef]
- Gontier, Y.; Taivainen, A.; Fontao, L.; Sonnenberg, A.; van der Flier, A.; Carpen, O.; Faulkner, G.; Borradori, L. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. J. Cell Sci. 2005, 118, 3739–3749. [CrossRef]
- Ivandic, B.T.; Mastitsky, S.E.; Schönsiegel, F.; Bekeredjian, R.; Eils, R.; Frey, N.; Katus, H.A.; Brors, B. Whole-genome analysis of gene expression associates the ubiquitin-proteasome system with the cardiomyopathy phenotype in disease-sensitized congenic mouse strains. Cardiovasc. Res. 2012, 94, 87–95. [CrossRef]
- Osio, A.; Tan, L.; Chen, S.N.; Lombardi, R.; Nagueh, S.F.; Shete, S.; Roberts, R.; Willerson, J.T.; Marian, A.J. Myozenin 2 Is a Novel Gene for Human Hypertrophic Cardiomyopathy. Circ. Res. 2007, 100, 766–768. [CrossRef]
- Guo, X.; Fan, C.; Wang, Y.; Wang, M.; Cai, C.; Yang, Y.; Zhao, S.; Duan, F.; Li, Y. Genetic anticipation in a special form of hypertrophic cardiomyopathy with sudden cardiac death in a family with 74 members across 5 generations. Medicine 2017, 96, e6249. [CrossRef]
- Ruggiero, A.; Chen, S.N.; Lombardi, R.; Rodriguez, G.; Marian, A.J. Pathogenesis of hypertrophic cardiomyopathy caused by myozenin 2 mutations is independent of calcineurin activity. Cardiovasc. Res. 2012, 97, 44–54. [CrossRef]
- Otey, C.A.; Dixon, R.; Stack, C.; Goicoechea, S.M. Cytoplasmic Ig-domain proteins: Cytoskeletal regulators with a role in human disease. Cell Motil. Cytoskelet. 2009, 66, 618–634. [CrossRef]
- Bang, M.-L.; Mudry, R.E.; McElhinny, A.S.; Trombitás, K.; Geach, A.J.; Yamasaki, R.; Sorimachi, H.; Granzier, H.; Gregorio, C.C.; Labeit, S. Myopalladin, a Novel 145-Kilodalton Sarcomeric Protein with Multiple Roles in Z-Disc and I-Band Protein Assemblies. J. Cell Biol. 2001, 153, 413–428. [CrossRef]
- Purevjav, E.; Arimura, T.; Augustin, S.; Huby, A.-C.; Takagi, K.; Nunoda, S.; Kearney, D.L.; Taylor, M.D.; Terasaki, F.; Bos, J.M.; et al. Molecular basis for clinical heterogeneity in inherited cardiomyopathies due to myopalladin mutations. Hum. Mol. Genet. 2012, 21, 2039–2053. [CrossRef]
- Filomena, M.C.; Yamamoto, D.L.; Carullo, P.; Medvedev, R.; Ghisleni, A.; Piroddi, N.; Scellini, B.; Crispino, R.; D’Autilia, F.; Zhang, J.; et al. Myopalladin knockout mice develop cardiac dilation and show a maladaptive response to mechanical pressure overload. eLife 2021, 10. [CrossRef]
- Cronin, H.; Crinion, D.; Kerins, D.; Fahy, G.; Vaughan, C.J. Inferolateral T wave inversion in athletes: phenotype-genotype correlation. Ir. J. Med Sci. (1971 -) 2020, 189, 1283–1287. [CrossRef]
- Refaat, M.M.; Hassanieh, S.; Ballout, J.A.; Zakka, P.; Hotait, M.; Khalil, A.; Bitar, F.; Arabi, M.; Arnaout, S.; Skouri, H.; et al. Non-familial cardiomyopathies in Lebanon: exome sequencing results for five idiopathic cases. BMC Med Genom. 2019, 12, 1–11. [CrossRef]
- Hernandez, D.A.; Bennett, C.M.; Dunina-Barkovskaya, L.; Wedig, T.; Capetanaki, Y.; Herrmann, H.; Conover, G.M. Nebulette is a powerful cytolinker organizing desmin and actin in mouse hearts. Mol. Biol. Cell 2016, 27, 3869–3882. [CrossRef]
- Arimura, T.; Nakamura, T.; Hiroi, S.; Satoh, M.; Takahashi, M.; Ohbuchi, N.; Ueda, K.; Nouchi, T.; Yamaguchi, N.; Akai, J.; et al. Characterization of the human nebulette gene: a polymorphism in an actin-binding motif is associated with nonfamilial idiopathic dilated cardiomyopathy. Hum. Genet. 2000, 107, 440–451. [CrossRef]
- Maiellaro-Rafferty, K.; Wansapura, J.; Mendsaikhan, U.; Osinska, H.; James, J.; Taylor; Robbins, J.; Kranias, E.; Towbin, J.; Purevjav, E. Altered regional cardiac wall mechanics are associated with differential cardiomyocyte calcium handling due to nebulette mutations in preclinical inherited dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2013, 60, 151–160. [CrossRef]
- Purevjav, E.; Varela, J.; Morgado, M.; Kearney, D.L.; Li, H.; Taylor, M.D.; Arimura, T.; Moncman, C.L.; McKenna, W.; Murphy, R.T.; et al. Nebulette Mutations Are Associated With Dilated Cardiomyopathy and Endocardial Fibroelastosis. Circ. 2010, 56, 1493–1502. [CrossRef]
- Mastrototaro, G.; Liang, X.; Li, X.; Carullo, P.; Piroddi, N.; Tesi, C.; Gu, Y.; Dalton, N.D.; Peterson, K.L.; Poggesi, C.; et al. Nebulette knockout mice have normal cardiac function, but show Z-line widening and up-regulation of cardiac stress markers. Cardiovasc. Res. 2015, 107, 216–225. [CrossRef]
- Perrot, A.; Tomasov, P.; Villard, E.; Faludi, R.; Melacini, P.; Lossie, J.; Lohmann, N.; Richard, P.; De Bortoli, M.; Angelini, A.; et al. Mutations in NEBL encoding the cardiac Z-disk protein nebulette are associated with various cardiomyopathies. Arch. Med Sci. 2016, 2, 263–278. [CrossRef]
- Lee, A.; Hakuno, F.; Northcott, P.; Pessin, J.E.; Adcock, M.R. Nexilin, a Cardiomyopathy-Associated F-Actin Binding Protein, Binds and Regulates IRS1 Signaling in Skeletal Muscle Cells. PLOS ONE 2013, 8, e55634. [CrossRef]
- Chopra, N.; Knollmann, B.C. Genetics of sudden cardiac death syndromes. Curr. Opin. Cardiol. 2011, 26, 196–203. [CrossRef]
- Liu, C.; Spinozzi, S.; Chen, J.-Y.; Fang, X.; Feng, W.; Perkins, G.; Cattaneo, P.; Guimarães-Camboa, N.; Dalton, N.D.; Peterson, K.L.; et al. Nexilin Is a New Component of Junctional Membrane Complexes Required for Cardiac T-Tubule Formation. Circulation 2019, 140, 55–66. [CrossRef]
- Spinozzi S, Liu C, Chen Z ’e, Feng W, Zhang L, Ouyang K, et al. Nexilin Is Necessary for Maintaining the Transverse-Axial Tubular System in Adult Cardiomyocytes. Circ Heart Fail. 2020;13: e006935. [CrossRef]
- Aherrahrou, Z.; Schlossarek, S.; Stoelting, S.; Klinger, M.; Geertz, B.; Weinberger, F.; Kessler, T.; Aherrahrou, R.; Moreth, K.; Bekeredjian, R.; et al. Knock-out of nexilin in mice leads to dilated cardiomyopathy and endomyocardial fibroelastosis. Basic Res. Cardiol. 2015, 111, 6. [CrossRef]
- Kean, A.C.; Helm, B.M.; Vatta, M.; Ayers, M.D.; Parent, J.J.; Darragh, R.K. Clinical characterisation of a novel SCN5A variant associated with progressive malignant arrhythmia and dilated cardiomyopathy. Cardiol. Young- 2019, 29, 1257–1263. [CrossRef]
- Zhang, X.-L.; Xie, J.; Lan, R.-F.; Kang, L.-N.; Wang, L.; Xu, W.; Xu, B. Genetic Basis and Genotype-Phenotype Correlations in Han Chinese Patients with Idiopathic Dilated Cardiomyopathy. Sci. Rep. 2020, 10, 1–8. [CrossRef]
- Bruyndonckx, L.; Vogelzang, J.L.; Bugiani, M.; Straver, B.; Kuipers, I.M.; Onland, W.; Nannenberg, E.A.; Clur, S.; van der Crabben, S.N.&. Childhood onset nexilin dilated cardiomyopathy: A heterozygous and a homozygous case. Am. J. Med Genet. Part A 2021, 185, 2464–2470. [CrossRef]
- Johansson, J.; Frykholm, C.; Ericson, K.; Kazamia, K.; Lindberg, A.; Mulaiese, N.; Falck, G.; Gustafsson, P.; Lidéus, S.; Gudmundsson, S.; et al. Loss of Nexilin function leads to a recessive lethal fetal cardiomyopathy characterized by cardiomegaly and endocardial fibroelastosis. Am. J. Med Genet. Part A 2022, 188, 1676–1687. [CrossRef]
- Hermida, A.; Ader, F.; Millat, G.; Jedraszak, G.; Maury, P.; Cador, R.; Catalan, P.-A.; Clerici, G.; Combes, N.; De Groote, P.; et al. NEXN Gene in Cardiomyopathies and Sudden Cardiac Deaths: Prevalence, Phenotypic Expression, and Prognosis. Circ. Genom. Precis. Med. 2024, 17, e004285. [CrossRef]
- Schott, J.-J.; Benson, D.W.; Basson, C.T.; Pease, W.; Silberbach, G.M.; Moak, J.P.; Maron, B.J.; Seidman, C.E.; Seidman, J.G. Congenital Heart Disease Caused by Mutations in the Transcription Factor NKX2-5. Science 1998, 281, 108–111. [CrossRef]
- Jay PY, Harris BS, Maguire CT, Buerger A, Wakimoto H, Tanaka M, et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113: 1130–1137.
- Brody, M.J.; Cho, E.; Mysliwiec, M.R.; Kim, T.-G.; Carlson, C.D.; Lee, K.-H.; Lee, Y. Lrrc10 is a novel cardiac-specific target gene of Nkx2-5 and GATA4. J. Mol. Cell. Cardiol. 2013, 62, 237–246. [CrossRef]
- McElhinney, D.B.; Geiger, E.; Blinder, J.; Benson, D.W.; Goldmuntz, E. NKX2.5mutations in patients with congenital heart disease. J Am Coll Cardiol. 2003, 42, 1650–1655. [CrossRef]
- Hanley, A.; Walsh, K.A.; Joyce, C.; McLellan, M.A.; Clauss, S.; Hagen, A.; Shea, M.A.; Tucker, N.R.; Lin, H.; Fahy, G.J.; et al. Mutation of a common amino acid in NKX2.5 results in dilated cardiomyopathy in two large families. BMC Med Genet. 2016, 17, 1–7. [CrossRef]
- Costa, M.W.; Guo, G.; Wolstein, O.; Vale, M.; Castro, M.L.; Wang, L.; Otway, R.; Riek, P.; Cochrane, N.; Furtado, M.; et al. Functional Characterization of a Novel Mutation in NKX2-5 Associated With Congenital Heart Disease and Adult-Onset Cardiomyopathy. Circ. Cardiovasc. Genet. 2013, 6, 238–247. [CrossRef]
- Yuan, F.; Qiu, X.-B.; Li, R.-G.; Qu, X.-K.; Wang, J.; Xu, Y.-J.; Liu, X.; Fang, W.-Y.; Yang, Y.-Q.; Liao, D.-N. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int. J. Mol. Med. 2014, 35, 478–486. [CrossRef]
- Xu, J.-H.; Gu, J.-Y.; Guo, Y.-H.; Zhang, H.; Qiu, X.-B.; Li, R.-G.; Shi, H.-Y.; Liu, H.; Yang, X.-X.; Xu, Y.-J.; et al. Prevalence and Spectrum of NKX2-5 Mutations Associated With Sporadic Adult-Onset Dilated Cardiomyopathy. Int. Hear. J. 2017, 58, 521–529. [CrossRef]
- Sveinbjornsson G, Olafsdottir EF, Thorolfsdottir RB, Davidsson OB, Helgadottir A, Jonasdottir A, et al. Variants in NKX2-5 and FLNC Cause Dilated Cardiomyopathy and Sudden Cardiac Death. Circ Genom Precis Med. 2018;11: e002151. [CrossRef]
- Bang, M.-L.; Centner, T.; Fornoff, F.; Geach, A.J.; Gotthardt, M.; McNabb, M.; Witt, C.C.; Labeit, D.; Gregorio, C.C.; Granzier, H.; et al. The Complete Gene Sequence of Titin, Expression of an Unusual ≈700-kDa Titin Isoform, and Its Interaction With Obscurin Identify a Novel Z-Line to I-Band Linking System. Circ. Res. 2001, 89, 1065–1072. [CrossRef]
- Borisov, A.B.; Sutter, S.B.; Kontrogianni-Konstantopoulos, A.; Bloch, R.J.; Westfall, M.V.; Russell, M.W. Essential role of obscurin in cardiac myofibrillogenesis and hypertrophic response: evidence from small interfering RNA-mediated gene silencing. Histochem Cell Biol. 2005, 125, 227–238. [CrossRef]
- Hu, L.-Y.R.; Kontrogianni-Konstantopoulos, A. The kinase domains of obscurin interact with intercellular adhesion proteins. FASEB J. 2013, 27, 2001–2012. [CrossRef]
- Marston, S. Obscurin variants and inherited cardiomyopathies. Biophys. Rev. 2017, 9, 239–243. [CrossRef]
- Arimura, T.; Matsumoto, Y.; Okazaki, O.; Hayashi, T.; Takahashi, M.; Inagaki, N.; Hinohara, K.; Ashizawa, N.; Yano, K.; Kimura, A. Structural analysis of obscurin gene in hypertrophic cardiomyopathy. Biochem. Biophys. Res. Commun. 2007, 362, 281–287. [CrossRef]
- Marston S, Montgiraud C, Munster AB, Copeland O ’neal, Choi O, Dos Remedios C, et al. OBSCN Mutations Associated with Dilated Cardiomyopathy and Haploinsufficiency. PLoS One. 2015;10: e0138568.
- Xia, H.; Winokur, S.T.; Kuo, W.-L.; Altherr, M.R.; Bredt, D.S. Actinin-associated LIM Protein: Identification of a Domain Interaction between PDZ and Spectrin-like Repeat Motifs. J. Cell Biol. 1997, 139, 507–515. [CrossRef]
- Lorenzen-Schmidt, I.; McCulloch, A.D.; Omens, J.H. Deficiency of Actinin-Associated LIM Protein Alters Regional Right Ventricular Function and Hypertrophic Remodeling. Ann. Biomed. Eng. 2005, 33, 888–896. [CrossRef]
- Bagnall, R.D.; Yeates, L.; Semsarian, C. Analysis of the Z-disc genes PDLIM3 and MYPN in Patients with Hypertrophic Cardiomyopathy. Int. J. Cardiol. 2010, 145, 601–602. [CrossRef]
- Lopes, L.; Murphy, C.; Syrris, P.; Dalageorgou, C.; McKenna, W.; Elliott, P.; Plagnol, V. Use of high-throughput targeted exome-sequencing to screen for copy number variation in hypertrophic cardiomyopathy. Eur. J. Med Genet. 2015, 58, 611–616. [CrossRef]
- Rosa-Ferreira, C.; Munro, S. Arl8 and SKIP Act Together to Link Lysosomes to Kinesin-1. Dev. Cell 2011, 21, 1171–1178. [CrossRef]
- Khatter, D.; Raina, V.B.; Dwivedi, D.; Sindhwani, A.; Bahl, S.; Sharma, M. The small GTPase Arl8b regulates assembly of the mammalian HOPS complex to lysosomes. J. Cell Sci. 2015, 128, 1746–1761. [CrossRef]
- Almacellas, E.; Pelletier, J.; Day, C.; Ambrosio, S.; Tauler, A.; Mauvezin, C. Lysosomal degradation ensures accurate chromosomal segregation to prevent chromosomal instability. Autophagy 2020, 17, 796–813. [CrossRef]
- Muhammad E, Levitas A, Singh SR, Braiman A, Ofir R, Etzion S, et al. PLEKHM2 mutation leads to abnormal localization of lysosomes, impaired autophagy flux and associates with recessive dilated cardiomyopathy and left ventricular noncompaction. Hum Mol Genet. 2015;24: 7227–7240. [CrossRef]
- Cibi, D.M.; Bi-Lin, K.W.; Shekeran, S.G.; Sandireddy, R.; Tee, N.; Singh, A.; Wu, Y.; Srinivasan, D.K.; Kovalik, J.-P.; Ghosh, S.; et al. Prdm16 Deficiency Leads to Age-Dependent Cardiac Hypertrophy, Adverse Remodeling, Mitochondrial Dysfunction, and Heart Failure. Cell Rep. 2020, 33, 108288. [CrossRef]
- Bjork, B.C.; Turbe-Doan, A.; Prysak, M.; Herron, B.J.; Beier, D.R. Prdm16 is required for normal palatogenesis in mice. Hum. Mol. Genet. 2009, 19, 774–789. [CrossRef]
- Liu, C.; Shao, N.-Y. The Differences in the Developmental Stages of the Cardiomyocytes and Endothelial Cells in Human and Mouse Embryos at the Single-Cell Level. Int. J. Mol. Sci. 2024, 25, 3240. [CrossRef]
- Arndt, A.-K.; Schafer, S.; Drenckhahn, J.-D.; Sabeh, M.K.; Plovie, E.R.; Caliebe, A.; Klopocki, E.; Musso, G.; Werdich, A.A.; Kalwa, H.; et al. Fine Mapping of the 1p36 Deletion Syndrome Identifies Mutation of PRDM16 as a Cause of Cardiomyopathy. Am. J. Hum. Genet. 2013, 93, 67–77. [CrossRef]
- Long, P.A.; Evans, J.M.; Olson, T.M. Diagnostic Yield of Whole Exome Sequencing in Pediatric Dilated Cardiomyopathy. J. Cardiovasc. Dev. Dis. 2017, 4, 11. [CrossRef]
- Li, D.; Parks, S.B.; Kushner, J.D.; Nauman, D.; Burgess, D.; Ludwigsen, S.; Partain, J.; Nixon, R.R.; Allen, C.N.; Irwin, R.P.; et al. Mutations of Presenilin Genes in Dilated Cardiomyopathy and Heart Failure. Am. J. Hum. Genet. 2006, 79, 1030–1039. [CrossRef]
- Takeda, T.; Asahi, M.; Yamaguchi, O.; Hikoso, S.; Nakayama, H.; Kusakari, Y.; Kawai, M.; Hongo, K.; Higuchi, Y.; Kashiwase, K.; et al. Presenilin 2 regulates the systolic function of heart by modulating Ca2+signaling. FASEB J. 2005, 19, 2069–2071. [CrossRef]
- Sciarretta, S.; Forte, M.; Frati, G.; Sadoshima, J. New Insights Into the Role of mTOR Signaling in the Cardiovascular System. Circ. Res. 2018, 122, 489–505. [CrossRef]
- Jain, P.K.; Jayappa, S.; Sairam, T.; Mittal, A.; Paul, S.; Rao, V.J.; Chittora, H.; Kashyap, D.K.; Palakodeti, D.; Thangaraj, K.; et al. Ribosomal protein S6 kinase beta-1 gene variants cause hypertrophic cardiomyopathy. J. Med Genet. 2021, 59, 984–992. [CrossRef]
- Grossman, W. The role of sarcoplasmic reticulum proteins in heart disease: introduction.. Ann. New York Acad. Sci. 1998, 853, 207–208. [CrossRef]
- Yano M, Ikeda Y, Matsuzaki M. Altered intracellular Ca2+ handling in heart failure. J Clin Invest. 2005;115: 556–564.
- Kohno, M.; Kobayashi, S.; Yamamoto, T.; Yoshitomi, R.; Kajii, T.; Fujii, S.; Nakamura, Y.; Kato, T.; Uchinoumi, H.; Oda, T.; et al. Enhancing calmodulin binding to cardiac ryanodine receptor completely inhibits pressure-overload induced hypertrophic signaling. Commun. Biol. 2020, 3, 1–15. [CrossRef]
- Stanczyk, P.J.; Seidel, M.; White, J.; Viero, C.; George, C.H.; Zissimopoulos, S.; Lai, F.A. Association of cardiac myosin-binding protein-C with the ryanodine receptor channel – putative retrograde regulation?. J. Cell Sci. 2018, 131, jcs210443. [CrossRef]
- Fujino N, Ino H, Hayashi K, Uchiyama K, Nagata M, Konno T, et al. Abstract 915: A Novel Missense Mutation in Cardiac Ryanodine Receptor Gene as a Possible Cause of Hypertrophic Cardiomyopathy: Evidence From Familial Analysis. Circulation. 2006 [cited 17 Jul 2024]. Available: https://www.ahajournals.org/doi/10.1161/circ.114.suppl_18.II_165-b.
- Tang, Y.; Tian, X.; Wang, R.; Fill, M.; Chen, S.W. Abnormal Termination of Ca 2+ Release Is a Common Defect of RyR2 Mutations Associated With Cardiomyopathies. Circ. Res. 2012, 110, 968–977. [CrossRef]
- Xu, J.; Li, Z.; Ren, X.; Dong, M.; Li, J.; Shi, X.; Zhang, Y.; Xie, W.; Sun, Z.; Liu, X.; et al. Investigation of Pathogenic Genes in Chinese sporadic Hypertrophic Cardiomyopathy Patients by Whole Exome Sequencing. Sci. Rep. 2015, 5, 16609–16609. [CrossRef]
- Alvarado, F.J.; Bos, J.M.; Yuchi, Z.; Valdivia, C.R.; Hernández, J.J.; Zhao, Y.-T.; Henderlong, D.S.; Chen, Y.; Booher, T.R.; Marcou, C.A.; et al. Cardiac hypertrophy and arrhythmia in mice induced by a mutation in ryanodine receptor 2. JCI Insight. 2019, 4. [CrossRef]
- Krishnaswamy, S.M.; Arunachal, G.; Singh, K.G.; Thomson, V.S.; George, P.; Rao, S.; Danda, S. Investigation of mutation spectrum amongst patients with familial primary cardiomyopathy using targeted NGS in Indian population. J. Appl. Genet. 2024, 1–14. [CrossRef]
- Ervasti, J.M.; Campbell, K.P. Membrane organization of the dystrophin-glycoprotein complex. Cell 1991, 66, 1121–1131. [CrossRef]
- Way M, Pope B, Cross RA, Kendrick-Jones J, Weeds AG. Expression of the N-terminal domain of dystrophin in E. coli and demonstration of binding to F-actin. FEBS Lett. 1992;301: 243–245. [CrossRef]
- Tsubata S, Bowles KR, Vatta M, Zintz C, Titus J, Muhonen L, et al. Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J Clin Invest. 2000;106: 655–662.
- Kärkkäinen S, Miettinen R, Tuomainen P, Kärkkäinen P, Heliö T, Reissell E, et al. A novel mutation, Arg71Thr, in the delta-sarcoglycan gene is associated with dilated cardiomyopathy. J Mol Med . 2003;81: 795–800. [CrossRef]
- Campbell, M.D.; Witcher, M.; Gopal, A.; Michele, D.E. Dilated cardiomyopathy mutations in δ-sarcoglycan exert a dominant-negative effect on cardiac myocyte mechanical stability. Am J Physiol Heart Circ Physiol. 2016, 310, H1140–H1150. [CrossRef]
- Kirk, E.P.; Sunde, M.; Costa, M.; Rankin, S.A.; Wolstein, O.; Castro, M.L.; Butler, T.L.; Hyun, C.; Guo, G.; Otway, R.; et al. Mutations in Cardiac T-Box Factor Gene TBX20 Are Associated with Diverse Cardiac Pathologies, Including Defects of Septation and Valvulogenesis and Cardiomyopathy. Am. J. Hum. Genet. 2007, 81, 280–291. [CrossRef]
- Qian, L.; Mohapatra, B.; Akasaka, T.; Liu, J.; Ocorr, K.; Towbin, J.A.; Bodmer, R. Transcription factor neuromancer/TBX20 is required for cardiac function in Drosophila with implications for human heart disease. Proc. Natl. Acad. Sci. 2008, 105, 19833–19838. [CrossRef]
- Chen, Y.; Xiao, D.; Zhang, L.; Cai, C.-L.; Li, B.-Y.; Liu, Y. The Role of Tbx20 in Cardiovascular Development and Function. Front. Cell Dev. Biol. 2021, 9. [CrossRef]
- Giri, P.; Mukhopadhyay, A.; Gupta, M.; Mohapatra, B. Dilated cardiomyopathy: a new insight into the rare but common cause of heart failure. Hear. Fail. Rev. 2021, 27, 431–454. [CrossRef]
- Zhao C-M, Bing-Sun, Song H-M, Wang J, Xu W-J, Jiang J-F, et al. TBX20 loss-of-function mutation associated with familial dilated cardiomyopathy. Clin Chem Lab Med. 2016;54: 325–332.
- Mittal, A.; Sharma, R.; Prasad, R.; Bahl, A.; Khullar, M. Role of cardiac TBX20 in dilated cardiomyopathy. Mol. Cell. Biochem. 2016, 414, 129–136. [CrossRef]
- Amor-Salamanca, A.; Rodríguez, A.S.; Rasoul, H.; Rodríguez-Palomares, J.F.; Moldovan, O.; Hey, T.M.; Delgado, M.G.; Cuenca, D.L.; Campos, D.d.C.; Basurte-Elorz, M.T.; et al. Role of TBX20 Truncating Variants in Dilated Cardiomyopathy and Left Ventricular Noncompaction. Circ. Genom. Precis. Med. 2024, 17, e004404–e004404. [CrossRef]
- Knöll, R.; Hoshijima, M.; Hoffman, H.M.; Person, V.; Lorenzen-Schmidt, I.; Bang, M.-L.; Hayashi, T.; Shiga, N.; Yasukawa, H.; Schaper, W.; et al. The Cardiac Mechanical Stretch Sensor Machinery Involves a Z Disc Complex that Is Defective in a Subset of Human Dilated Cardiomyopathy. Cell 2002, 111, 943–955. [CrossRef]
- Hayashi, T.; Arimura, T.; Itoh-Satoh, M.; Ueda, K.; Hohda, S.; Inagaki, N.; Takahashi, M.; Hori, H.; Yasunami, M.; Nishi, H.; et al. Tcap gene mutations in hypertrophic cardiomyopathy and dilated cardiomyopathy. Circ. 2004, 44, 2192–2201. [CrossRef]
- Martins E, Sousa A, Canedo P, Leite S, Pinto R, Campelo M, et al. Genetic variants identified by target next-generation sequencing in heart transplant patients with dilated cardiomyopathy. Rev Port Cardiol. 2019;38: 441–447. [CrossRef]
- Nguyen, T.V.; Vu, M.T.T.; Do, T.N.P.; Tran, T.H.N.; Huynh, B.N.T.; Le, L.A.; Pham, N.T.N.; Nguyen, T.M.N.; Le, N.H.P.; Nguyen, V.P.; et al. Genetic Determinants and Genotype-Phenotype Correlations in Vietnamese Patients With Dilated Cardiomyopathy. Circ. J. 2021, 85, 1469–1478. [CrossRef]
- Graycar JL, Miller DA, Arrick BA, Lyons RM, Moses HL, Derynck R. Human transforming growth factor-beta 3: recombinant expression, purification, and biological activities in comparison with transforming growth factors-beta 1 and -beta 2. Mol Endocrinol. 1989;3: 1977–1986. [CrossRef]
- Rampazzo, A.; Nava, A.; Danieli, G.A.; Buja, G.; Daliento, L.; Fasoli, G.; Scognamiglio, R.; Corrado, D.; Thlene, G. The gene for arrhythmogenic right ventricular cardiomyopathy maps to chromosome 14q23–q24. Hum. Mol. Genet. 1994, 3, 959–962. [CrossRef]
- Rampazzo, A.; Beffagna, G.; Nava, A.; Occhi, G.; Bauce, B.; Noiato, M.; Basso, C.; Frigo, G.; Thiene, G.; Towbin, J.; et al. Arrhythmogenic right ventricular cardiomyopathy type 1 (ARVD1): confirmation of locus assignment and mutation screening of four candidate genes. Eur. J. Hum. Genet. 2003, 11, 69–76. [CrossRef]
- Beffagna G, Occhi G, Nava A, Vitiello L, Ditadi A, Basso C, et al. Regulatory mutations in transforming growth factor-beta3 gene cause arrhythmogenic right ventricular cardiomyopathy type 1. Cardiovasc Res. 2005;65: 366–373. [CrossRef]
- Rampazzo A. Regulatory mutations in transforming growth factor-β3 gene involved in arrhythmogenic right ventricular cardiomyopathy: AUTHOR’S RETROSPECTIVE. Cardiovasc Res. 2012;96: 191–194. [CrossRef]
- Bao, J.-R.; Wang, J.-Z.; Yao, Y.; Wang, Y.-L.; Fan, X.-H.; Sun, K.; Zhang, S.; Hui, R.-T.; Song, L. Screening of pathogenic genes in Chinese patients with arrhythmogenic right ventricular cardiomyopathy. Chin Med J. 2013, 126, 4238–41. [CrossRef]
- Vermij, S.H.; Abriel, H.; van Veen, T.A.B. Refining the molecular organization of the cardiac intercalated disc. Cardiovasc. Res. 2017, 113, 259–275. [CrossRef]
- Roberts, J.D. TJP1 Mutations in Arrhythmogenic Cardiomyopathy. Circ. Genom. Precis. Med. 2018, 11, e002337. [CrossRef]
- De Bortoli, M.; Postma, A.V.; Poloni, G.; Calore, M.; Minervini, G.; Mazzotti, E.; Rigato, I.; Ebert, M.; Lorenzon, A.; Vazza, G.; et al. Whole-Exome Sequencing Identifies Pathogenic Variants in TJP1 Gene Associated With Arrhythmogenic Cardiomyopathy. Circ. Genom. Precis. Med. 2018, 11, e002123. [CrossRef]
- Zhao, Y.; Meng, X.-M.; Wei, Y.-J.; Zhao, X.-W.; Liu, D.-Q.; Cao, H.-Q.; Liew, C.-C.; Ding, J.-F. Cloning and characterization of a novel cardiac-specific kinase that interacts specifically with cardiac troponin I. J. Mol. Med. 2003, 81, 297–304. [CrossRef]
- Wheeler, F.C.; Tang, H.; Marks, O.A.; Hadnott, T.N.; Chu, P.-L.; Mao, L.; Rockman, H.A.; Marchuk, D.A. Tnni3k Modifies Disease Progression in Murine Models of Cardiomyopathy. PLOS Genet. 2009, 5, e1000647. [CrossRef]
- Theis, J.L.; Zimmermann, M.T.; Larsen, B.T.; Rybakova, I.N.; Long, P.A.; Evans, J.M.; Middha, S.; de Andrade, M.; Moss, R.L.; Wieben, E.D.; et al. TNNI3K mutation in familial syndrome of conduction system disease, atrial tachyarrhythmia and dilated cardiomyopathy. Hum. Mol. Genet. 2014, 23, 5793–5804. [CrossRef]
- Podliesna, S.; Delanne, J.; Miller, L.; Tester, D.J.; Uzunyan, M.; Yano, S.; Klerk, M.; Cannon, B.C.; Khongphatthanayothin, A.; Laurent, G.; et al. Supraventricular tachycardias, conduction disease, and cardiomyopathy in 3 families with the same rare variant in TNNI3K (p.Glu768Lys). Hear. Rhythm. 2018, 16, 98–105. [CrossRef]
- Fan L-L, Huang H, Jin J-Y, Li J-J, Chen Y-Q, Zhao S-P, et al. Whole exome sequencing identifies a novel mutation (c.333 + 2T > C) of TNNI3K in a Chinese family with dilated cardiomyopathy and cardiac conduction disease. Gene. 2018;648: 63–67. [CrossRef]
- Pham, C.; Andrzejczyk, K.; Jurgens, S.J.; Deprez, R.L.; Palm, K.C.; Vermeer, A.M.; Nijman, J.; Christiaans, I.; Barge-Schaapveld, D.Q.; van Dessel, P.F.; et al. Genetic Burden of TNNI3K in Diagnostic Testing of Patients With Dilated Cardiomyopathy and Supraventricular Arrhythmias. Circ. Genom. Precis. Med. 2023, 16, 328–336. [CrossRef]
- Chen, S.N.; Czernuszewicz, G.; Lombardi, R.; Jin, J.; Willerson, J.; Marian, A.J. Abstract 18430: Human Molecular Genetic and Functional Studies Identify TRIM63, Encoding Muscle RING Finger Protein 1, as a Novel Gene for Human Hypertrophic Cardiomyopathy. Circulation 2012, 126. [CrossRef]
- Su, M.; Wang, J.; Kang, L.; Wang, Y.; Zou, Y.; Feng, X.; Wang, D.; Ahmad, F.; Zhou, X.; Hui, R.; et al. Rare Variants in Genes Encoding MuRF1 and MuRF2 Are Modifiers of Hypertrophic Cardiomyopathy. Int. J. Mol. Sci. 2014, 15, 9302–9313. [CrossRef]
- Witt, C.C.; Witt, S.H.; Lerche, S.; Labeit, D.; Back, W.; Labeit, S. Cooperative control of striated muscle mass and metabolism by MuRF1 and MuRF2. EMBO J. 2007, 27, 350–360. [CrossRef]
- Andreeva, S.; Chumakova, O.; Karelkina, E.; Lebedeva, V.; Lubimtseva, T.; Semenov, A.; Nikitin, A.; Speshilov, G.; Kozyreva, A.; Sokolnikova, P.; et al. Case Report: Two New Cases of Autosomal-Recessive Hypertrophic Cardiomyopathy Associated With TRIM63-Compound Heterozygous Variant. Front. Genet. 2022, 13, 743472. [CrossRef]
- Moiseyeva, E.; Weller, P.; Zhidkova, N.; Corben, E.; Patel, B.; Jasinska, I.; Koteliansky, V.; Critchley, D. Organization of the human gene encoding the cytoskeletal protein vinculin and the sequence of the vinculin promoter.. J. Biol. Chem. 1993, 268, 4318–4325. [CrossRef]
- Vasile VC, Will ML, Ommen SR, Edwards WD, Olson TM, Ackerman MJ. Identification of a metavinculin missense mutation, R975W, associated with both hypertrophic and dilated cardiomyopathy. Mol Genet Metab. 2006;87: 169–174. [CrossRef]
- Wells, Q.S.; Ausborn, N.L.; Funke, B.H.; Pfotenhauer, J.P.; Fredi, J.L.; Baxter, S.; DiSalvo, T.G.; Hong, C.C. Familial Dilated Cardiomyopathy Associated with Congenital Defects in the Setting of a Novel VCL Mutation (Lys815Arg) in Conjunction with a Known MYPBC3 Variant. Cardiogenetics 2011, 1, e10–e10. [CrossRef]
- Mazzarotto, F.; Tayal, U.; Buchan, R.J.; Midwinter, W.; Wilk, A.; Whiffin, N.; Govind, R.; Mazaika, E.; de Marvao, A.; Dawes, T.J.; et al. Reevaluating the Genetic Contribution of Monogenic Dilated Cardiomyopathy. Circulation 2020, 141, 387–398. [CrossRef]
- Hawley, M.H.; Almontashiri, N.; Biesecker, L.G.; Berger, N.; Chung, W.K.; Garcia, J.; Grebe, T.A.; Kelly, M.A.; Lebo, M.S.; Macaya, D.; et al. An assessment of the role of vinculin loss of function variants in inherited cardiomyopathy. Hum. Mutat. 2020, 41, 1577–1587. [CrossRef]
- Xu, W.; Baribault, H.; Adamson, E.D. Vinculin knockout results in heart and brain defects during embryonic development. Development 1998, 125, 327–337. [CrossRef]
- Zemljic-Harpf, A.E.; Ponrartana, S.; Avalos, R.T.; Jordan, M.C.; Roos, K.P.; Dalton, N.D.; Phan, V.Q.; Adamson, E.D.; Ross, R.S. Heterozygous Inactivation of the Vinculin Gene Predisposes to Stress-Induced Cardiomyopathy. Am. J. Pathol. 2004, 165, 1033–1044. [CrossRef]
- Marg S, Winkler U, Sestu M, Himmel M, Schönherr M, Bär J, et al. The vinculin-DeltaIn20/21 mouse: characteristics of a constitutive, actin-binding deficient splice variant of vinculin. PLoS One. 2010;5: e11530.
- Zemljic-Harpf, A.E.; Miller, J.C.; Henderson, S.A.; Wright, A.T.; Manso, A.M.; Elsherif, L.; Dalton, N.D.; Thor, A.K.; Perkins, G.A.; McCulloch, A.D.; et al. Cardiac-Myocyte-Specific Excision of the Vinculin Gene Disrupts Cellular Junctions, Causing Sudden Death or Dilated Cardiomyopathy. Mol. Cell. Biol. 2007, 27, 7522–7537. [CrossRef]
- Vogel, B.; Meder, B.; Just, S.; Laufer, C.; Berger, I.; Weber, S.; Katus, H.A.; Rottbauer, W. In-vivo characterization of human dilated cardiomyopathy genes in zebrafish. Biochem. Biophys. Res. Commun. 2009, 390, 516–522. [CrossRef]
- Cheng, F.; Miao, L.; Wu, Q.; Gong, X.; Xiong, J.; Zhang, J. Vinculin b deficiency causes epicardial hyperplasia and coronary vessel disorganization in zebrafish. Development 2016, 143, 3522–3531. [CrossRef]
- Maartens AP, Wellmann J, Wictome E, Klapholz B, Green H, Brown NH. Drosophila vinculin is more harmful when hyperactive than absent, and can circumvent integrin to form adhesion complexes. J Cell Sci. 2016;129: 4354–4365. [CrossRef]
- Liu, Y.; Li, X.; Zhao, X.; Dong, J.; Zhang, C.; Lin, T. Establishment of a human iPSC (ZZUNEUi026-A) from a dilated cardiomyopathy patient carrying heterozygous Vinculin (c. 625A > T) mutant. Stem Cell Res. 2022, 62, 102812. [CrossRef]
- Fatkin, D.; Johnson, R. Are Double Mutations Double Trouble?. Circ. Cardiovasc. Genet. 2017, 10. [CrossRef]
- Gray, M.P.; Fatkin, D.; Ingles, J.; Robertson, E.N.; A Figtree, G. Genetic testing in cardiovascular disease. The Medical Journal of Australia 2024, 220, 428–434. [CrossRef]
- Hershberger, R.E.; Givertz, M.M.; Ho, C.Y.; Judge, D.P.; Kantor, P.F.; McBride, K.L.; Morales, A.; Taylor, M.R.; Vatta, M.; Ware, S.M. Genetic Evaluation of Cardiomyopathy—A Heart Failure Society of America Practice Guideline. J. Card. Fail. 2018, 24, 281–302. [CrossRef]
- Monaco, I.; Santacroce, R.; Casavecchia, G.; Correale, M.; Bottigliero, D.; Cordisco, G.; Leccese, A.; Di Biase, M.; Margaglione, M.; Brunetti, N.D. Double de novo mutations in dilated cardiomyopathy with cardiac arrest. J. Electrocardiol. 2018, 53, 40–43. [CrossRef]
| Gene | Phenotype Association |
|---|---|
| ACTC1 | HCM |
| BAG3 | DCM |
| DES | DCM |
| DSC2 | ARVC |
| DSG2 | ARVC |
| DSP | ARVC |
| FLNC | DCM |
| LMNA | DCM |
| MYBPC3 | HCM |
| MYH7 | DCM; HCM |
| MYL2 | HCM |
| MYL3 | HCM |
| PKP2 | ARVC |
| RBM20 | DCM |
| SCN5A | DCM |
| TMEM43 | ARVC |
| TNNC1 | DCM |
| TNNI3 | HCM |
| TNNT2 | DCM; HCM |
| TPM1 | HCM |
| TTN | DCM |
| ClinGen [17] | GeneReview [23] | EMQN [15,16] | Expert Consensus Statement [25] | ClinGen [17] | GeneReview [23] | EMQN [15,16] | Expert Consensus Statement [25] | ClinGen [17] | GeneReview [23] | EMQN [15,16] | Expert Consensus Statement [25] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Hereditary | ARVC | DCM | HCM | |||||||||
| ABCC9 | AD | not associated | not associated | - | not associated | limited | limited | - | not associated | not associated | not associated | - | not associated |
| ACTN2 | AD | not associated | not associated | not associated | not associated | not associated | moderate | moderate | moderate | not associated | moderate | not associated | moderate |
| ANKRD1 | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | limited | - | - |
| CALR3 | AD | not associated | not associated | - | - | not associated | not associated | - | - | not associated | limited | - | - |
| CSRP3 | AD, SD | not associated | not associated | - | not associated | limited | limited | - | not associated | moderate | moderate | - | moderate |
| CTF1 | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| DTNA | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| EYA4 | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| GATAD1 | AR | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| ILK | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| JPH2 | AD, SD | not associated | not associated | not associated | not associated | moderate | moderate | moderate | moderate | moderate | moderate | not associated | moderate |
| KLF10 | AD | not associated | not associated | - | - | not associated | not associated | - | - | limited | limited | - | - |
| KLHL24 | AR | not associated | - | - | - | not associated | - | - | - | moderate | - | - | - |
| LAMA4 | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| LDB3 | AD | not associated | not associated | - | not associated | limited | limited | - | not associated | not associated | not associated | - | not associated |
| MYH6 | AD | not associated | not associated | - | - | limited | limited | - | - | limited | limited | - | - |
| MYLK2 | AD | not associated | not associated | - | - | not associated | not associated | - | - | not associated | limited | - | - |
| MYOM1 | AD | not associated | not associated | - | - | not associated | not associated | - | - | not associated | limited | - | - |
| MYOZ2 | AD | not associated | not associated | - | - | not associated | not associated | - | - | not associated | limited | - | - |
| MYPN | AD | not associated | not associated | - | - | not associated | limited | - | - | not associated | limited | - | - |
| NEBL | AD | not associated | not associated | - | - | limited | limited | - | -- | not associated | not associated | - | - |
| NEXN | AD | not associated | not associated | - | - | moderate | moderate | - | - | limited | limited | - | - |
| NKX2-5 | AD | not associated | not associated | - | - | limited | limited | - | not associated | not associated | - | - | |
| OBSCN | AD | not associated | not associated | - | limited | limited | - | - | limited | not associated | - | - | |
| PDLIM3 | AD | not associated | not associated | - | - | not associated | not associated | - | - | limited | limited | - | - |
| PLEKHM2 | AR | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| PRDM16 | AD | not associated | not associated | - | limited | limited | - | - | not associated | not associated | - | - | |
| PSEN2 | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| RPS6KB1 | AD | not associated | not associated | - | - | not associated | not associated | - | - | limited | not associated | - | - |
| RYR2 | AD | not associated | not associated | - | - | not associated | not associated | - | - | limited | limited | - | - |
| SGCD | AD | not associated | not associated | - | - | limited | limited | - | not associated | not associated | - | - | |
| TBX20 | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | not associated | - | - |
| TCAP | AD | not associated | not associated | - | - | limited | limited | - | - | not associated | limited | - | - |
| TNNI3K | AD | not associated | not associated | - | limited | limited | - | - | not associated | not associated | - | - | |
| TRIM63 | AD, AR | not associated | not associated | - | - | not associated | not associated | - | - | moderate | limited | - | - |
| VCL | AD | not associated | not associated | - | - | moderate | moderate | - | - | not associated | limited | - | - |
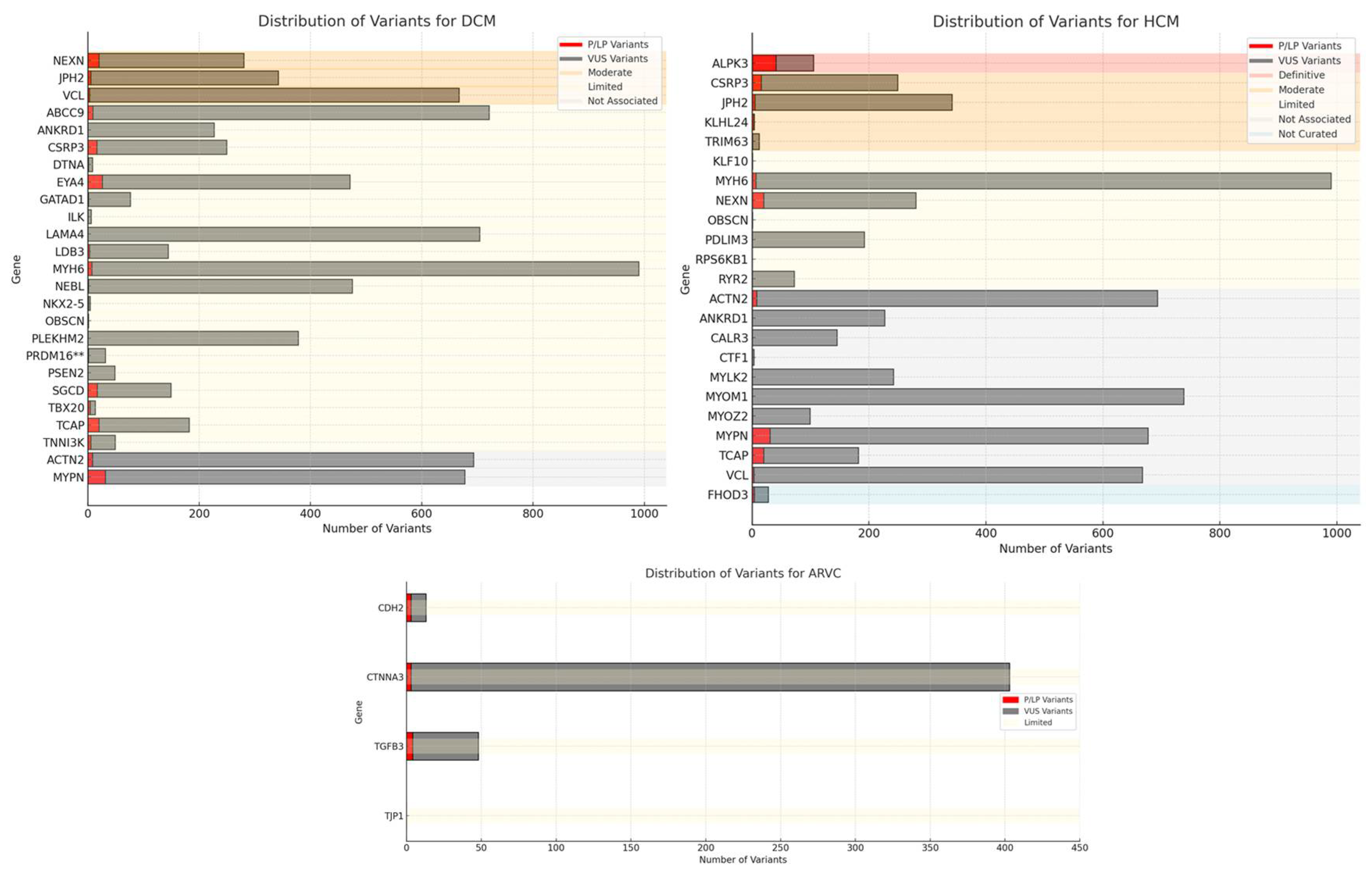
| Gene | Protein | ARVC | DCM | HCM | N. P/LP Variants | N. VUS | Phenotype Submission | Hereditary |
|---|---|---|---|---|---|---|---|---|
| ABCC9 | ATP Binding Cassette Subfamily C Member 9 | not associated | limited | not associated | 9 | 712 | DCM | AD |
| ACTN2 | Alpha-actinin-2 | not associated | not associated | not associated | 8 | 685 | DCM, HCM | AD |
| ANKRD1 | Ankyrin Repeat Domain 1 | not associated | limited | not associated | 0 | 227 | DCM, HCM | AD |
| CALR3 | Calreticulin 3 | not associated | not associated | not associated | 0 | 145 | HCM | AD |
| CDH2 | Cadherin 2 | limited | not associated | not associated | 3 | 10 | ARVC | AD |
| CSRP3 | Cysteine And Glycine Rich Protein 3 | not associated | limited | moderate | 16 | 233 | DCM, HCM | AD, SD |
| CTF1 | Cardiotrophin 1 | not associated | limited | not associated | 0 | 3 | HCM | AD |
| CTNNA3 | Catenin Alpha 3 | limited | not associated | not associated | 3 | 400 | ARVC | AD |
| DTNA | Dystrobrevin Alpha | not associated | limited | not associated | 1 | 7 | DCM | AD |
| EYA4 | EYA Transcriptional Coactivator and Phosphatase 4 | not associated | limited | not associated | 26 | 445 | DCM | AD |
| FHOD3 | Formin Homology 2 Domain Containing 3 | not curated | not curated | not curated | 4 | 24 | HCM | na |
| GATAD1 | GATA Zinc Finger Domain Containing 1 | not associated | limited | not associated | 1 | 75 | DCM | AR |
| ILK | Integrin Linked Kinase | not associated | limited | not associated | 0 | 6 | DCM | AD |
| JPH2 | Junctophilin 2 | not associated | moderate | moderate | 5 | 337 | DCM, HCM | AD, SD |
| KLF10 | Kruppel -Like Factor 1 | not associated | not associated | limited | 0 | 1 | HCM | AD |
| KLHL24 | Kelch Like Family Member 24 | not associated | not associated | moderate | 3 | 1 | HCM | AR |
| LAMA4 | Laminin Subunit Alpha 4 | not associated | limited | not associated | 0 | 704 | DCM | AD |
| LDB3 | LIM Domain Binding 3 | not associated | limited | not associated | 3 | 141 | DCM | AD |
| MYH6 | Myosin Heavy Chain 6 | not associated | limited | limited | 7 | 983 | DCM, HCM | AD |
| MYLK2 | Myosin Light Chain Kinase 2 | not associated | not associated | not associated | 0 | 242 | HCM | AD |
| MYOM1 | Myomesin 1 | not associated | not associated | not associated | 0 | 738 | HCM | AD |
| MYOZ2 | Myozenin 2 | not associated | not associated | not associated | 1 | 98 | HCM | AD |
| MYPN | Myopalladin | not associated | not associated | not associated | 31 | 646 | DCM, HCM | AD |
| NEBL | Nebulette | not associated | limited | not associated | 1 | 474 | DCM | AD |
| NEXN | Nexilin F-Actin Binding Protein | not associated | moderate | limited | 20 | 260 | DCM, HCM | AD |
| NKX2-5 | NK2 Homeobox 5 | not associated | limited | not associated | 1 | 3 | DCM | AD |
| OBSCN | Obscurin | not associated | limited | limited | 0 | 1 | DCM, HCM | AD |
| PDLIM3 | PDZ And LIM Domain 3 | not associated | not associated | limited | 0 | 192 | HCM | AD |
| PLEKHM2 | Pleckstrin Homology and RUN Domain Containing M2 | not associated | limited | not associated | 0 | 378 | DCM | AR |
| PRDM16 | PR/SET Domain 16 | not associated | limited | not associated | 1 | 30 | DCM | AD |
| PSEN2 | Presenilin 2 | not associated | limited | not associated | 0 | 48 | DCM | AD |
| RPS6KB1 | Ribosomal Protein S6 Kinase B1 | not associated | not associated | limited | 0 | 0 | HCM | AD |
| RYR2 | Ryanodine Receptor 2 | not associated | not associated | limited | 0 | 72 | HCM | AD |
| SGCD | Sarcoglycan Delta | not associated | limited | not associated | 17 | 132 | DCM | AD |
| TBX20 | T-Box Transcription Factor 20 | not associated | limited | not associated | 4 | 9 | DCM | AD |
| TCAP | Titin-Cap | not associated | limited | not associated | 20 | 162 | DCM, HCM | AD |
| TGFB3 | Transforming Growth Factor Beta 3 | limited | not associated | not associated | 4 | 44 | ARVC | AD |
| TJP1 | Tight Junction Protein 1 | limited | not associated | not associated | 0 | 0 | ARVC | AD |
| TNNI3K | TNNI3 Interacting Kinase | not associated | limited | not associated | 5 | 44 | DCM | AD |
| TRIM63 | Tripartite Motif Containing 63 | not associated | not associated | moderate | 1 | 11 | HCM | AD, AR |
| VCL | Vinculin | not associated | moderate | not associated | 3 | 664 | DCM, HCM | AD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





