Submitted:
19 July 2024
Posted:
22 July 2024
You are already at the latest version
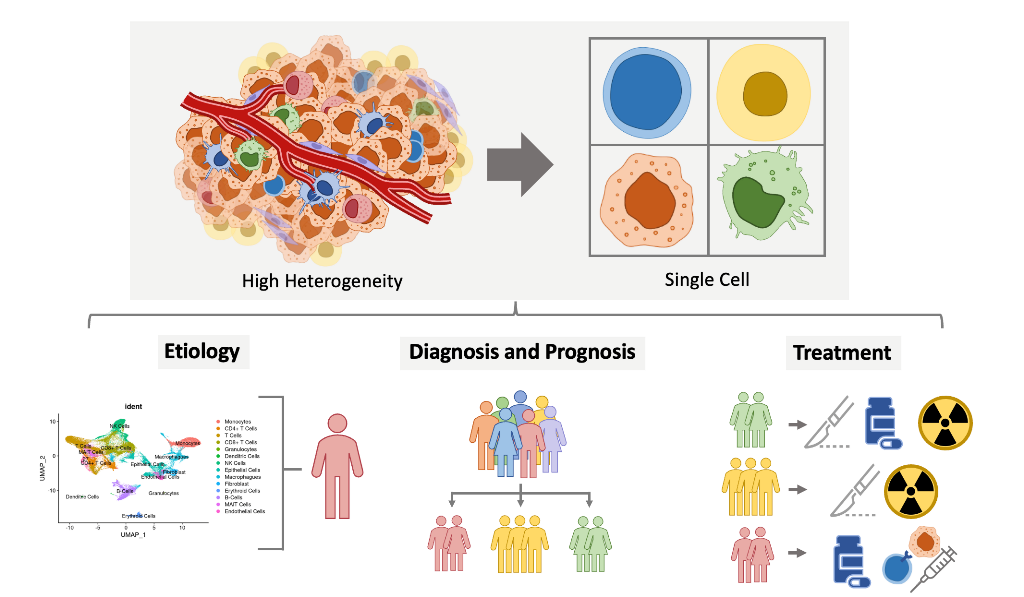
Abstract
Keywords:
1. Introduction
2. HNSCC Etiology
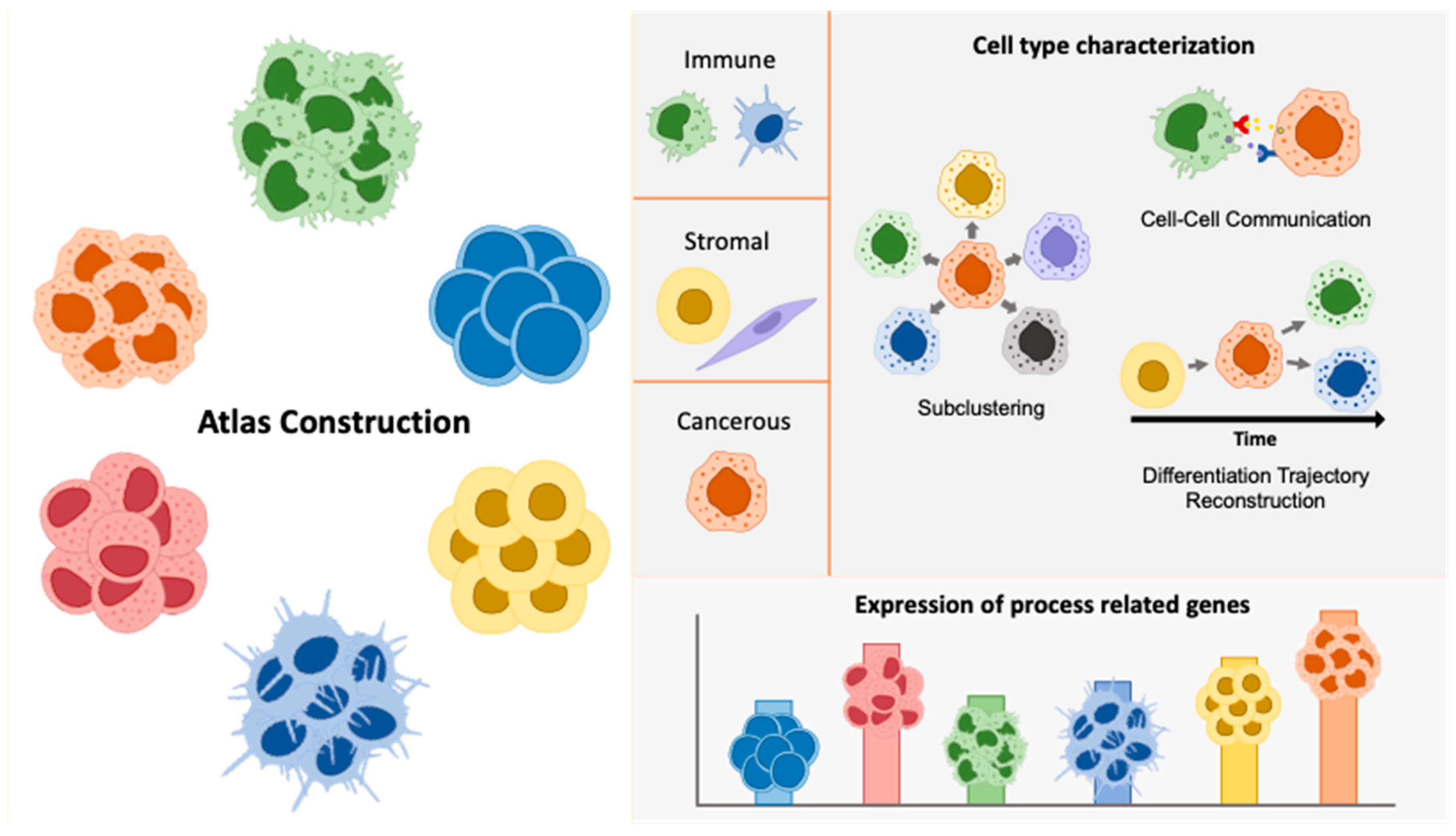
3.1. Atlas Construction
3.2. Cell Type Specific Characterization
3.2.1. Tumor Cell Characterization and Crosstalk with Stromal Compartments
3.2.2. Immune Landscape
3.3. Expression of Process Related Genes
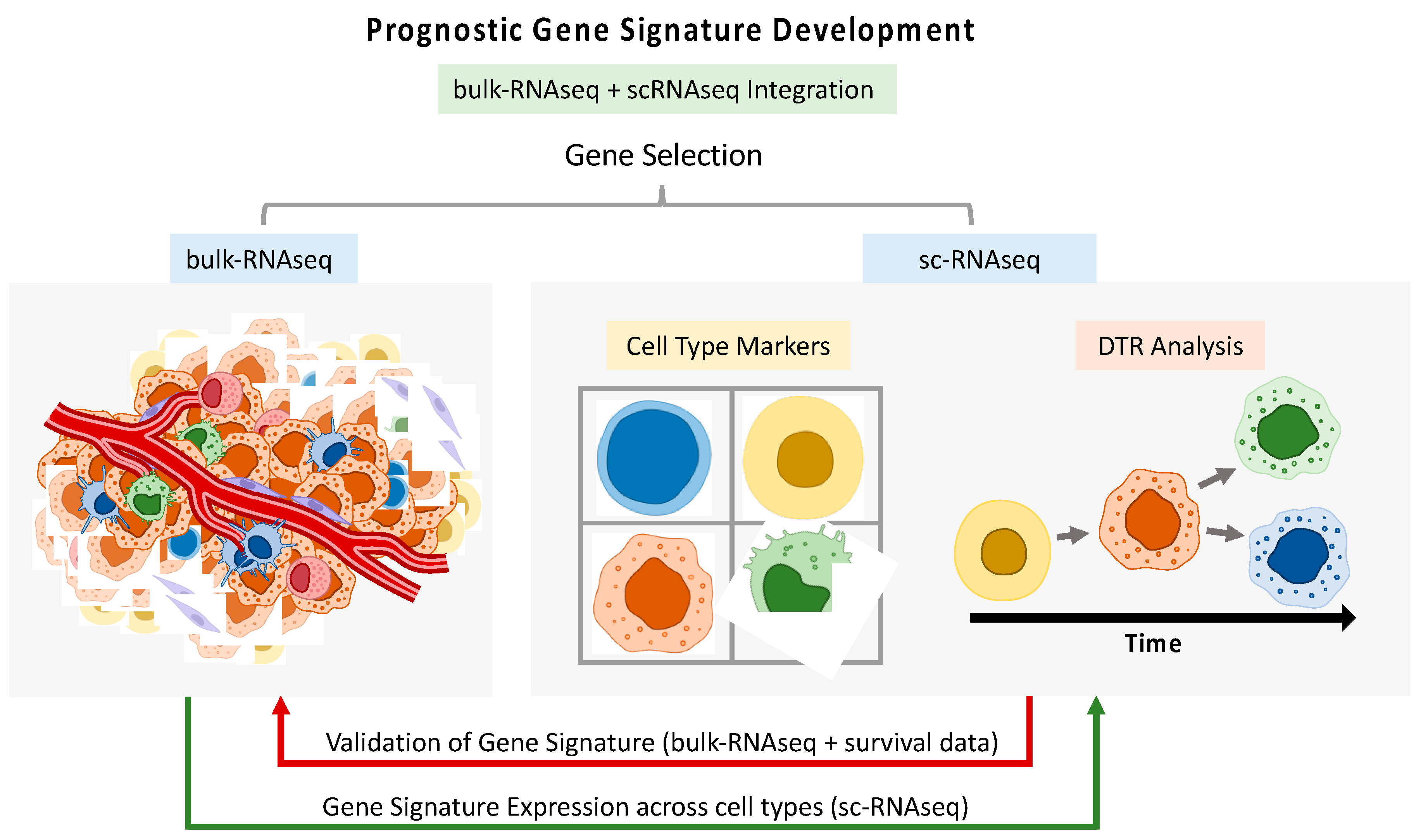
4. HNSCC Diagnosis and Prognosis
5. HNSCC Treatment
6. Outlook
7. Conclusions
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J. Clin. 2018, 68, 394–424, . [CrossRef]
- Jawa, Y.; Yadav, P.; Gupta, S.; Mathan, S.V.; Pandey, J.; Saxena, A.K.; Kateriya, S.; Tiku, A.B.; Mondal, N.; Bhattacharya, J.; et al. Current Insights and Advancements in Head and Neck Cancer: Emerging Biomarkers and Therapeutics with Cues from Single Cell and 3D Model Omics Profiling. Front. Oncol. 2021, 11, . [CrossRef]
- Cai, Z.; Tang, B.; Chen, L.; Lei, W. Mast cell marker gene signature in head and neck squamous cell carcinoma. BMC Cancer 2022, 22, 1–16, . [CrossRef]
- The Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120, . [CrossRef]
- Edwards, N.J.; Oberti, M.; Thangudu, R.R.; Cai, S.; McGarvey, P.B.; Jacob, S.; Madhavan, S.; Ketchum, K.A. The CPTAC Data Portal: A Resource for Cancer Proteomics Research. J. Proteome Res. 2015, 14, 2707–2713, . [CrossRef]
- Stampe, H.; Jakobsen, K.K.; Bendtsen, S.K.; Grønhøj, C.; von Buchwald, C. Systematic review on the current knowledge and use of single-cell RNA sequencing in head and neck cancer. APMIS 2021, 129, 619–625, . [CrossRef]
- Huang, Z.-D.; Liu, Z.-Z.; Liu, Y.-Y.; Fu, Y.-C.; Lin, L.-L.; Hu, C.; Gu, H.-Y.; Wei, R.-X. Molecular Subtypes Based on Cell Differentiation Trajectories in Head and Neck Squamous Cell Carcinoma: Differential Prognosis and Immunotherapeutic Responses. Front. Immunol. 2021, 12, 791621, . [CrossRef]
- Qi, Z.; Barrett, T.; Parikh, A.S.; Tirosh, I.; Puram, S.V. Single-cell sequencing and its applications in head and neck cancer. Oral Oncol. 2019, 99, 104441–104441, . [CrossRef]
- Cao, S.; Wang, J.R.; Ji, S.; Yang, P.; Dai, Y.; Guo, S.; Montierth, M.D.; Shen, J.P.; Zhao, X.; Chen, J.; et al. Estimation of tumor cell total mRNA expression in 15 cancer types predicts disease progression. Nat. Biotechnol. 2022, 40, 1624–1633, . [CrossRef]
- Choi, J.-H.; Lee, B.-S.; Jang, J.Y.; Lee, Y.S.; Kim, H.J.; Roh, J.; Shin, Y.S.; Woo, H.G.; Kim, C.-H. Single-cell transcriptome profiling of the stepwise progression of head and neck cancer. Nat. Commun. 2023, 14, 1–13, . [CrossRef]
- Yu, X.; Chen, Y.A.; Conejo-Garcia, J.R.; Chung, C.H.; Wang, X. Estimation of immune cell content in tumor using single-cell RNA-seq reference data. BMC Cancer 2019, 19, 1–11, . [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24, doi:10.1016/j.cell.2017.10.044.
- Puram, S.V.; Mints, M.; Pal, A.; Qi, Z.; Reeb, A.; Gelev, K.; Barrett, T.F.; Gerndt, S.; Liu, P.; Parikh, A.S.; et al. Cellular states are coupled to genomic and viral heterogeneity in HPV-related oropharyngeal carcinoma. Nat. Genet. 2023, 55, 640–650, . [CrossRef]
- Kürten, C.H.L.; Kulkarni, A.; Cillo, A.R.; Santos, P.M.; Roble, A.K.; Onkar, S.; Reeder, C.; Lang, S.; Chen, X.; Duvvuri, U.; et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat. Commun. 2021, 12, 1–16, . [CrossRef]
- Song, L.; Zhang, S.; Yu, S.; Ma, F.; Wang, B.; Zhang, C.; Sun, J.; Mao, X.; Wei, L. Cellular heterogeneity landscape in laryngeal squamous cell carcinoma. Int. J. Cancer 2020, 147, 2879–2890, . [CrossRef]
- Bill, R.; Wirapati, P.; Messemaker, M.; Roh, W.; Zitti, B.; Duval, F.; Kiss, M.; Park, J.C.; Saal, T.M.; Hoelzl, J.; et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science 2023, 381, 515–524, . [CrossRef]
- Cillo, A.R.; Kürten, C.H.; Tabib, T.; Qi, Z.; Onkar, S.; Wang, T.; Liu, A.; Duvvuri, U.; Kim, S.; Soose, R.J.; et al. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 2020, 52, 183–199.e9, . [CrossRef]
- Janjic, B.M.; Kulkarni, A.; Ferris, R.L.; Vujanovic, L.; Vujanovic, N.L. Human B Cells Mediate Innate Anti-Cancer Cytotoxicity Through Concurrent Engagement of Multiple TNF Superfamily Ligands. Front. Immunol. 2022, 13, 837842, . [CrossRef]
- Huynh, N.C.-N.; Huang, T.-T.; Nguyen, C.T.-K.; Lin, F.-K. Comprehensive Integrated Single-Cell Whole Transcriptome Analysis Revealed the p-EMT Tumor Cells—CAFs Communication in Oral Squamous Cell Carcinoma. Int. J. Mol. Sci. 2022, 23, 6470, . [CrossRef]
- Xiao, M.; Zhang, X.; Zhang, D.; Deng, S.; Zheng, A.; Du, F.; Shen, J.; Yue, L.; Yi, T.; Xiao, Z.; et al. Complex interaction and heterogeneity among cancer stem cells in head and neck squamous cell carcinoma revealed by single-cell sequencing. Front. Immunol. 2022, 13, 1050951, . [CrossRef]
- Chen, J.; Yang, J.; Li, H.; Yang, Z.; Zhang, X.; Li, X.; Wang, J.; Zhang, Y.; Chen, S.; Song, M. Single-Cell Transcriptomics Reveal the Intratumoral Landscape of Infiltrated T-Cell Subpopulations in Oral Squamous Cell Carcinoma. Mol. Oncol. 2021, 15, 866–886.
- Wieland, A.; Patel, M.R.; Cardenas, M.A.; Eberhardt, C.S.; Hudson, W.H.; Obeng, R.C.; Griffith, C.C.; Wang, X.; Chen, Z.G.; Kissick, H.T.; et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature 2021, 597, 274–278, . [CrossRef]
- Eberhardt, C.S.; Kissick, H.T.; Patel, M.R.; Cardenas, M.A.; Prokhnevska, N.; Obeng, R.C.; Nasti, T.H.; Griffith, C.C.; Im, S.J.; Wang, X.; et al. Functional HPV-specific PD-1+ stem-like CD8 T cells in head and neck cancer. Nature 2021, 597, 279–284, . [CrossRef]
- Dohmen, J.; Baranovskii, A.; Ronen, J.; Uyar, B.; Franke, V.; Akalin, A. Identifying tumor cells at the single-cell level using machine learning. Genome Biol. 2022, 23, 1–23, . [CrossRef]
- Yu, X.; Wang, Z.; Zeng, T. Essential gene expression pattern of head and neck squamous cell carcinoma revealed by tumor-specific expression rule based on single-cell RNA sequencing. Biochim. et Biophys. Acta (BBA) - Mol. Basis Dis. 2020, 1866, 165791, . [CrossRef]
- Zhang, Q.; Wang, Y.; Xia, C.; Ding, L.; Pu, Y.; Hu, X.; Cai, H.; Hu, Q. Integrated analysis of single-cell RNA-seq and bulk RNA-seq reveals distinct cancer-associated fibroblasts in head and neck squamous cell carcinoma. Ann. Transl. Med. 2021, 9, 1017–1017, . [CrossRef]
- Meng, L.; Lu, H.; Li, Y.; Zhao, J.; He, S.; Wang, Z.; Shen, J.; Huang, H.; Xiao, J.; Sooranna, S.R.; et al. Human papillomavirus infection can alter the level of tumour stemness and T cell infiltration in patients with head and neck squamous cell carcinoma. Front. Immunol. 2022, 13, 1013542, . [CrossRef]
- Chen, S.M.Y.; Krinsky, A.L.; Woolaver, R.A.; Wang, X.; Chen, Z.; Wang, J.H. Tumor immune microenvironment in head and neck cancers. Mol. Carcinog. 2020, 59, 766–774, . [CrossRef]
- Canning, M.; Guo, G.; Yu, M.; Myint, C.; Groves, M.W.; Byrd, J.K.; Cui, Y. Heterogeneity of the Head and Neck Squamous Cell Carcinoma Immune Landscape and Its Impact on Immunotherapy. Front. Cell Dev. Biol. 2019, 7, 52, . [CrossRef]
- Gameiro, S.F.; Evans, A.M.; Mymryk, J.S. The Tumor Immune Microenvironments of HPV+ and HPV- Head and Neck Cancers. WIREs Mech Dis 2022, 14, e1539.
- Bonaventura, P.; Shekarian, T.; Alcazer, V.; Valladeau-Guilemond, J.; Valsesia-Wittmann, S.; Amigorena, S.; Caux, C.; Depil, S. Cold Tumors: A Therapeutic Challenge for Immunotherapy. Front. Immunol. 2019, 10, 168, . [CrossRef]
- Huang, Y.; Liu, H.; Liu, X.; Li, N.; Bai, H.; Guo, C.; Xu, T.; Zhu, L.; Liu, C.; Xiao, J. The Chemokines Initiating and Maintaining Immune Hot Phenotype Are Prognostic in ICB of HNSCC. Front. Genet. 2022, 13, 820065, . [CrossRef]
- Li, S.; Wang, Y.; Sun, R.; Franceschi, D.; Pan, H.; Wei, C.; Ogbuehi, A.C.; Lethaus, B.; Savkovic, V.; Gaus, S.; et al. Single-Cell Transcriptome Analysis Reveals Different Immune Signatures in HPV- and HPV + Driven Human Head and Neck Squamous Cell Carcinoma. J. Immunol. Res. 2022, 2022, 1–19, . [CrossRef]
- Jiang, Y.; Zhang, S.; Tang, L.; Li, R.; Zhai, J.; Luo, S.; Peng, Y.; Chen, X.; Wei, L. Single-cell RNA sequencing reveals TCR+ macrophages in HPV-related head and neck squamous cell carcinoma. Front. Immunol. 2022, 13, 1030222, . [CrossRef]
- Liu, F.; Tang, L.; Li, Q.; Chen, L.; Pan, Y.; Yin, Z.; He, J.; Tian, J. Single-cell transcriptomics uncover the key ferroptosis regulators contribute to cancer progression in head and neck squamous cell carcinoma. Front. Mol. Biosci. 2022, 9, 962742, . [CrossRef]
- Jiang, X.; Ke, J.; Jia, L.; An, X.; Ma, H.; Li, Z.; Yuan, W. A novel cuproptosis-related gene signature of prognosis and immune microenvironment in head and neck squamous cell carcinoma cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 203–218, . [CrossRef]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908, . [CrossRef]
- Sun, D.; Wang, J.; Han, Y.; Dong, X.; Ge, J.; Zheng, R.; Shi, X.; Wang, B.; Li, Z.; Ren, P.; et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021, 49, D1420–D1430, . [CrossRef]
- Zhang, Z.; Liu, R.; Jin, R.; Fan, Y.; Li, T.; Shuai, Y.; Li, X.; Wang, X.; Luo, J. Integrating Clinical and Genetic Analysis of Perineural Invasion in Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2019, 9, 434, . [CrossRef]
- Liu, H.; Li, Y. Potential roles of Cornichon Family AMPA Receptor Auxiliary Protein 4 (CNIH4) in head and neck squamous cell carcinoma. Cancer Biomarkers 2022, 35, 439–450, . [CrossRef]
- Hegde, M.; Daimary, U.D.; Jose, S.; Sajeev, A.; Chinnathambi, A.; Alharbi, S.A.; Shakibaei, M.; Kunnumakkara, A.B. Differential Expression of Genes Regulating Store-operated Calcium Entry in Conjunction With Mitochondrial Dynamics as Potential Biomarkers for Cancer: A Single-Cell RNA Analysis. Front. Genet. 2022, 13, 866473, . [CrossRef]
- Messerschmidt, C.; Obermayer, B.; Klinghammer, K.; Ochsenreither, S.; Treue, D.; Stenzinger, A.; Glimm, H.; Fröhling, S.; Kindler, T.; Brandts, C.H.; et al. Distinct Immune Evasion in APOBEC-enriched, HPV-negative HNSCC. Int. J. Cancer 2020, 147, 2293–2302.
- Weng, J.; Fan, H.; Liu, H.; Tang, S.; Zheng, Y. YTHDC1 Promotes Stemness Maintenance and Malignant Progression in Head and Neck Squamous Cell Carcinoma. Stem Cells Int. 2022, 2022, 1–13, . [CrossRef]
- Hu, C.; Fan, J.; He, G.; Dong, C.; Zhou, S.; Zheng, Y. Signal peptidase complex catalytic subunit SEC11A upregulation is a biomarker of poor prognosis in patients with head and neck squamous cell carcinoma. PLOS ONE 2022, 17, e0269166, . [CrossRef]
- Carofino, B.L.; Dinshaw, K.M.; Ho, P.Y.; Cataisson, C.; Michalowski, A.M.; Ryscavage, A.; Alkhas, A.; Wong, N.W.; Koparde, V.; Yuspa, S.H. Head and neck squamous cancer progression is marked by CLIC4 attenuation in tumor epithelium and reciprocal stromal upregulation of miR-142-3p, a novel post-transcriptional regulator of CLIC4. Oncotarget 2019, 10, 7251–7275, . [CrossRef]
- Yorozu, A.; Sekiguchi, S.; Takasawa, A.; Okazaki, F.; Niinuma, T.; Kitajima, H.; Yamamoto, E.; Kai, M.; Toyota, M.; Hatanaka, Y.; et al. CXCL12 is expressed by skeletal muscle cells in tongue oral squamous cell carcinoma. Cancer Med. 2022, 12, 5953–5963, . [CrossRef]
- Zhang, S.; Zhang, W.; Zhang, J. 8-Gene signature related to CD8+ T cell infiltration by integrating single-cell and bulk RNA-sequencing in head and neck squamous cell carcinoma. Front. Genet. 2022, 13, 938611, . [CrossRef]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386, doi:10.1002/ijc.29210.
- Kang, X.; Chen, Y.; Yi, B.; Yan, X.; Jiang, C.; Chen, B.; Lu, L.; Sun, Y.; Shi, R. An integrative microenvironment approach for laryngeal carcinoma: the role of immune/methylation/autophagy signatures on disease clinical prognosis and single-cell genotypes. J. Cancer 2021, 12, 4148–4171, . [CrossRef]
- Chen, Q.; Chu, L.; Li, X.; Li, H.; Zhang, Y.; Cao, Q.; Zhuang, Q. Investigation of an FGFR-Signaling-Related Prognostic Model and Immune Landscape in Head and Neck Squamous Cell Carcinoma. Front. Cell Dev. Biol. 2021, 9, 801715, . [CrossRef]
- Huang, C.; Liang, Y.; Dong, Y.; Huang, L.; Li, A.; Du, R.; Huang, H. Novel prognostic matrisome-related gene signature of head and neck squamous cell carcinoma. Front. Cell Dev. Biol. 2022, 10, 884590, . [CrossRef]
- Qi, Z.; Liu, Y.; Mints, M.; Mullins, R.; Sample, R.; Law, T.; Barrett, T.; Mazul, A.L.; Jackson, R.S.; Kang, S.Y.; et al. Single-Cell Deconvolution of Head and Neck Squamous Cell Carcinoma. Cancers 2021, 13, 1230, . [CrossRef]
- Yin, J.; Zheng, S.; He, X.; Huang, Y.; Hu, L.; Qin, F.; Zhong, L.; Li, S.; Hu, W.; Zhu, J. Identification of molecular classification and gene signature for predicting prognosis and immunotherapy response in HNSCC using cell differentiation trajectories. Sci. Rep. 2022, 12, 1–17, . [CrossRef]
- Han, P.; Tan, L.; Ouyang, Q.; Yu, P.; Shi, X.; Hu, J.; Wei, W.; Lu, Z.; Ji, Q.; Qu, N.; et al. Development and validation of a gene model predicting lymph node metastasis and prognosis of oral squamous cell carcinoma based on single-cell and bulk RNA-seq analysis. J. Oral Pathol. Med. 2022, 52, 389–401, . [CrossRef]
- Wang, J.; Sun, H.-C.; Cao, C.; Hu, J.-D.; Qian, J.; Jiang, T.; Jiang, W.-B.; Zhou, S.; Qiu, X.-W.; Wang, H.-L. Identification and validation of a novel signature based on cell-cell communication in head and neck squamous cell carcinoma by integrated analysis of single-cell transcriptome and bulk RNA-sequencing. Front. Oncol. 2023, 13, 1136729, . [CrossRef]
- Caudell, J.J.; Gillison, M.L.; Maghami, E.; Spencer, S.; Pfister, D.G.; Adkins, D.; Birkeland, A.C.; Brizel, D.M.; Busse, P.M.; Cmelak, A.J.; et al. NCCN Guidelines® Insights: Head and Neck Cancers, Version 1.2022. J. Natl. Compr. Canc. Netw. 2022, 20, 224–234.
- Pulte, D.; Brenner, H. Changes in Survival in Head and Neck Cancers in the Late 20th and Early 21st Century: A Period Analysis. Oncologist 2010, 15, 994–1001, doi:10.1634/theoncologist.2009-0289.
- Davis-Marcisak, E.F.; Sherman, T.D.; Orugunta, P.; Stein-O'Brien, G.L.; Puram, S.V.; Torres, E.T.R.; Hopkins, A.C.; Jaffee, E.M.; Favorov, A.V.; Afsari, B.; et al. Differential Variation Analysis Enables Detection of Tumor Heterogeneity Using Single-Cell RNA-Sequencing Data. Cancer Res. 2019, 79, 5102–5112, . [CrossRef]
- Obradovic, A.; Graves, D.; Korrer, M.; Wang, Y.; Roy, S.; Naveed, A.; Xu, Y.; Luginbuhl, A.; Curry, J.; Gibson, M.; et al. Immunostimulatory Cancer-Associated Fibroblast Subpopulations Can Predict Immunotherapy Response in Head and Neck Cancer. Clin. Cancer Res. 2022, 28, 2094–2109, . [CrossRef]
- Song, H.; Lou, C.; Ma, J.; Gong, Q.; Tian, Z.; You, Y.; Ren, G.; Guo, W.; Wang, Y.; He, K.; et al. Single-Cell Transcriptome Analysis Reveals Changes of Tumor Immune Microenvironment in Oral Squamous Cell Carcinoma After Chemotherapy. Front. Cell Dev. Biol. 2022, 10, 914120, . [CrossRef]
- Kim, S.S.; Shen, S.; Miyauchi, S.; Sanders, P.D.; Franiak-Pietryga, I.; Mell, L.; Gutkind, J.S.; Cohen, E.E.; Califano, J.A.; Sharabi, A.B. B Cells Improve Overall Survival in HPV-Associated Squamous Cell Carcinomas and Are Activated by Radiation and PD-1 Blockade. Clin. Cancer Res. 2020, 26, 3345–3359, . [CrossRef]
- Gong, W.; Donnelly, C.R.; Heath, B.R.; Bellile, E.; Donnelly, L.A.; Taner, H.F.; Broses, L.; Brenner, J.C.; Chinn, S.B.; Ji, R.-R.; et al. Cancer-specific type-I interferon receptor signaling promotes cancer stemness and effector CD8+ T-cell exhaustion. OncoImmunology 2021, 10, 1997385, . [CrossRef]
- Zhou, L.; Zeng, Z.; Egloff, A.M.; Zhang, F.; Guo, F.; Campbell, K.M.; Du, P.; Fu, J.; Zolkind, P.; Ma, X.; et al. Checkpoint blockade-induced CD8+ T cell differentiation in head and neck cancer responders. J. Immunother. Cancer 2022, 10, e004034, . [CrossRef]
- Chen, J.; Li, K.; Chen, J.; Wang, X.; Ling, R.; Cheng, M.; Chen, Z.; Chen, F.; He, Q.; Li, S.; et al. Aberrant Translation Regulated by METTL1/WDR4-Mediated TRNA N7-Methylguanosine Modification Drives Head and Neck Squamous Cell Carcinoma Progression. Cancer Commun. 2022, 42, 223–244.
- Aran, D.; Looney, A.P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R.P.; Wolters, P.J.; Abate, A.R.; et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019, 20, 163–172, . [CrossRef]
- Andreatta, M.; Berenstein, A.J.; Carmona, S.J. scGate: marker-based purification of cell types from heterogeneous single-cell RNA-seq datasets. Bioinformatics 2022, 38, 2642–2644, . [CrossRef]
- Zappia, L.; Theis, F.J. Over 1000 tools reveal trends in the single-cell RNA-seq analysis landscape. Genome Biol. 2021, 22, 1–18, . [CrossRef]
- Heumos, L.; Schaar, A.C.; Lance, C.; Litinetskaya, A.; Drost, F.; Zappia, L.; Lücken, M.D.; Strobl, D.C.; Henao, J.; Curion, F.; et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 2023, 24, 550–572, . [CrossRef]
- Du, P.; Fan, R.; Zhang, N.; Wu, C.; Zhang, Y. Advances in Integrated Multi-omics Analysis for Drug-Target Identification. Biomolecules 2024, 14, 692, . [CrossRef]
- Murphy, A.E.; Skene, N.G. A balanced measure shows superior performance of pseudobulk methods in single-cell RNA-sequencing analysis. Nat. Commun. 2022, 13, 1–4, . [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-R.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296, . [CrossRef]
- Andreatta, M.; Hérault, L.; Gueguen, P.; Gfeller, D.; Berenstein, A.J.; Carmona, S.J. Semi-supervised integration of single-cell transcriptomics data. Nat. Commun. 2024, 15, 1–13, . [CrossRef]
- Shapiro, G.K. HPV Vaccination: An Underused Strategy for the Prevention of Cancer. Curr. Oncol. 2022, 29, 3780–3792, . [CrossRef]
- Vermeulen, M.C.; Pearse, R.; Young-Pearse, T.; Mostafavi, S. Mosaic loss of Chromosome Y in aged human microglia. Genome Res. 2022, 32, 1795–1807, . [CrossRef]
- Hollows, R.; Wei, W.; Cazier, J.; Mehanna, H.; Parry, G.; Halford, G.; Murray, P. Association between loss of Y chromosome and poor prognosis in male head and neck squamous cell carcinoma. Head Neck 2019, 41, 993–1006, . [CrossRef]
| Accession Number | Year | Publication | Data type | Tissue types | Sample Number | Cell Number |
|---|---|---|---|---|---|---|
| GSE234933 | 2023 | Bill R, Wirapati P, Messemaker M, Roh W et al. CXCL9:SPP1 macrophage polarity identifies a network of cellular programs that control human cancers. Science 2023 Aug 4;381(6657):515-524 | scRNAseq | Primary tumor Local recurrence Distant metastasis |
52 | 87399 |
| GSE182227 | 2022 | Puram SV, Mints M, Pal A, Qi Z et al. Cellular states are coupled to genomic and viral heterogeneity in HPV-related oropharyngeal carcinoma. Nat Genet2023 Apr;55(4):640-650 | scRNAseq | Primary tumor Normal tissue |
24 | 70970 |
| GSE139324 | 2019 | Cillo AR, Kürten CHL, Tabib T, Qi Z et al. Immune Landscape of Viral- and Carcinogen-Driven Head and Neck Cancer. Immunity 2020 Jan 14;52(1):183-199.e9. | scRNAseq | Peripheral/Intra-tumoral CD45+ populations | 63 | 131224 |
| GSE164690 | 2021 | Kürten CHL, Kulkarni A, Cillo AR, Santos PM et al. Investigating immune and non-immune cell interactions in head and neck tumors by single-cell RNA sequencing. Nat Commun 2021 Dec 17;12(1):7338 | scRNAseq | Primary tumor Pheripheral Blood Leucocytes |
51 | 134606 |
| GSE103322 | 2017 | Puram SV, Tirosh I, Parikh AS, Patel AP et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017 Dec 14;171(7):1611-1624.e24. | scRNAseq | Primary tumor | 18 | 5902 |
| GSE181919 | 2022 | Choi JH, Lee BS, Jang JY, Lee YS et al. Single-cell transcriptome profiling of the stepwise progression of head and neck cancer. Nat Commun 2023 Feb 24;14(1):1055. | scRNAseq | Primary tumor Normal tissue Leukoplakia Lymph-node metastasis |
37 | 54239 |
| GSE173647 | 2022 | --- | scRNAseq | Primary tumor | 2 | 13903 |
| GSE195832 | 2022 | Obradovic A, Graves D, Korrer M, Wang Y et al. Immunostimulatory Cancer-Associated Fibroblast Subpopulations Can Predict Immunotherapy Response in Head and Neck Cancer. Clin Cancer Res 2022 May 13;28(10):2094-2109. | scRNAseq | Primary tumor | 8 | 22906 |
| GSE140042 | 2021 | --- | scRNAseq | Primary tumor Lymph-node metastasis |
9 | --- |
| GSE200996 | 2022 | Luoma AM, Suo S, Wang Y, Gunasti L et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 2022 Aug 4;185(16):2918-2935.e29. | scRNAseq + scTCR | Peripheral/Intra-tumoral CD45+ populations | 204 | 74557 |
| GSE153559 | 2020 | Wieland A, Patel MR, Cardenas MA, Eberhardt CS et al. Defining HPV-specific B cell responses in patients with head and neck cancer. Nature 2021 Sep;597(7875):274-278. | scRNAseq | B cells Primary tumor Lymph-node metastasis Pheriphery |
7 | 8271 |
| GSE180268 | 2021 | Eberhardt CS, Kissick HT, Patel MR, Cardenas MA et al. Functional HPV-specific PD-1(+) stem-like CD8 T cells in head and neck cancer. Nature 2021 Sep;597(7875):279-284. | scRNAseq | TILs Primary tumor/Lymph-node metastasis | 39 | --- |
| GSE162025 | 2020 | Liu Y, He S, Wang XL, Peng W et al. Tumour heterogeneity and intercellular networks of nasopharyngeal carcinoma at single cell resolution. Nat Commun2021 Feb 2;12(1):741 | scRNAseq + scTCR | Primary tumor/Pheripheral Blood Leucocytes | 40 | 176447 |
| GSE150321 | 2020 | Song L, Zhang S, Yu S, Ma F et al. Cellular heterogeneity landscape in laryngeal squamous cell carcinoma. Int J Cancer 2020 Nov 15;147(10):2879-2890. | scRNAseq | Primary tumor | 2 | 12985 |
| GSE213047 | 2022 | Lin M, Sade-Feldman M, Wirth L, Lawrence MS et al. Single-cell transcriptomic profiling for inferring tumor origin and mechanisms of therapeutic resistance. NPJ Precis Oncol 2022 Oct 10;6(1):71. | scRNAseq | Primary tumor/Normal tissue/Lymph-node metastasis | 3 | 11470 |
| GSE172577 | 2021 | Peng Y, Xiao L, Rong H, Ou Z et al. Single-cell profiling of tumor-infiltrating TCF1/TCF7(+) T cells reveals a T lymphocyte subset associated with tertiary lymphoid structures/organs and a superior prognosis in oral cancer. Oral Oncol2021 Aug;119:105348. | scRNAseq | Primary tumor | 6 | --- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





