Submitted:
06 July 2024
Posted:
09 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
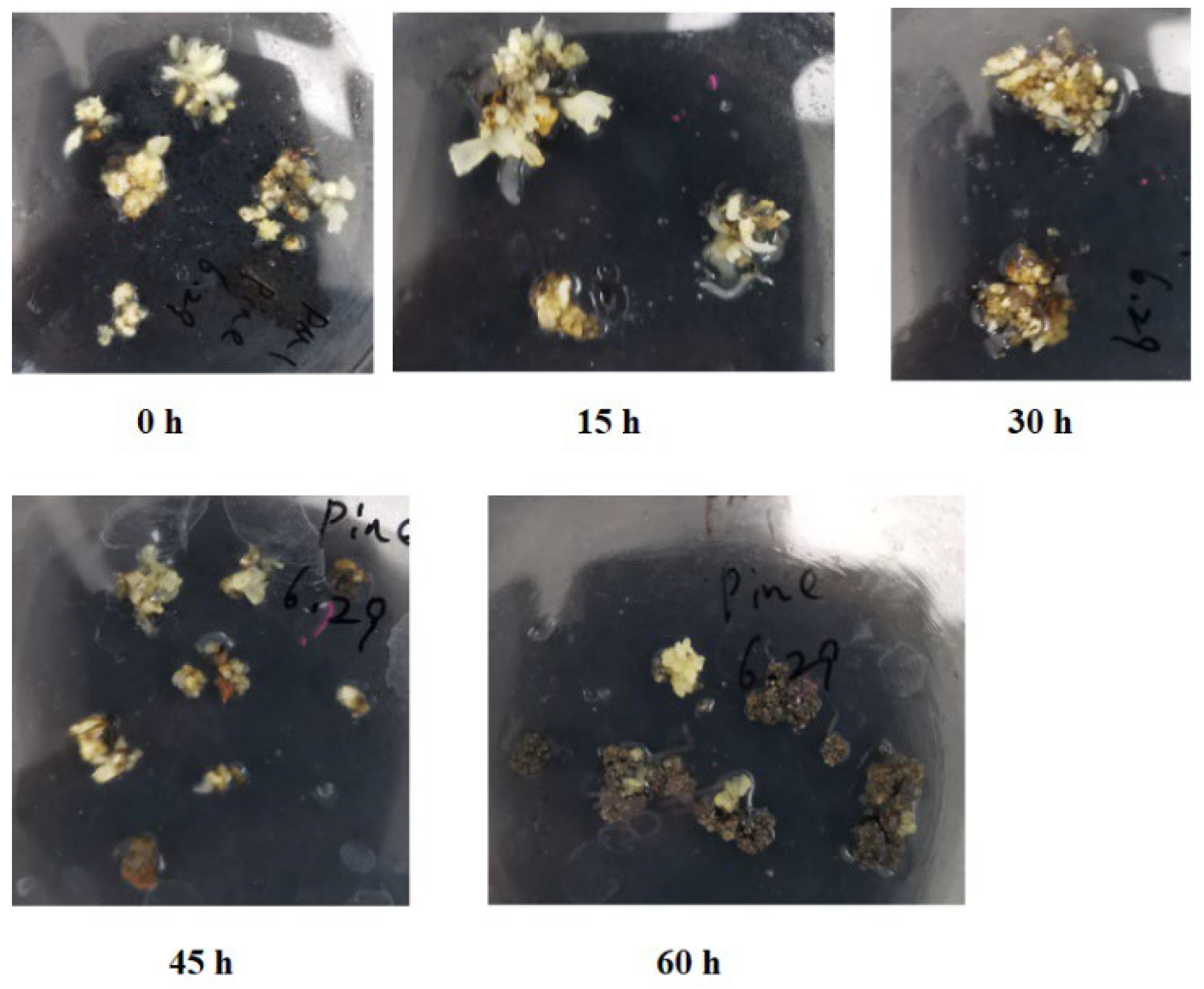
2.1. Phenotypic Characteristics of Mango Callus Infected by Agrobacterium
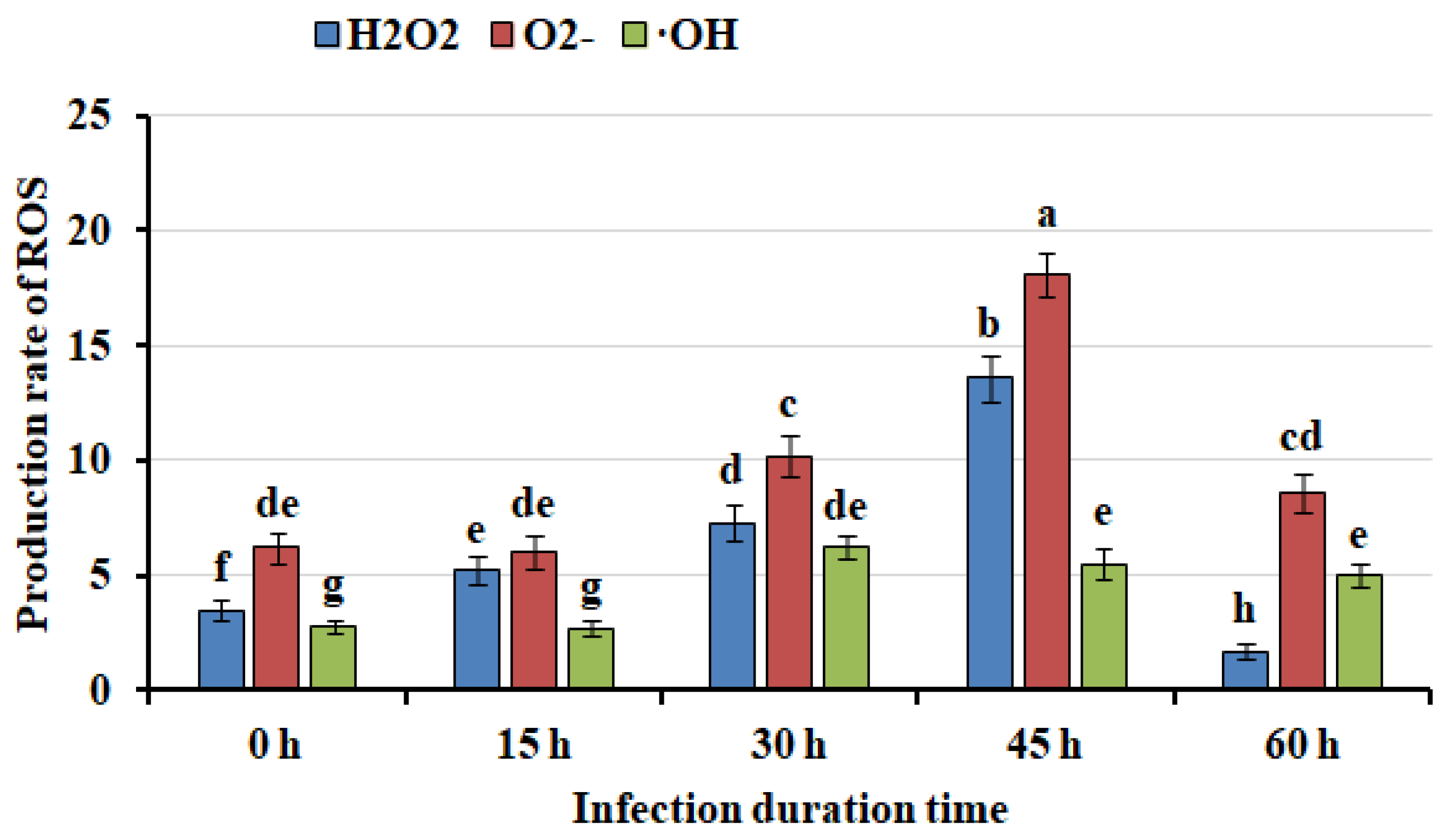
2.2. Mango Callus Infected by Agrobacterium Contained More Reactive Oxygen Species (ROS) than Control
2.3. Gallic Acid Was up Regulated after Mango Callus Was Infected
2.4. The Expressions of ROS-Related Genes and Gallic Acid Synthesis Genes Were up Regulated in Callus Infected by Agrobacterium
2.5. Verification the Expressions of Genes Participating in Immune Responses in Mango Callus Infected by Agrobacterium
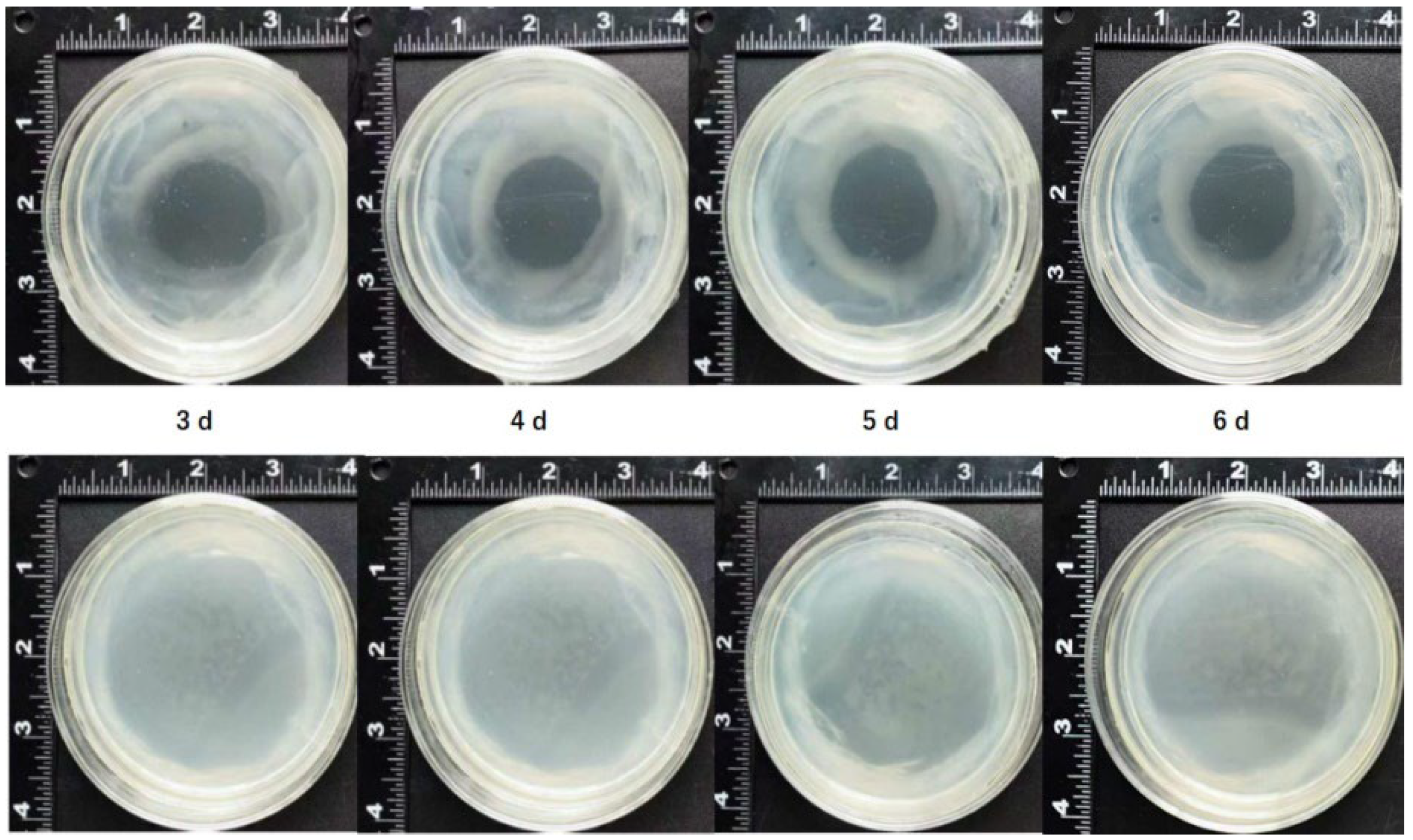
2.6. Gallic Acid Can Inhibit the Growth of Agrobacterium
2.7. Agrobacterium Harboring Type III Secretion Gene Cluster (T3SS) and Effector Gene AvrPto Had Higher Infection Efficiency than Control Agrobacterium
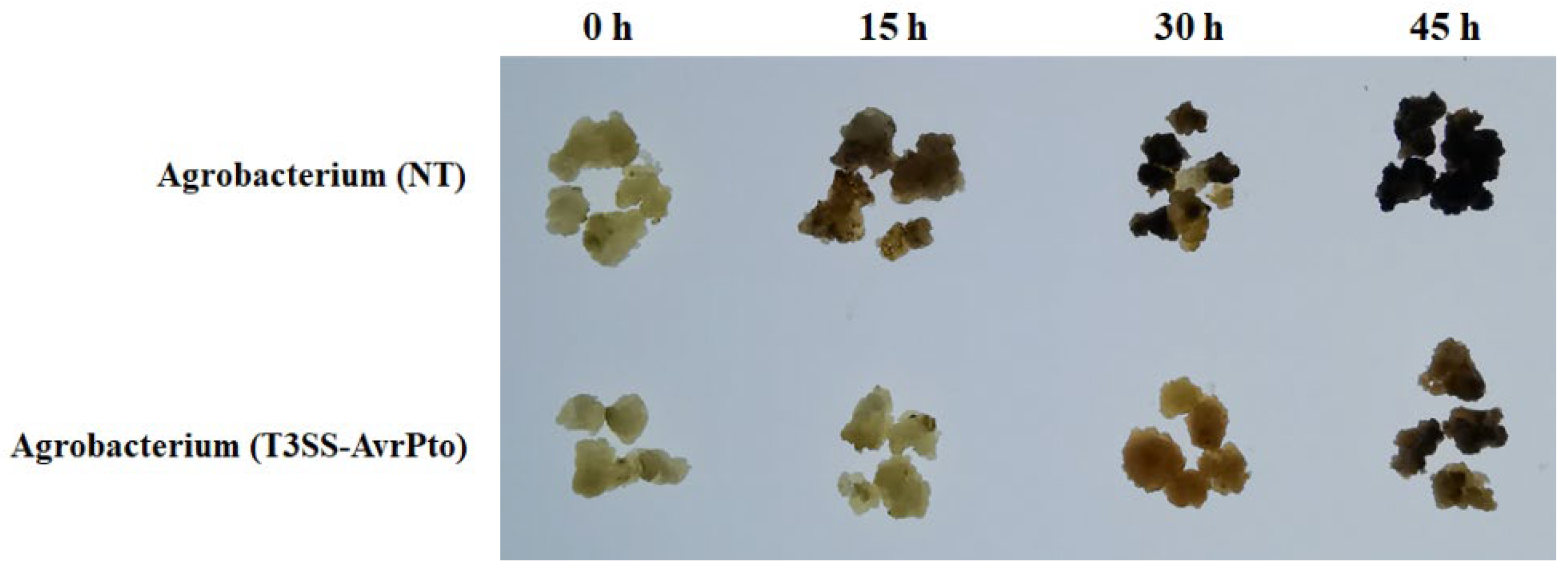
2.8. Mango Callus Infected by Agrobacterium (T3SS-AvrPto) Brown and Necrotize Slower than Those Infected by Control Agrobacterium
2.9. Mango Callus Infected by Agrobacterium (T3SS-AvrPto) Produced Less ROS than Those Infected by Agrobacterium (NV)
2.10. QRT-PCR Analysis of the Genes Up-Regulated in Mango Callus Infected by Agrobacterium (T3SS-AvrPto)
3. Discussion
4. Materials and Methods
4.1. Construction Agrobacterium Strains
4.2. Mango Embryogenic Callus Induction
4.3. Mango Callus Transformation
4.4. Metabolome and Transcriptome Experiments
4.5. QRT-PCR
4.6. Confocal Observation
5.7. Determination of ROS Contents in Mitochondria Isolated from Mango Callus
Author Contributions
Funding
Data Availability Statement
References
- Rai AC, Halon E, Zemach H, Zviran T, Sisai I, Philosoph-Hadas S, Meir S, Cohen Y, Irihimovitch V. Characterization of two ethephon-induced IDA-like genes from mango, and elucidation of their involvement in regulating organ abscission. Genes (Basel), 2021, 12, 439. [CrossRef]
- Denisov Y, Glick S, Zviran T, Ish-Shalom M, Levin A, Faigenboim A, Cohen Y, Irihimovitch V. Distinct organ-specific and temporal expression profiles of auxin-related genes during mango fruitlet drop. Plant Physiol Biochem., 2017, 115:439-448. [CrossRef]
- Njie A, Zhang W, Dong X, Lu C, Pan X, Liu Q. Effect of melatonin on fruit quality via decay inhibition and enhancement of antioxidative enzyme activities and genes expression of two mango cultivars during cold storage. Foods, 2022, 11, 3209. [CrossRef]
- Zhou Y, Liu J, Zhuo Q, Zhang K, Yan J, Tang B, Wei X, Lin L, Liu K. Exogenous glutathione maintains the postharvest quality of mango fruit by modulating the ascorbate-glutathione cycle. PeerJ., 2023, 11: e15902. [CrossRef]
- Yan T, Hu C, Que Y, Song Y, Lu D, Gu J, Ren Y, He J. Chitosan coating enriched with biosynthetic CuO NPs: Effects on postharvest decay and quality of mango fruit. International Journal of Biological Macromolecules, 2023, 253:126668. [CrossRef]
- Bally ISE, Lu P, Johnson PR. Mango Breeding. Jain SM, Priyadarshan PM (eds. T: Breeding Plantation Tree Crops, 2009.
- Litz RE, Hormaza JI. Mangifera indica mango. In: Litz RE, Pliego-Alfaro F, Hormaza JI (eds) Biotechnology of fruit and nut crops, 2nd edn. 2020.
- Petri C, Litz RE, Singh SK, Hormaza JI. In vitro culture and genetic transformation in mango. C. Kole (ed.), The mango genome, compendium of plant genomes, Springer Nature Switzerland AG, 2021, pp 131-151.
- Pandey K, Karthik K, Singh SK, Vinod, Sreevathsa R, Srivastav M. Amenability of an Agrobacterium tumefaciens-mediated shoot apical meristem-targeted in planta transformation strategy in Mango (Mangifera indica L.). GM Crops & Food, 2022, 13, 342–354. [CrossRef]
- Adjei MO, Zhao H, Tao X, Yang L, Deng S, Li X, Mao X, Li S, Huang J, Luo R, Gao A, Ma J. Using a protoplast transformation system to enable functional studies in Mangifera indica L. International Journal of Molecular Sciences, 2023, 24: 11984. [CrossRef]
- Chen X, He S, Wang Z, Zhai Y, Guo W, Li X. Production of transgenic periclinal chimeras in pumpkin - a tool for revealing cell fates of L1 meristem. Plant biology, 2023, 26, 126–139. [CrossRef]
- Song X, Xie Y, Tian X, Wang N, Zhou Y, Xie Z, Ye J, Deng X. CRISPR/Cas9 editing characteristics of multiple transgenic generations in Fortunella hindsii, an early flowering mini-citrus. Scientia Horticulturae, 2023, 321: 112236. [CrossRef]
- Aida R, Sasaki K, Yoshioka S, Noda N. Distribution of cell layers in floral organs of chrysanthemum analyzed with periclinal chimeras carrying a transgene encoding fluorescent protein. Plant Cell Reports, 2020. [CrossRef]
- Raza G, Singh MB, Bhalla PL. Somatic embryogenesis and plant regeneration from commercial soybean cultivars. Plants, 2020, 9: 38. [CrossRef]
- Li M, Wang D, Long X, Hao Z, Lu Y, Zhou Y, Peng Y, Cheng T, Shi J, Chen J. Agrobacterium, 8: genetic transformation of embryogenic callus in a Liriodendron hybrid (L. Chinense × L. Tulipifera). Frontiers in Plant Science, 2022, 13, 2022. [CrossRef]
- Nakajima I, Endo M, Haji T, Moriguchi T, Yamamoto T. Embryogenic callus induction and Agrobacterium-mediated genetic transformation of ‘Shine Muscat’ grape. Plant Biotechnology, 2020, 37: 185-194. [CrossRef]
- Chen Z, Debernardi JM, Dubcovsky J, Gallavotti A. Recent advances in crop transformation technologies. Nat Plants, 2022, 8, 1343–1351.
- Christie, PJ. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Bophys. Acta, 2004, 1694: 219-234. [CrossRef]
- Raman V, Rojas CM, Vasudevan B, Dunning K, Kolape J, Oh S, Yun J, Yang L, Li G, Pant BD, Jiang Q, Mysore KS. Agrobacterium, 2: a type III secretion system delivers Pseudomonas effectors into plant cells to enhance transformation. Nature Communications, 2022, 13, 2022. [CrossRef]
- Büttner, D. Behind the lines–actions of bacterial type III effector proteins in plant cells. FEMS Microbiol. Rev., 2016, 40: 894-937. [CrossRef]
- Tang D, Wang G, Zhou J. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell, 2017, 29: 618-637.
- Zhang D, Ren L, Chen G, Zhang J, Reed BM, Shen X. ROS-induced oxidative stress and apoptosis-like event directly affect the cell viability of cryopreserved embryogenic callus in Agapanthus praecox. Plant Cell Rep, 2015, 34, 1499–513. [CrossRef]
- Agita A, Alsagaff MT. Inflammation, immunity, and hypertension. Acta Med Indones, 2017, 49, 158–165.
- Pitsili E, Phukan UJ, Coll NS. Cell death in plant immunity. Cold Spring Harb Perspect Biol 2020, 12: a036483. [CrossRef]
- Zavaliev R, Mohan R, Chen T, Dong X. Formation of NPR1 condensates promotes cell survival during plant immune response. Cell, 2020, 182, 1093–1108. [CrossRef]
- Peng T, Tao X, Xia Z, Hu S, Xue J, Zhu Q, Pan X, Zhang Q, Li S. Pathogen hijacks programmed cell death signaling by arginine ADPR-deacylization of caspases. Molecular Cell, 2022, 82: 1806-1820. [CrossRef]
- Yang F, Li G, Felix G, Albert M, Guo M. Engineered Agrobacterium improves transformation by mitigating plant immunity detection. New Phytologist, 2022, 237, 2493–2504. [CrossRef]
- Li X, Pan SQ. Agrobacterium, e: VirE2 protein into host cells via clathrin-mediated endocytosis. Science Advances, 2017, 3, 2017. [CrossRef]
- Huang HC, Schuurink R, Denny TP, Atkinson MM, Baker CJ, Yucel I, Hutcheson SW, Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J. Bacteriol., 1988, 170, 4748–4756. [CrossRef]
- Muir RM, Ibanez AM, Uratsu SL, Ingham ES, Leslie CA, McGranahan GH, Batra N, Goyal S, Joseph J, Jemmis ED, Dandekar AM. Mechanism of gallic acid biosynthesis in bacteria (Escherichia coli) and walnut (Juglans regia). Plant Mol Biol, 2011, 75:555-565. [CrossRef]
- Zhu Y, Kun Lin K, Zhang X, Ma H, Yang L, Wei L, Yang L, Jiang M. Antibacterial effect of gallic acid in UV-C light treatment against Escherichia coli O157:H7 and the underlying mechanism. Food and Bioprocess Technology, 2023. [CrossRef]
- Ilie CI, Oprea E, Geana EI, Spoiala A, Buleandra M, Pircalabioru G, Badea IA, Ficai D, Andronescu E, Ficai A, Ditu LM. Bee pollen extracts: Chemical composition, antioxidant properties, and effect on the growth of selected probiotic and pathogenic bacteria. Antioxidants (Basel), 2022, 11(5). [CrossRef]
- Delplace F, Huard-Chauveau C, Berthome R, Roby D. Network organization of the plant immune system: from pathogen perception to robust defense induction. The Plant Journal, 2022, 109: 447-470. [CrossRef]
- Lovelace AH, Dorhmi S, Hulin MT, Li Y, Mansfield JW, Ma W. Effector identification in plant pathogens. Phytopathology, 2023, 113, 637–650. [CrossRef]
- Ngou BPM, Ding P, Jones JDG. Thirty years of resistance: Zig-zag through the plant immune system. The Plant Cell, 2022, 34: 1447-1478. [CrossRef]
- Liu T, Cao L, Cheng Y, Ji J, Wei Y, Wang C, Duan K. MKK4/5-MPK3/6 cascade regulates Agrobacterium-mediated transformation by modulating plant immunity in Arabidopsis. Frontiers in Plant Science, 2021, 12: 731690. [CrossRef]
- Cai J, Chen T, Wang Y, Qin G, Tian S. SlREM1 triggers cell death by activating an oxidative burst and other regulators. Plant Physiology, 2020, 183, 717–732. [CrossRef]
- Ding L, Li Y, Wu Y, Li T, Geng R, Cao J, Zhang W, Tan X. Plant disease resistance-related signaling pathways: recent progress and future prospects. International Journal of Molecular Sciences, 2022, 23: 16200. [CrossRef]
- Zhou JM, Zhang Y. Plant immunity: danger perception and signaling. Cell, 2020, 181: 978-989. [CrossRef]
- Tiwari M, Mishra AK, Chakrabarty D. Agrobacterium, 3: gene transfer: recent advancements and layered immunity in plants. Planta, 2022, 256, 2022. [CrossRef]
- Wise AA, Liu Z, Binns AN. in Agrobacterium Protocols (ed. Kan Wang) 43-54 (Humana Press, 2006).
- Xu W, Wu H, Luo C, Li L, Liang Q, Ma X, Yao Q, Gao Y, Wang S. Callus proliferation, early somatic embryogenesis and cytological observation of mango ‘Jinhuang’. Molecular Plant Breeding, 2019, 17, 3001–3008 (In Chinese language), (In Chinese language).
- Iqbal A, Qiang D, Xiangru W, Huiping G, Hengheng Z, Xiling Z, Meizhen S. Integrative physiological, transcriptome and metabolome analysis reveals the involvement of carbon and flavonoid biosynthesis in low phosphorus tolerance in cotton. Plant Physiology and Biochemistry, 2023, 196:302-317. [CrossRef]
- Huang and Hwang, Arabidopsis RETICULON-LIKE4 (RTNLB4) protein participates in Agrobacterium infection and VirB2 peptide-induced plant defense response. International Journal of Molecular Sciences, 2020, 21: 1722. [CrossRef]
- Vanlerberghe GC, McIntosh L. Lower growth temperature increases alternative pathway capacity and alternative oxidase protein in tobacco. Plant Physiol., 1992, 100: 115-119. [CrossRef]
- Patterson BD, Mackae EA, Ferguson IB. Estimation of hydrogen peroxide in plant extracts using Titanium. Anal. Biochem., 1984, 139: 487-492. [CrossRef]
- Wang AG, Luo GH. Quantitative relation between the reaction of hydroxylamine and superoxide anion radicals in plants. Plant Physiol. Commun., 1990, 6:55-57.
- Simontacchi M, Puntarulo S. Oxygen radical generation by isolated microsomes from soybean seedling. Plant Physiol., 1992, 100: 1263-1268. [CrossRef]
- Nash, T. The colorimetric estimation of formaldehyde by means of the Hantzsch reaction, Biochem. J., 1953, 55: 416-421. [CrossRef]
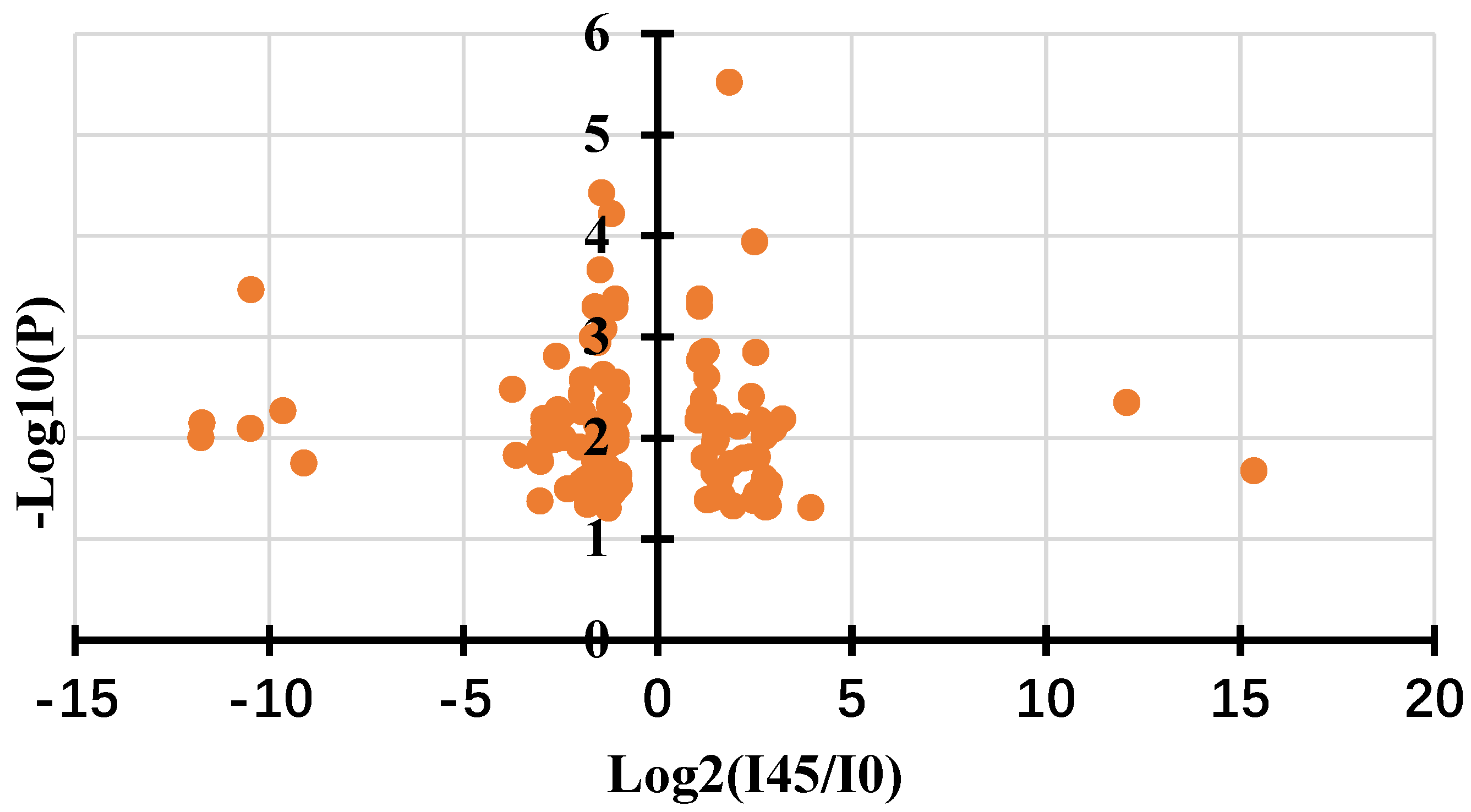
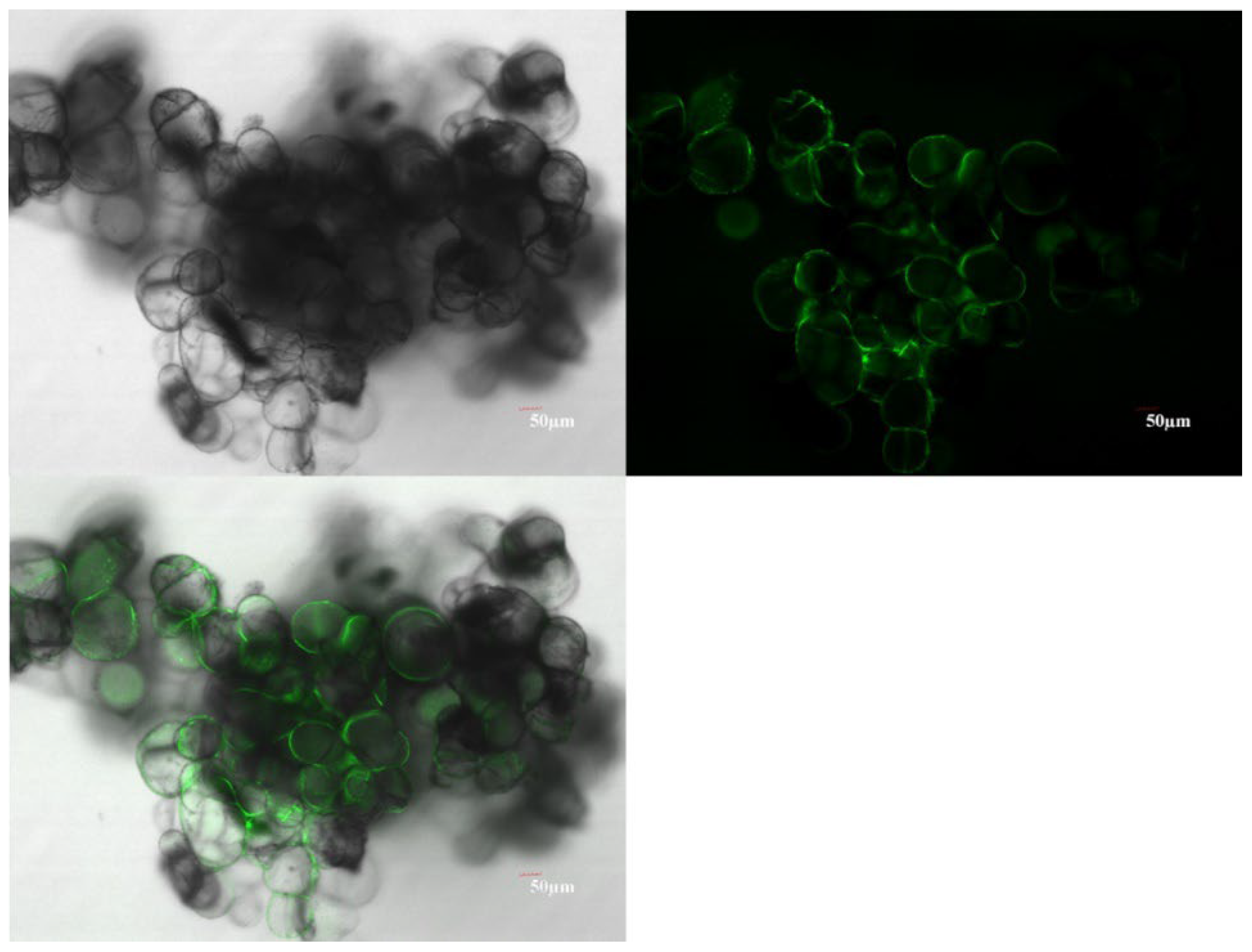

| Index | compounds | Fold | VIP value |
| Gallic acid | Organic acid | 7.1 | 1.41 |
| Phosphoenolpyruvic acid | Organic acid | 2.9 | 1.22 |
| D-erythrose-4-phosphate | Phosphorylated derivatives of erythritol | 4.3 | 1.36 |
| Ubiquinone | Liposoluble quinone compound | 5.2 | 1.25 |
| Dihydrotricetin | Flavonoids | 2.3 | 1.26 |
| Dihydrokaempferol | Flavonoid | 1.8 | 1.19 |
| Cyanidin | Antioxidant | 2.5 | 1.28 |
| Query | Symbol | Log2(I45/I0) | Gene annotation | Subject | |
|---|---|---|---|---|---|
| LOC123211694 | DAHPS1 | 5.2 | Mangifera indica 3-oxoacyl-[acyl-carrier-protein] synthase | XM_044646917.1 | |
| LOC123214892 | DAHPS2 | 5.5 | Mangifera indica 3-oxoacyl-[acyl-carrier-protein] synthase II | XM_044641225.1 | |
| LOC123210632 | DHQD1 | 3 | Mangifera indica bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase | XM_044644036.1 | |
| LOC123236405 | DHQD2 | 3.7 | Mangifera indica bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase | XM_044644035.1 | |
| LOC123207068 | SHD1 | 2.4 | Mangifera indica bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase | XM_044644037.1 | |
| LOC123215035 | SHD2 | 2.2 | Mangifera indica bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase | XM_044644039.1 | |
| LOC123225839 | CAT1 | 2.9 | Mangifera indica catalase isozyme 1 | XM_044655447.1 | |
| LOC123228490 | CAT2 | 2.6 | Mangifera indica catalase isozyme 1 | XM_044655447.1 | |
| LOC123217301 | GOX1 | 1.7 | Mangifera indica peroxisomal (S)-2-hydroxy-acid oxidase-like | XM_044639087.1 | |
| LOC123227499 | GOX2 | 1.9 | Mangifera indica peroxisomal (S)-2-hydroxy-acid oxidase-like | XM_044618393.1 | |
| LOC123204838 | FOX1 | 3.4 | Mangifera indica FAD-linked sulfhydryl oxidase ERV1-like | XM_044652170.1 | |
| LOC123227490 | FOX2 | 3.8 | Mangifera indica FAD-linked sulfhydryl oxidase ERV1-like | XM_044625447.1 | |
| LOC123206396 | POD1 | 2.8 | Mangifera indica peroxidase A2-like | XM_044652117.1 | |
| LOC123208403 | POD2 | 2.5 | Mangifera indica lignin-forming anionic peroxidase-like | XM_044627935.1 | |
| LOC123217403 | NADO1 | 3.2 | Mangifera indica respiratory burst oxidase homolog protein D-like | XM_044606776.1 | |
| LOC123228596 | NADO2 | 3.6 | Mangifera indica respiratory burst oxidase homolog protein A-like | XM_044630849.1 | |
| LOC123217494 | FLS2-1 | 2.1 | Mangifera indica LRR receptor-like serine/threonine-protein kinase FLS2 | XM_044604788.1 | |
| LOC123227485 | MPK3-1 | 1.7 | Mangifera indica mitogen-activated protein kinase kinase 3-like | XM_044610886.1 | |
| LOC123210475 | MPK6-1 | 2.6 | Mangifera indica mitogen-activated protein kinase kinase 6 | XM_044607731.1 | |
| LOC123210574 | WRKY22-1 | 1.5 | Mangifera indica WRKY transcription factor 22-like | XM_044646893.1 | |
| LOC123226579 | NHL10-1 | 3.8 | Arabidopsis thaliana Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family (NDR1) | XM_044626069.1 |
| Gene | 0 h | 15 h | 30 h | 45 h | 60 h |
| MiDAHPS1 | 1.0±0.1 | 1.5±0.1 | 1.9±0.1 | 5.0±0.3 | 6.0±0.5 |
| MiDHQD1 | 0.6±0.1 | 0.6±0.1 | 0.8±0.1 | 1.9±0.2 | 2.0±0.2 |
| MiSHD1 | 1.5±0.1 | 1.7±0.1 | 1.9±0.2 | 3.6±0.2 | 3.8±0.3 |
| MiCAT1 | 0.8±0.1 | 0.8±0.1 | 1.2±0.1 | 1.9±0.2 | 1.8±0.2 |
| MiGOX1 | 2.3±0.2 | 2.5±0.2 | 2.8±0.2 | 3.9±0.3 | 3.5±0.3 |
| MiFOX1 | 1.6±0.1 | 1.7±0.1 | 2.1±0.2 | 5.6±0.3 | 5.0±0.3 |
| MiPOD1 | 3.1±0.2 | 3.2±0.2 | 3.5±0.2 | 7.3±0.5 | 6.8±0.5 |
| MiNADO1 | 1.2±0.1 | 1.2±0.1 | 1.8±0.1 | 3.6±0.2 | 3.2±0.2 |
| MiFLS2-1 | 0.8±0.1 | 0.8±0.1 | 1.1±0.1 | 1.5±0.1 | 3.9±0.2 |
| MiPK3-1 | 5.2±0.3 | 5.5±0.3 | 5.8±0.3 | 8.5±0.5 | 8.8±0.5 |
| MiPK6-1 | 3.3±0.2 | 3.5±0.2 | 3.6±0.3 | 7.2±0.5 | 7.5±0.5 |
| MiWRKY22-1 | 1.6±0.1 | 1.9±0.2 | 2.1±0.2 | 2.6±0.2 | 6.0±0.5 |
| MiNHL10-1 | 2.6±0.2 | 2.6±0.2 | 2.8±0.2 | 8.5±0.5 | 7.2±0.5 |
| Agrobacterium | Infection time (h) | H2O2 (μmol/mg protein) | O2- (0.1 nmol/(min mg protein)) | ·OH (nmol/(min mg protein)) |
| NV | 0 h | 3.5±0.5 | 6.2±0.6 | 2.8±0.2 |
| NV | 15 h | 5.2±0.6 | 6.0±0.5 | 2.7±0.2 |
| NV | 30 h | 13.6±1.3 | 18.1±1.5 | 5.5±0.4 |
| NV | 45 h | 7.3±0.8 | 10.2±1.0 | 6.2±0.5 |
| NV | 60 h | 1.7±0.3 | 8.6±0.7 | 5.0±0.4 |
| T3SS-AvrPto | 0 h | 3.6±0.4 | 6.1±0.5 | 2.9±0.2 |
| T3SS-AvrPto | 15 h | 3.9±0.5 | 6.3±0.6 | 2.7±0.2 |
| T3SS-AvrPto | 30 h | 4.5±0.5 | 6.6±0.5 | 3.0±0.2 |
| T3SS-AvrPto | 45 h | 4.7±0.5 | 6.9±0.6 | 3.5±0.3 |
| T3SS-AvrPto | 60 h | 5.0±0.6 | 7.6±0.6 | 4.3±0.3 |
| Gene | 0 h (NV) | 15 h (NV) | 30 h (NV) | 45 h (NV) | 60 h (NV) | 0 h (T) | 15 h (T) | 30 h (T) | 45 h (T) | 60 h (T) |
| MiDAHPS1 | 1.0±0.1 | 1.5±0.1 | 1.9±0.1 | 5.0±0.3 | 6.0±0.5 | 1.1±0.1 | 1.3±0.1 | 1.7±0.1 | 5.0±0.3 | 6.0±0.5 |
| MiDHQD1 | 0.6±0.1 | 0.6±0.1 | 0.8±0.1 | 1.9±0.2 | 2.0±0.2 | 0.5±0.1 | 0.6±0.1 | 0.8±0.1 | 1.9±0.2 | 2.0±0.2 |
| MiSHD1 | 1.5±0.1 | 1.7±0.1 | 1.9±0.2 | 3.6±0.2 | 3.8±0.3 | 1.6±0.1 | 1.7±0.1 | 1.9±0.2 | 3.1±0.2 | 3.8±0.3 |
| MiCAT1 | 0.8±0.1 | 0.8±0.1 | 1.2±0.1 | 1.9±0.2 | 1.8±0.2 | 0.7±0.1 | 0.8±0.1 | 1.0±0.1 | 1.9±0.2 | 1.8±0.2 |
| MiGOX1 | 2.3±0.2 | 2.5±0.2 | 2.8±0.2 | 3.9±0.3 | 3.5±0.3 | 2.2±0.2 | 2.5±0.2 | 2.8±0.2 | 3.9±0.3 | 3.5±0.3 |
| MiFOX1 | 1.6±0.1 | 1.7±0.1 | 2.1±0.2 | 5.6±0.3 | 5.0±0.3 | 1.5±0.1 | 1.7±0.1 | 2.1±0.2 | 3.6±0.3 | 5.0±0.3 |
| MiPOD1 | 3.1±0.2 | 3.2±0.2 | 3.5±0.2 | 7.3±0.5 | 6.8±0.5 | 3.0±0.2 | 3.2±0.2 | 3.5±0.2 | 5.6±0.3 | 7.7±0.5 |
| MiNADO1 | 1.2±0.1 | 1.2±0.1 | 1.8±0.1 | 3.6±0.2 | 3.2±0.2 | 1.3±0.1 | 1.2±0.1 | 1.8±0.1 | 3.6±0.2 | 3.2±0.2 |
| MiFLS2-1 | 0.8±0.1 | 0.8±0.1 | 1.1±0.1 | 1.5±0.1 | 3.9±0.2 | 0.9±0.1 | 0.8±0.1 | 1.1±0.1 | 1.5±0.1 | 3.9±0.2 |
| MiPK3-1 | 5.2±0.3 | 5.5±0.3 | 5.8±0.3 | 8.5±0.5 | 8.8±0.5 | 5.1±0.3 | 5.5±0.3 | 5.8±0.3 | 7.6±0.5 | 8.8±0.5 |
| MiPK6-1 | 3.3±0.2 | 3.5±0.2 | 3.6±0.3 | 7.2±0.5 | 7.5±0.5 | 3.2±0.2 | 3.5±0.2 | 3.6±0.3 | 5.2±0.5 | 7.5±0.5 |
| MiWRKY22-1 | 1.6±0.1 | 1.9±0.2 | 2.1±0.2 | 2.6±0.2 | 6.0±0.5 | 1.5±0.1 | 1.7±0.2 | 2.1±0.2 | 2.6±0.2 | 4.5±0.5 |
| MiNHL10-1 | 2.6±0.2 | 2.6±0.2 | 2.8±0.2 | 8.5±0.5 | 7.2±0.5 | 2.6±0.2 | 2.6±0.2 | 2.8±0.2 | 3.5±0.5 | 5.3±0.5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




