Submitted:
04 July 2024
Posted:
05 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Genome Download and Collation
2.2. Gene Annotation and Functional Analysis
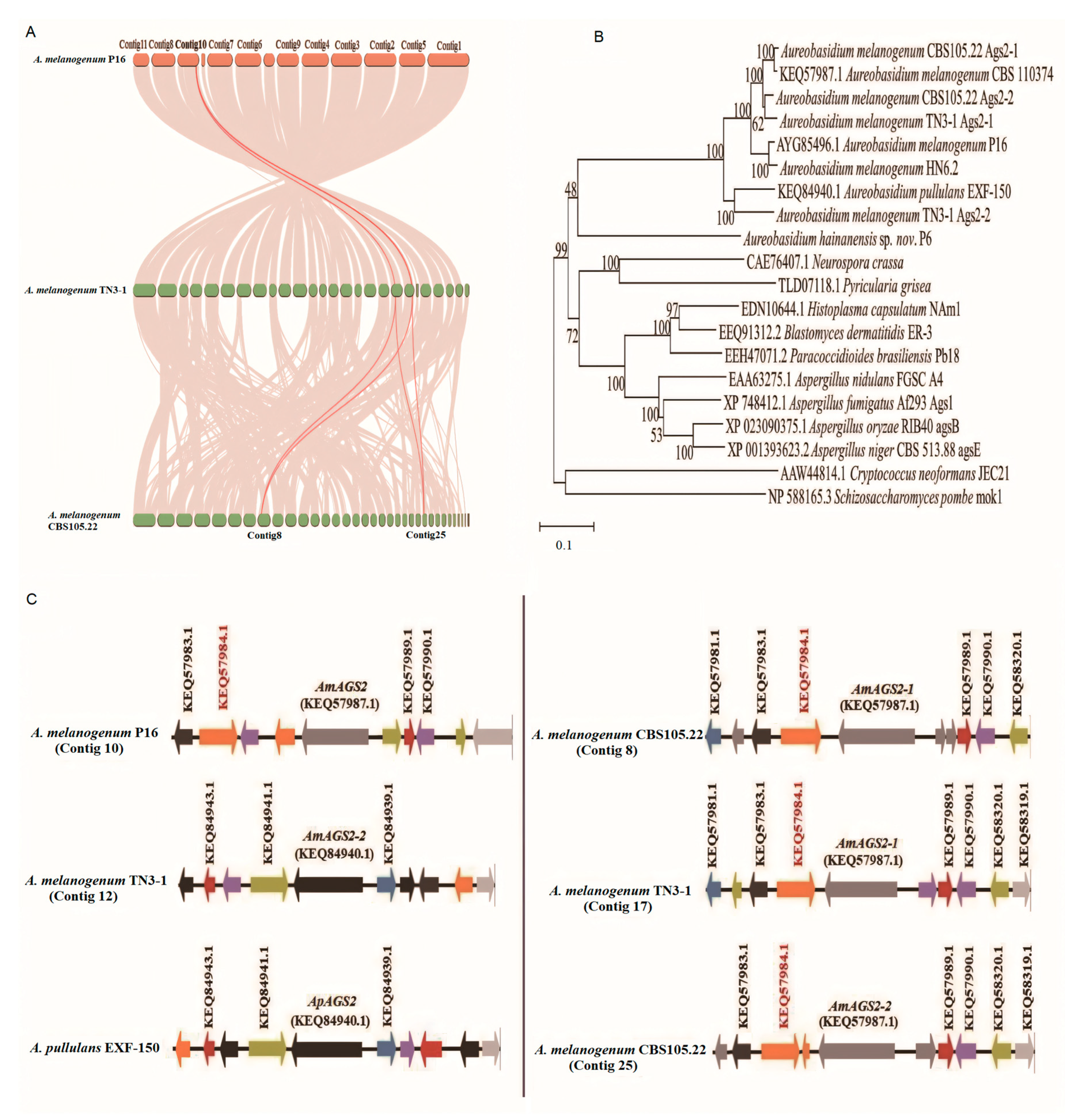
2.3. Gene Alignment and Genomic Collinearity Analysis
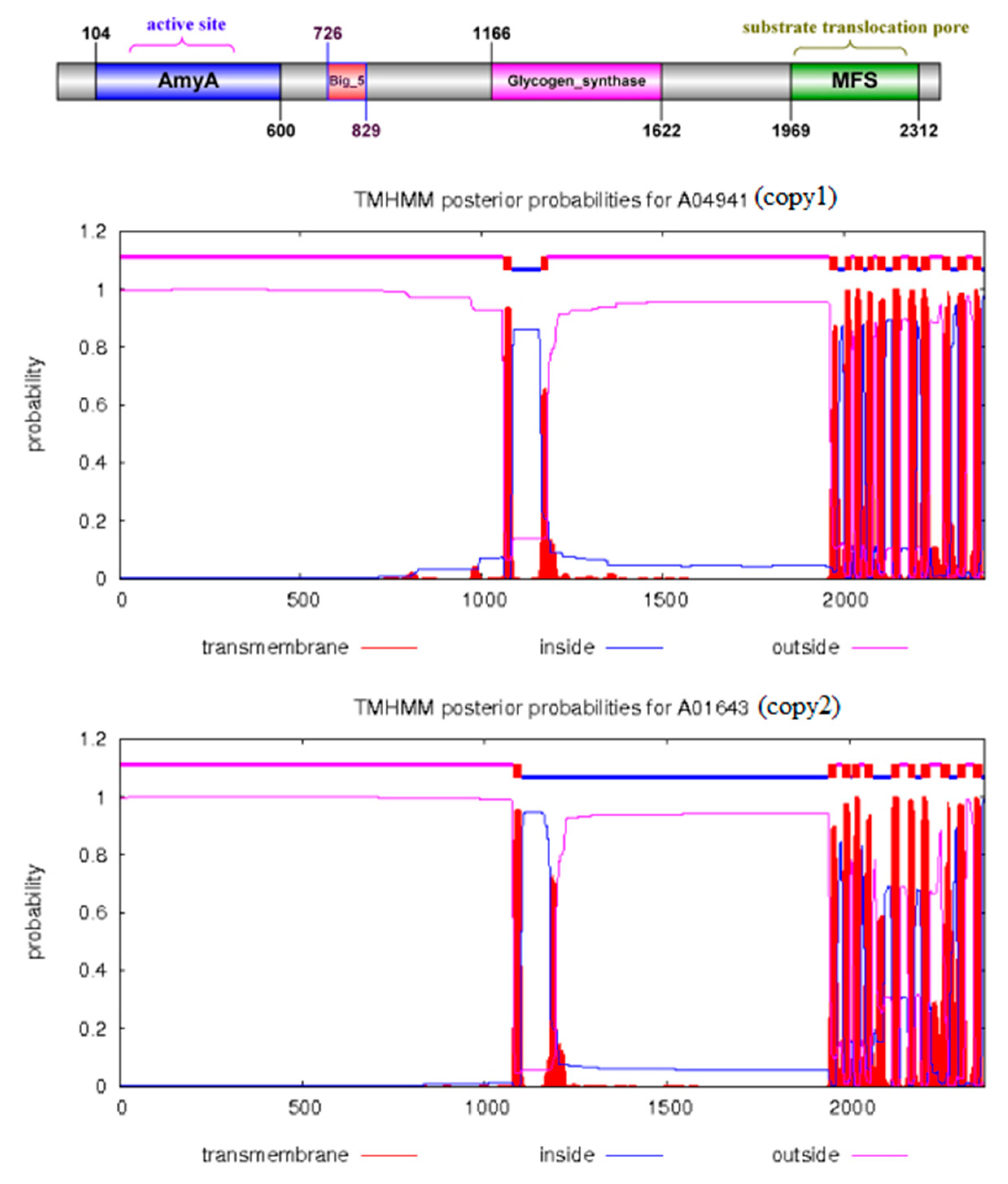
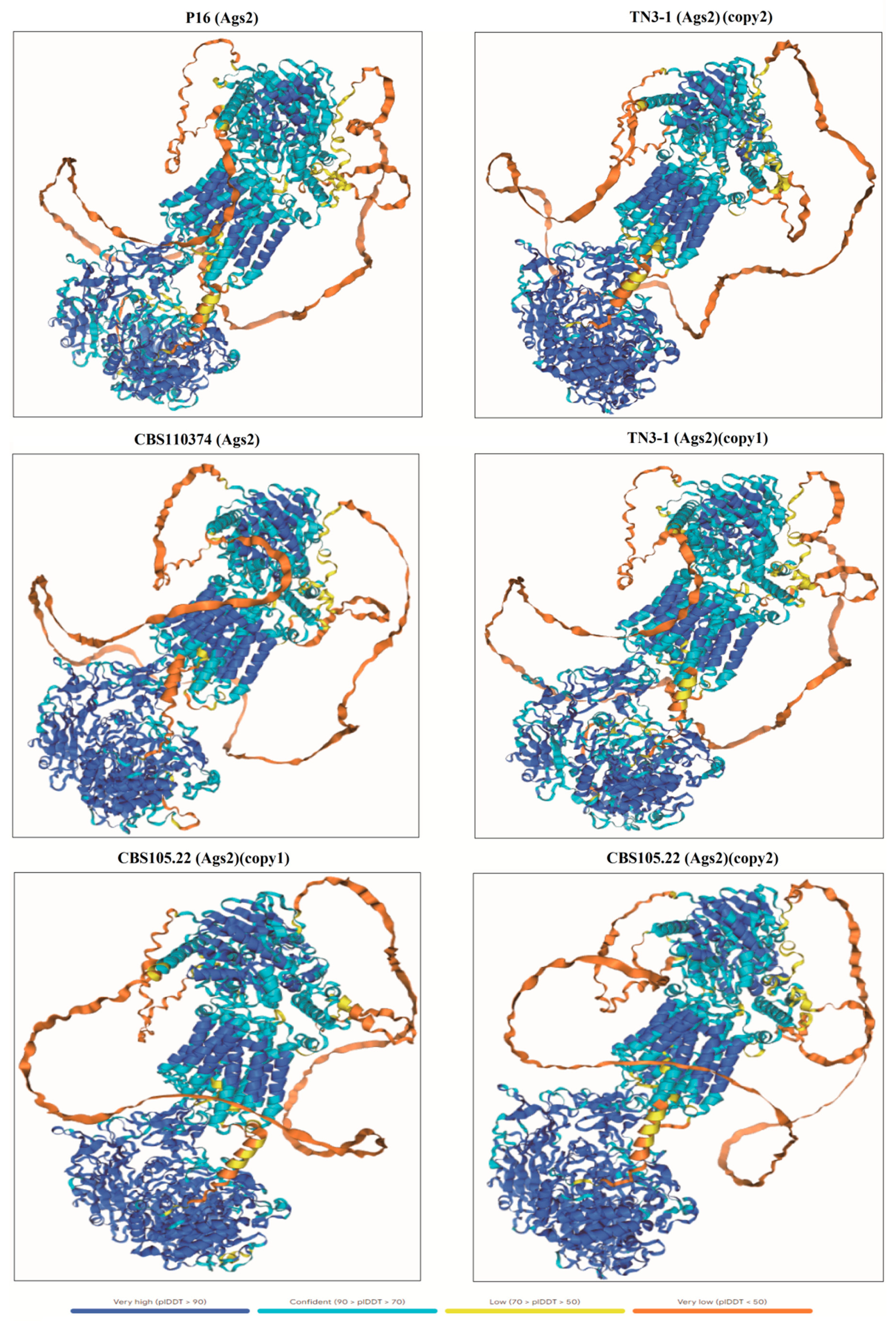
2.4. 3D Structure Prediction of the α-1,3-Glucan Synthase
2.5. Pan-Genome and Genome Evolution
2.6. Transcriptional Factor Analysis for Crucial Genes
3. Results
3.1. Genome Analysis of A. melanogenum TN3-1
3.2. TN3-1 Strain Had a Unique Pullulan Synthesis Gene
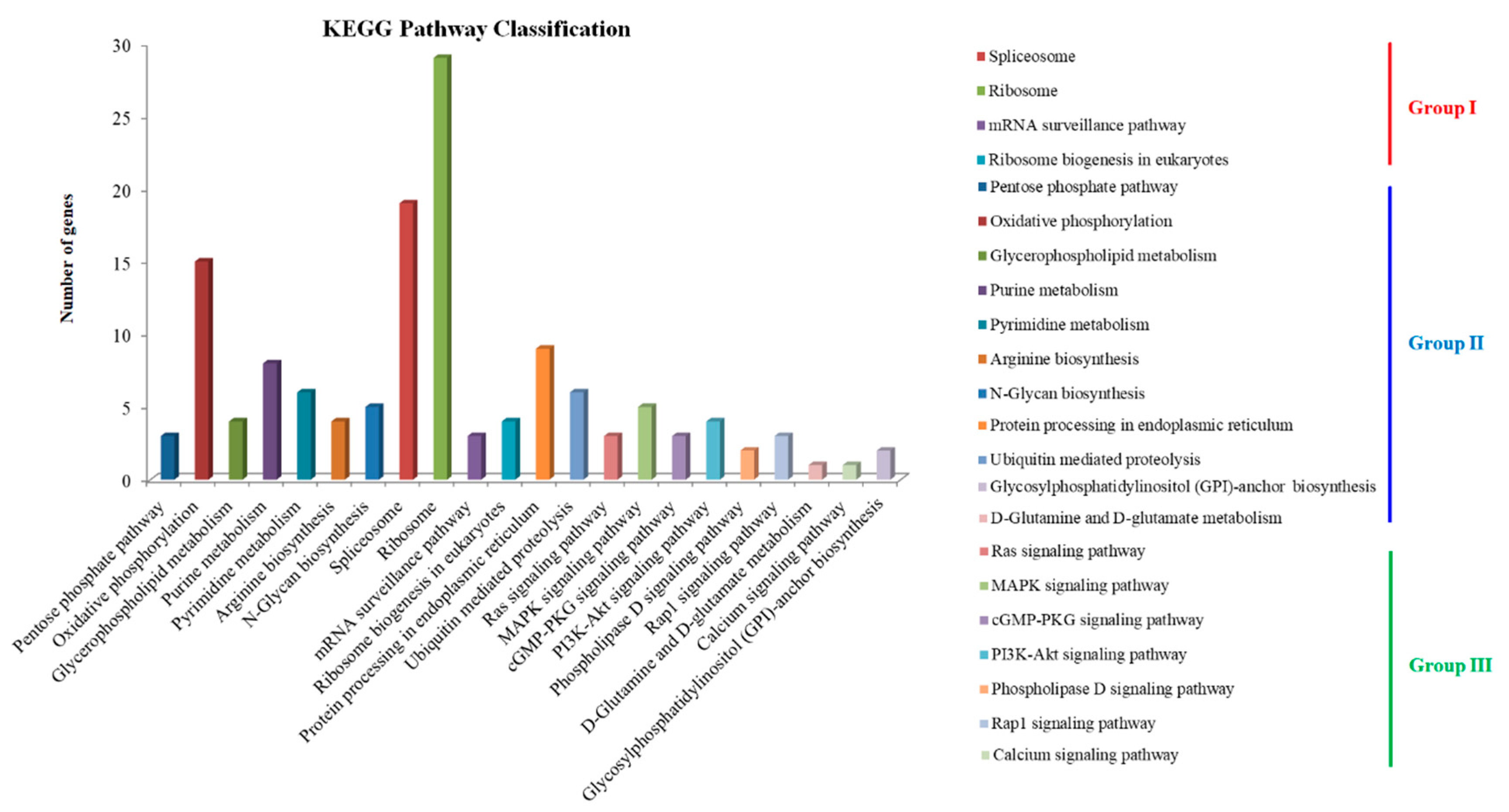
3.3. TN3-1 Strain Activated Distinct Pathways in Response to Environmental Changes
3.4. Efficient Energy Supply Provides Strong Assurance
4. Discussion
5. Conclusion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tiwari, S.; Patil, R.; Dubey, S.K.; Bahadur, P. Derivatization approaches and applications of pullulan. Adv. Colloid Interface Sci. 2019, 269, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liang, Z.; Liang, Z.; Lv, W.; Chen, M.; Zhao, Y. Advancements of Prussian blue-based nanoplatforms in biomedical fields: Progress and perspectives. J. Control. Release 2022, 351, 752–778. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.S.; Kaur, N.; Singh, D.; Purewal, S.S.; Kennedy, J.F. Pullulan in pharmaceutical and cosmeceutical formulations: A review. Int. J. Biol. Macromol. 2023, 231, 123353. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.-L.; Liu, G.-L.; Hu, Z.; Chi, Z.-M. Fungi in mangrove ecosystems and their potential applications. Crit. Rev. Biotechnol. 2020, 40, 852–864. [Google Scholar] [CrossRef]
- Chen, L.; Wei, X.; Liu, G.-L.; Hu, Z.; Chi, Z.-M.; Chi, Z. Glycerol, trehalose and vacuoles had relations to pullulan synthesis and osmotic tolerance by the whole genome duplicated strain Aureobasidium melanogenum TN3-1 isolated from natural honey. Int. J. Biol. Macromol. 2020, 165, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.J.; Liu, G.L.; Wei, X.; Wang, K.; Hu, Z.; Chi, Z.; Chi, Z.M. A multidomain alpha-Glucan synthetase 2 (AmAgs2) is the crucial enzyme for pullulan biosynthesis in Aureobasidium melanogenum P16. Int J Biol Macromol. 2020, 150, 1037–1045. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.-J.; Jiang, H.; Liu, G.-L.; Hu, Z.; Chi, Z.-M. Cell wall integrity is required for pullulan biosynthesis and glycogen accumulation in Aureobasidium melanogenum P16. Biochim. et Biophys. Acta (BBA) - Gen. Subj. 2018, 1862, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Yang, R.; Yang, S.; Guan, J.; Zhang, D.; Ma, Y.; Liu, H. Research progress of self-assembled nanogel and hybrid hydrogel systems based on pullulan derivatives. Drug Deliv. 2018, 25, 278–292. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.-J.; Chen, L.; Jiang, H.; Liu, G.-L.; Chi, Z.-M.; Hu, Z. High pullulan biosynthesis from high concentration of glucose by a hyperosmotic resistant, yeast-like fungal strain isolated from a natural comb-honey. Food Chem. 2019, 286, 123–128. [Google Scholar] [CrossRef]
- Jia, S.-L.; Zhang, M.; Liu, G.-L.; Chi, Z.-M.; Chi, Z. Novel chromosomes and genomes provide new insights into evolution and adaptation of the whole genome duplicated yeast-like fungus TN3-1 isolated from natural honey. Funct. Integr. Genom. 2023, 23, 1–21. [Google Scholar] [CrossRef]
- Gostinčar, C.; Ohm, R.A.; Kogej, T.; Sonjak, S.; Turk, M.; Zajc, J.; Zalar, P.; Grube, M.; Sun, H.; Han, J.; et al. Genome sequencing of four Aureobasidium pullulans varieties: biotechnological potential, stress tolerance, and description of new species. BMC Genom. 2014, 15, 549. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Wang, X.; Sun, Y.; Joseph, P.V.; Paterson, A.H. Detection of colinear blocks and synteny and evolutionary analyses based on utilization of MCScanX. Nat. Protoc. 2024, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Abramson, J.; Adler, J.; Dunger, J.; Evans, R.; Green, T.; Pritzel, A.; Ronneberger, O.; Willmore, L.; Ballard, A.J.; Bambrick, J.; et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 2024, 630, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a Versatile Software Package for Scalable and Robust Microbial Pangenome Analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, P.T.; Oliveira, J.; Pais, P.; Antunes, M.; Palma, M.; Cavalheiro, M.; Galocha, M.; Godinho, C.P.; Martins, L.C.; Bourbon, N.; et al. YEASTRACT+: a portal for cross-species comparative genomics of transcription regulation in yeasts. Nucleic Acids Res. 2020, 48, D642–D649. [Google Scholar] [CrossRef] [PubMed]
- Qi, C.Y.; Jia, S.L.; Wei, X.; Yang, G.; Chi, Z.; Liu, G.L.; Hu, Z.; Chi, Z.M. The differences between fungal alpha-glucan synthase determining pullulan synthesis and that controlling cell wall Alpha-1,3 glucan synthesis. Int J Biol Macromol. 2020, 162, 436–444. [Google Scholar] [CrossRef]
- lvarez-Carretero S, Kapli P, Yang Z. Beginner's Guide on the Use of PAML to Detect Positive Selection. Mol Biol Evol. 2023, 40, msad041. [Google Scholar]
- Vinod, P.K.; Sengupta, N.; Bhat, P.J.; Venkatesh, K.V. Integration of Global Signaling Pathways, cAMP-PKA, MAPK and TOR in the Regulation of FLO11. PLOS ONE 2008, 3, e1663. [Google Scholar] [CrossRef] [PubMed]
- Bastajian, N.; Friesen, H.; Andrews, B.J. Bck2 Acts through the MADS Box Protein Mcm1 to Activate Cell-Cycle-Regulated Genes in Budding Yeast. PLOS Genet. 2013, 9, e1003507. [Google Scholar] [CrossRef]
- Yang, G.; Liu, G.L.; Wang, S.J.; Chi, Z.M.; Chi, Z. Pullulan biosynthesis in yeast-like fungal cells is regulated by the transcriptional activator Msn2 and cAMP-PKA signaling pathway. Int J Biol Macromol. 2020, 157, 591–603. [Google Scholar] [CrossRef]
- Rødkær, S.; Færgeman, N. Glucose- and nitrogen sensing and regulatory mechanisms in Saccharomyces cerevisiae[J]. FEMS yeast research, 2014, 14(5): 683-696.
- Görner, W.; Durchschlag, E.; Wolf, J.; Brown, E.L.; Ammerer, G.; Ruis, H.; Schüller, C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002, 21, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.; Ji, H.; Boeke, J.D. Stress response factors drive regrowth of quiescent cells. Curr. Genet. 2018, 64, 807–810. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chi, Z.; Wang, G.-Y.; Wang, Z.-P.; Liu, G.-L.; Lee, C.-F.; Ma, Z.-C.; Chi, Z.-M. Taxonomy ofAureobasidiumspp. and biosynthesis and regulation of their extracellular polymers. Crit. Rev. Microbiol. 2013, 41, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005, 309–338. [Google Scholar] [CrossRef] [PubMed]
- Causton, H.C.; Ren, B.; Koh, S.S.; Harbison, C.T.; Kanin, E.; Jennings, E.G.; Lee, T.I.; True, H.L.; Lander, E.S.; Young, R.A. Remodeling of Yeast Genome Expression in Response to Environmental Changes. Mol. Biol. Cell 2001, 12, 323–337. [Google Scholar] [CrossRef] [PubMed]
- Hochstenbach F, Klis FM, Van d EH, et al. Identification of a putative alpha-glucan synthase essential for cell wall construction and morphogenesis in fission yeast. Proceedings of the National Academy of Sciences 1998, 95, 9161–9166. [Google Scholar] [CrossRef] [PubMed]
- Campbell, B.S.; Siddique, A.B.; McDougall, B.M.; Seviour, R.J. Which morphological forms of the fungus Aureobasidium pullulans are responsible for pullulan production? FEMS Microbiol Lett. 2004, 232, 225–228. [Google Scholar] [CrossRef]
- Estruch, F. Stress-controlled transcription factors, stress-induced genes and stress tolerance in budding yeast. FEMS Microbiol Rev. 2000, 24, 469–486. [Google Scholar] [CrossRef]
- Kottom, T.J.; Limper, A.H. The Pneumocystis Ace2 Transcription Factor Regulates Cell Wall-remodeling Genes and Organism Virulence. 2013, 288, 23893–23902. [CrossRef]
- Haberle, V.; Stark, A. Eukaryotic core promoters and the functional basis of transcription initiation. Nat. Rev. Mol. Cell Biol. 2018, 19, 621–637. [Google Scholar] [CrossRef]


| Strains | Accessions |
|---|---|
| Aureobasidium pullulans P25 | GCA_003574545.1 |
| Aureobasidium melanogenum P16 | GCA_019915885.1 |
| Aureobasidium melanogenum TN3-1 | GCA_017949655.1 |
| Aureobasidium melanogenum CBS105.22 | GCA_018290055.1 |
| Aureobasidium melanogenum P5 | GCA_025604605.1 |
| Aureobasidium pullulans var. namibiae CBS147.97 | GCF_000721765.1 |
| Aureobasidium pullulans var. subglaciale EXF-2481 | GCF_000721755.1 |
| Aureobasidium pullulans var. pullulans EXF-150 | GCF_000721785.1 |
| Aureobasidium melanogenum CBS 110374 | GCF_000721775.1 |
| Aureobasidium sp. P6 | GCA_003992365.1 |
| Aureobasidium melanogenum HN6.2 | GCA_002156615.1 |
| Aureobasidium sp. SLJ-2021a | GCA_019677095.1 |
| Aureobasidium zeae | GCA_017580825.2 |
| Saccharomyces cerevisiae S288C | GCF_000146045.2 |
| Schizosaccharomyces pombe 972h- | GCF_000002945.1 |
| Yarrowia lipolytica CLIB122 | GCF_000002525.2 |
| Aspergillus niger | GCF_000002855.4 |
| Blastomyces parvus UAMH130 | GCA_002572885.1 |
| Lepidopterella palustris CBS459.81 | GCA_001692735.1 |
| Strains | Max length (bp) | N50 length (bp) | GC (%) | Genome size (bp) | Gene numbers | Gene length (bp) |
|---|---|---|---|---|---|---|
| Strains | Gene percentage (%) | Gene average length (bp) | % of Genome (internal) | Specific genes | NRPS | T1PKS |
| P16 | 3,518,807 | 2,256,789 | 49.99 | 26,098,626 | 9,308 | 14,341,028 |
| TN3-1 | 4,274,497 | 2,187,685 | 50.04 | 51,646,945 | 17,915 | 20,405,225 |
| CBS105.22 | 4,273,107 | 2,075,361 | 49.99 | 48,644,395 | 17,817 | 27,348,966 |
| P16 | 54.95 | 1,524 | 45.05 | 1,138 | 1 | 2 |
| TN3-1 | 39.51 | 1,562 | 60.49 | 3,627 | 3 | 4 |
| CBS105.22 | 56.22 | 1,547 | 43.78 | 2,499 | 1 | 5 |
| Promoter | Strains | SWI4/6 | Promoter | Strains | MSN2 |
|---|---|---|---|---|---|
| FKS | P16 | - | PFK | P16 | - |
| TN3-1 | - | TN3-1 | - | ||
| CBS105.22 | + | CBS105.22 | + | ||
| Promoter | Strains | ACE2 | Promoter | Strains | MSN2 |
| AGS2 | P16 | + | PGM | P16 | + |
| TN3-1 | + | TN3-1 | + | ||
| CBS105.22 | - | CBS105.22 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





