Submitted:
28 June 2024
Posted:
01 July 2024
You are already at the latest version
Abstract
Keywords:
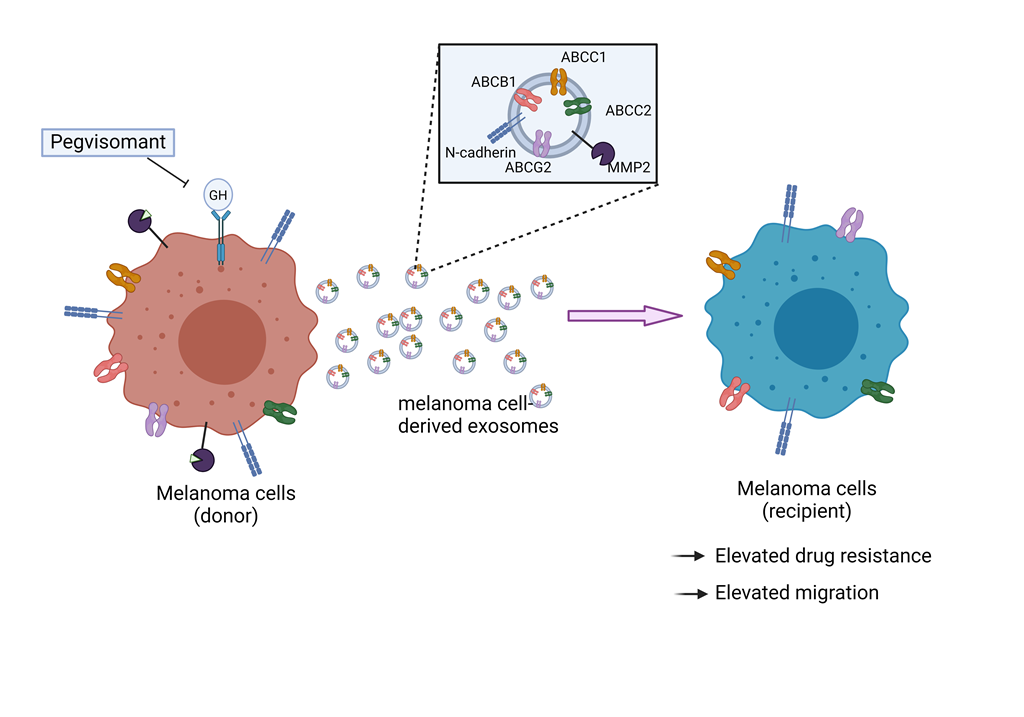
1. Introduction
2. Materials and Methods
- Cell culture and treatments: Human melanoma cell lines MALME-3M (HTB-64) and SK-MEL-28 (HTB-72) were obtained from American Type Culture Collection (ATCC, Manassas, VA), while SK-MEL-30 cell line was acquired from Creative Bioarray (Shirley, NY). Cells were grown and maintained in Iscove’s Modified Dulbecco’s Medium (IMDM), Eagle’s Minimum Essential Medium (EMEM), and Rosewell Park Memorial Institute (RPMI), media respectively supplemented with 10% fetal bovine serum (# 10-082-147, Thermo Fisher Scientific, Waltham, MA) and 100 U/ml penicillin-streptomycin (#15-140-22, Thermo Fisher Scientific, Waltham, MA). Cells were grown in a humidified incubator at 37 ˚C and 5% CO2. Recombinant human GH (#ABIN2017921, Antibodies-online, Pottstown, PA) at 50 ng/ml, doxorubicin at EC50 dosage, and Pegvisomant (Somavert, Pfizer) at 500 nM were added in the respective treatment media supplemented with 2% exosome-depleted fetal bovine serum (#EXO-FBS-250A-1, System Biosciences, Palo Alto, CA). The EC50 values were determined as 0.7 µM for Malme-3M, 1.5 µM for SK-MEL-28, and 2.8 µM for SK-MEL-30 (Supplementary Figure S1)
- Exosome isolation: The supernatant of the cell cultures from respective treatments were centrifuged at 3000g for 15 minutes at 4˚C to remove cell debris. Further, the supernatant was passed through the 0.22µm filter (Millipore Sigma, Burlington, MA) to remove relatively large vesicles. To effectively concentrate exosomes from large volumes, ultrafiltration was employed using Amicon Ultra 15 ml centrifugal filters (Millipore Sigma, Burlington, MA) [80]. Next, ExoQuick reagent was added to the supernatant in 1:5 ratio, according to the manufacturer’s instructions (Systems Biosciences, Palo Alto, CA) and incubated overnight at 4˚C with no rotation. Following incubation, the samples were centrifuged at 1,500 g for 30 minutes. The supernatant was aspirated, followed by a brief centrifugation step of 1,500g for 5 minutes to facilitate further removal of supernatant [81]. The final pellet was resuspended in phosphate buffered saline (PBS) for downstream analysis.
- Nanoparticle tracking analysis: Exosome labeling was conducted using EV tracker green NTA labeling kit (Systems Biosciences, Palo Alto, CA). Briefly, the pre-warmed reaction buffer was mixed with ExoGlowTM dye in a proportion of 5:1, and then 5 μl of the working solution was added to 200 μg of sample and then thoroughly mixed by pipetting. The samples were incubated at room temperature for 30 minutes while protected from light. Microscopic analysis was performed with Zetaview (Particle Matrix, Germany), equipped with a 520 nm laser, 550 nm long pass cutoff filter, and an sCOMS camera.
- Protein extraction and western blot: Protein extraction and western blot were performed as described previously [83]. Briefly, protein extraction was preformed using 1X RIPA buffer (#R-0278, Sigma Aldrich, St. Louis, MO) containing 1X HaltTM protease and phosphatase inhibitor cocktail (#78442, Thermo Fisher Scientific, Waltham, MA). Protein concentration was quantified using the Bradford assay (#B6916, Sigma Aldrich, St, Lou) and 30 μg of protein was loaded onto 4-16% gradient SDS-PAGE denaturing gels. Further, the proteins were transferred to the polyvinylidene fluoride membranes, blocked with 5% BSA solution in 1X TBST-T and probed using target specific antibodies. The exosomal markers in protein extracts from Malme-3M exosomes were determined using antibodies specific for CD63, CD9, CD81 (#EXOAB-CD63A-1, #EXOAB-CD9A-1, #EXOAB-CD81A-1 SBI, Palo Alto, CA). To determine the ABC transporters, EMT markers, and MMPs, protein extracts from Malme-3M exosomes were determined using antibodies specific for ABCC1, ABCC2, ABCB1, ABCG2, N-cadherin, E-cadherin, MMP2, MMP9 (#72202, #125595, #13342, #42078, #13116, #3195, #87809, #13667). β-actin (#4970, CST, Denver, MA) was used as a loading control. For detection, anti-rabbit IgG, HRP-linked secondary antibody (#7074, CST, Denver, MA) and SuperSignal West Femto Maximum Sensitivity Substrate (#34095, Thermo Fisher Scientific, Waltham, MA) were used.
- RNA extraction and RT-qPCR: RNA was extracted, and RT-qPCR was performed as previously described [83]. Briefly, total RNA was extracted using IBI Scientific total RNA extraction kit (Dubuque, IA), following manufacturer’s protocol. Up to 2000 ng of complementary DNA (cDNA) was synthesized from isolated exosomal RNA. Further, quantitative real time polymerase chain reaction (qRT-PCR) was performed using Applied Biosystems reagents (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s protocol. Primers used were GH (Forward: AGGAAACACAACAGAAATCC, Reverse: TTAGGAGGTCATAGACGTTG). The expression levels of differentially expressed RNAs were compared using 2-ΔΔCT method. β-actin (Forward: GACGACATGGAGAAAATCTG, Reverse: ATGATCTGGGTCATCTTCTC) was used as an internal control for the RNA. analysis
- Cell migration assay: Cells were seeded at 30,000 cells per well in 12-well plates. After 24 hours, a scratch wound was made using a 200 µl pipette tip along the midline of each well. Cultures were gently washed with PBS to remove loose cells. The cells were maintained in the respective media with Exocontrol (from PBS treated cells), ExoGH (from GH-treated cells), Exodoxo (from doxorubicin-treated cells), ExoGH+doxo (from cells treated with GH and doxorubicin), and ExoGH+doxo+Peg (from cells treated with GH, doxorubicin, and pegvisomant) for 24 hours. The scratch area was imaged at the start and end of the assay using a BioTek citation-3 microplate imager (Gen5v2.09.2 software) and quantified using ImageJ software. Three individual experiments were performed.
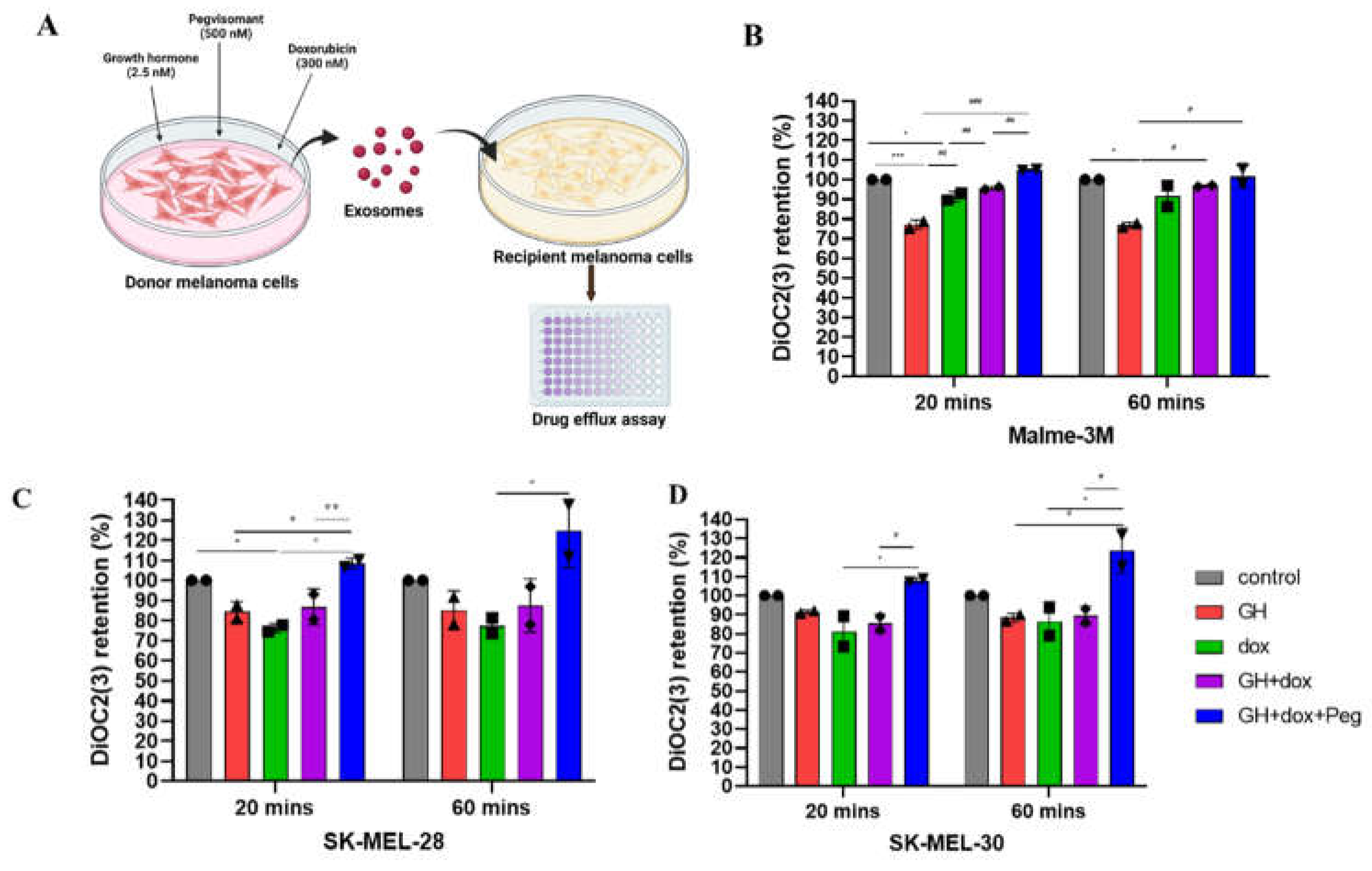
- Drug retention assay: Melanoma cells were treated for 12 hours with Exocontrol, ExoGH, Exodoxo, ExoGH+doxo, and ExoGH+doxo+Peg. On the day of assay, the cells were trypsinised, counted, suspended in cold DiOC2(3) dye on ice for 30 minutes (EMD Millipore, ECM 910). The cells were then centrifuged, supernatant was removed, and cell pellet were resuspended in cold efflux buffer. The resuspended cells were distributed in equal parts with one set serving as control with the other two parts kept at 37˚C water bath for 20 minutes and 60 minutes respectively. The cells were then washed, resuspended, and cells suspension were dispended into the wells of a black-walled 96-well plate, and fluorescence was measured using the fluorescent BioTek citation-3 microplate imager (Gen5v2.09.2 software) at an excitation wavelength of 485 nm and an emission wavelength of 530 nm. Two individual experiments were performed for each cell line.
- Statistical Analysis: For all experiments, analysis was performed by one-way or two-way ANOVA with Tukey’s multiple comparison test using GraphPad Prism 8.0 (GraphPad Software). P < 0.05 was considered statistically significant.
3. Results
3.1. GHR Antagonism Suppresses Melanoma Exosome Mediated Increase in Drug Efflux
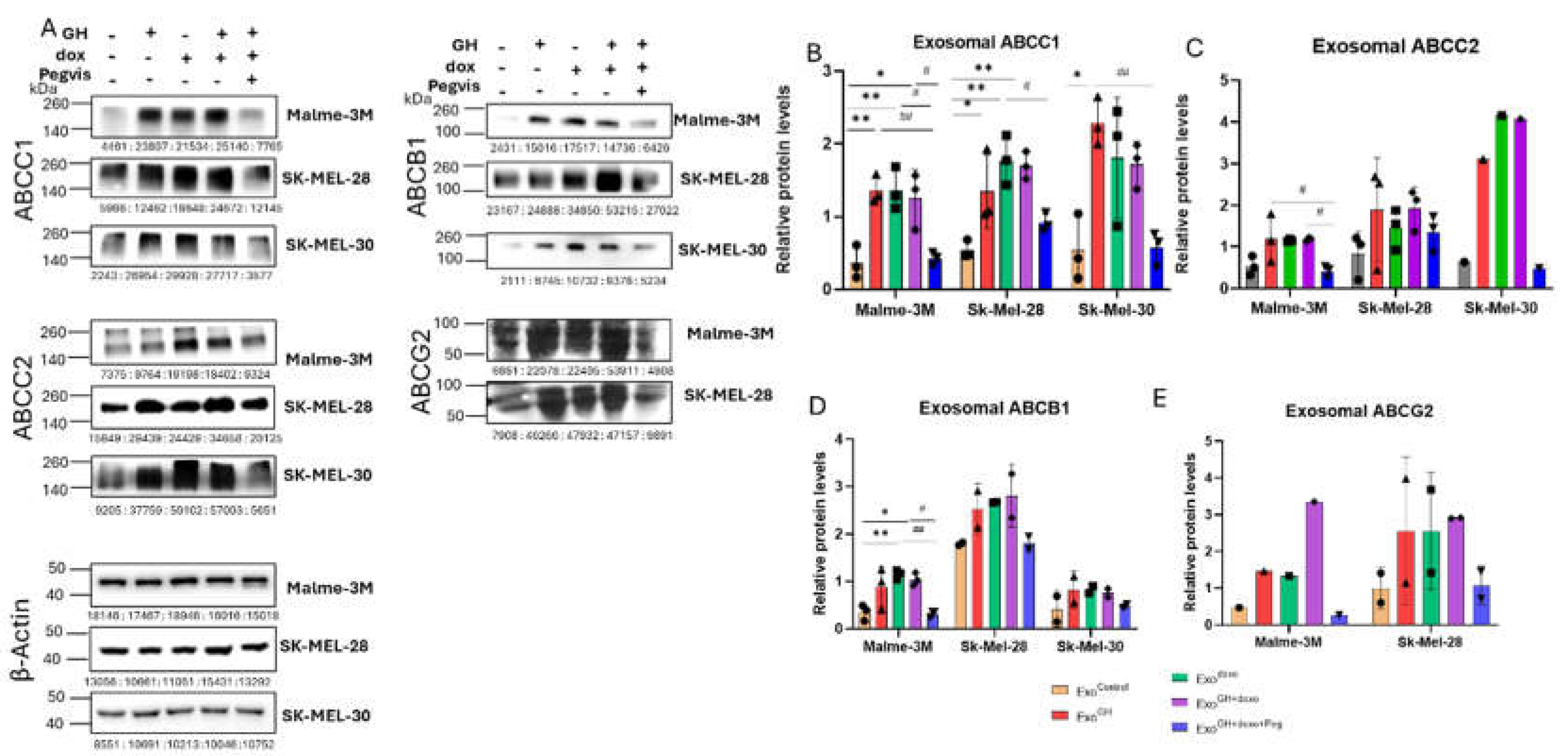
3.2. GH Elevates the Expression of ABC Transporters in Melanoma-Derived Exosomes and in Corresponding Recipient Cells
3.2.1. Effects of GH on ABC Transporters in Melanoma-Derived Exosomes
3.2.2. Effects of GH-Induced Melanoma-Derived Exosomes on Recipient Cells
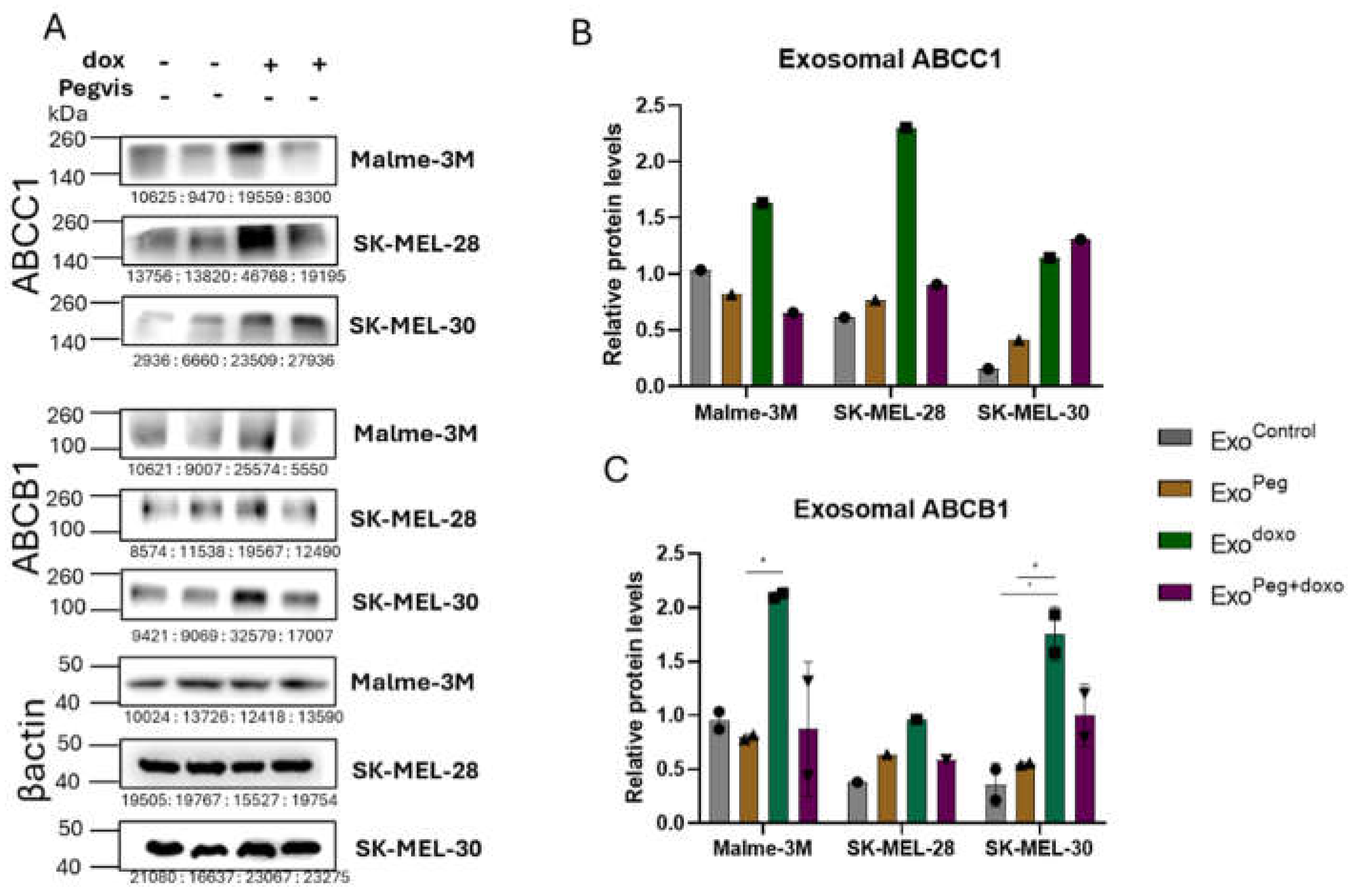
3.3. Blocking Autocrine/Paracrine GH Action Attenuates Exosomal ABC Transporter Levels
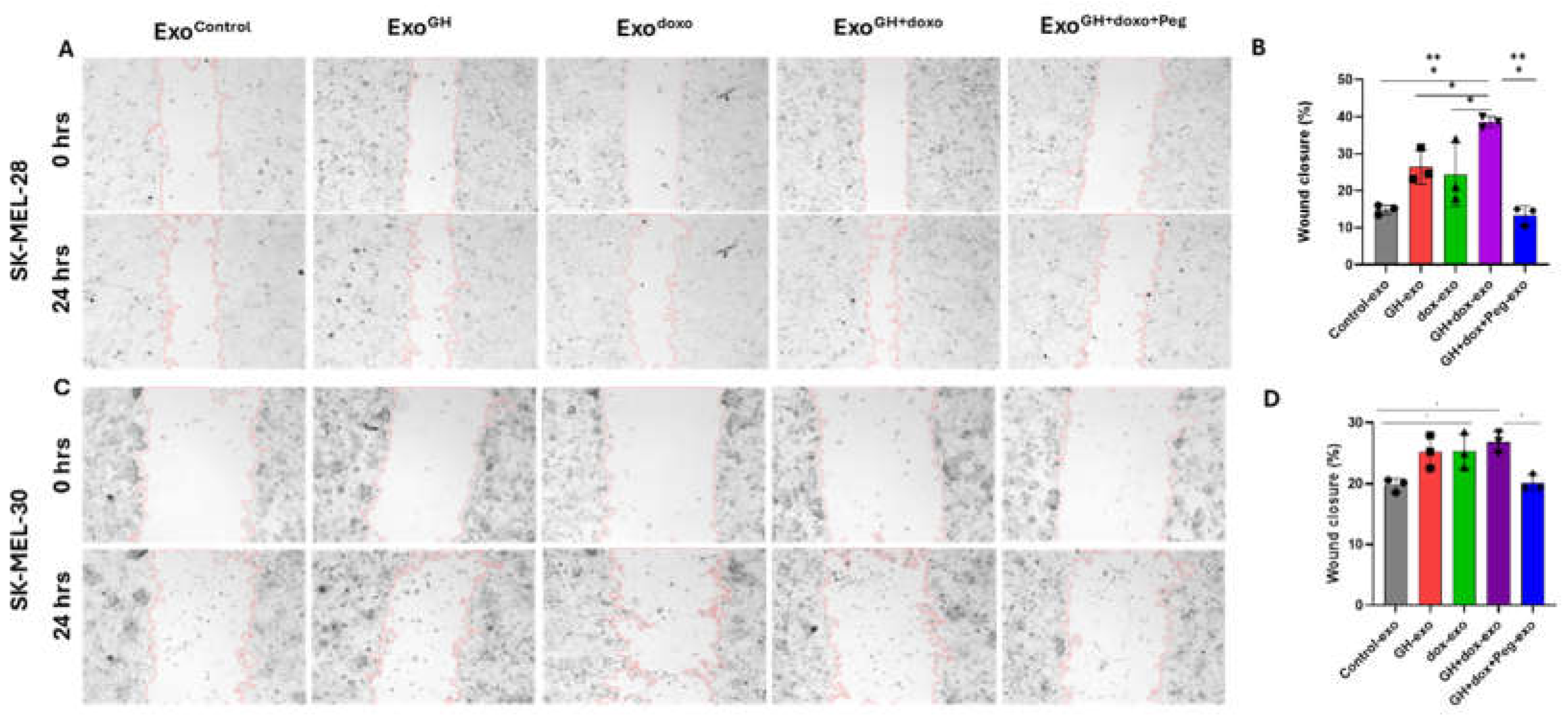
3.4. Pegvisomant Treatment of Donor Melanoma Cells Attenuates Exosomal EMT Inducing Effects
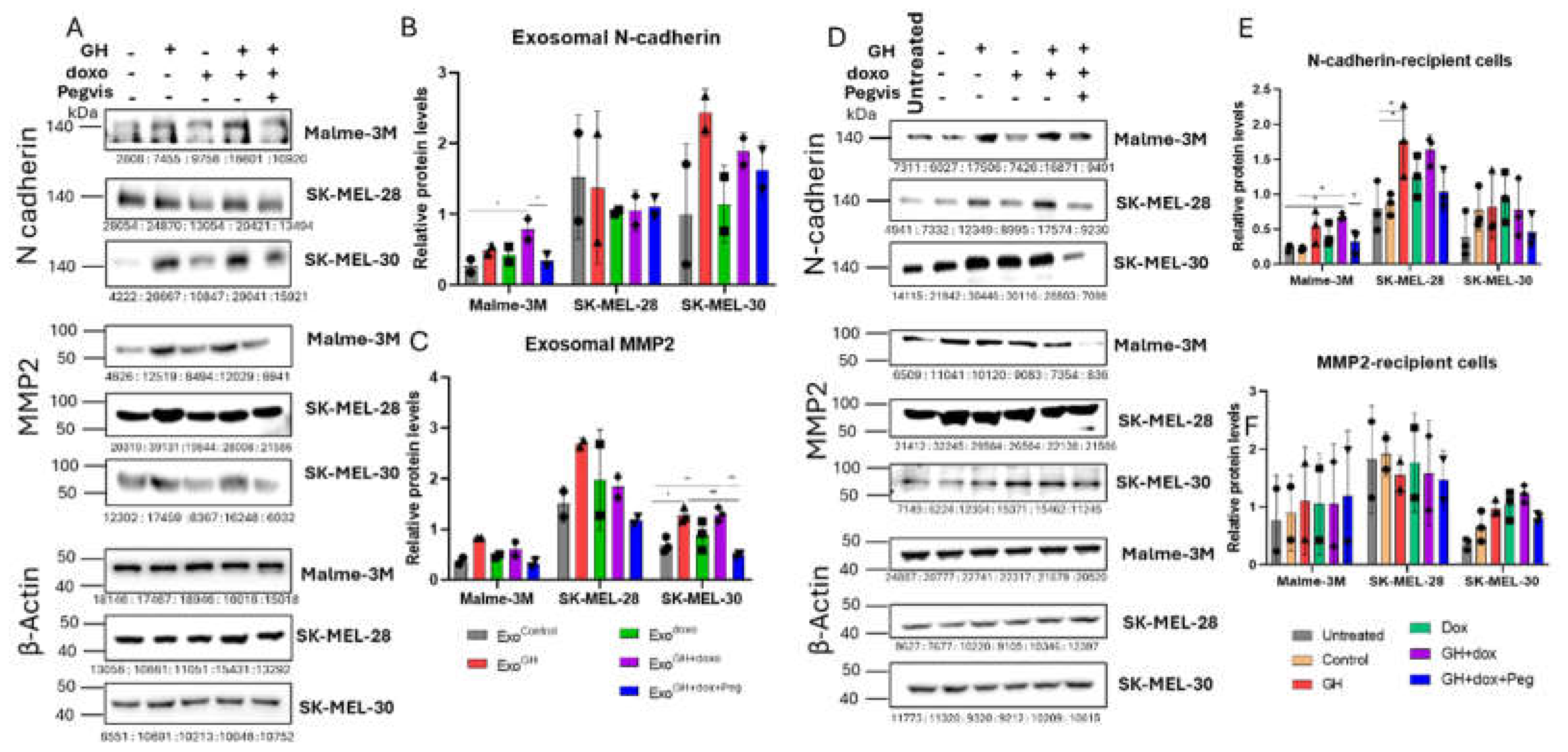
3.5. GH Elevates the Expression of N-cadherin and MMP2 in Melamona-Derived Exosomes and Only Transfers N-cadherin to Recipient Cells
3.5.1. Effects of GH on Cadherins and MMPs in Melanoma-Derived Exosomes
3.5.2. Effects of GH-induced Melanoma-Derived Exosomes on Cadherins and MMPs in Recipient Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Melanoma Skin Cancer Statistics. Available online: https://www.cancer.org/cancer/types/melanoma-skin-cancer/about/key-statistics.html (accessed on 18 June 2024).
- Kozar, I.; Margue, C.; Rothengatter, S.; Haan, C.; Kreis, S. Many Ways to Resistance: How Melanoma Cells Evade Targeted Therapies. Biochim. Biophys. Acta BBA - Rev. Cancer 2019, 1871, 313–322. [Google Scholar] [CrossRef]
- Kalal, B.S.; Upadhya, D.; Pai, V.R. Chemotherapy Resistance Mechanisms in Advanced Skin Cancer. Oncol. Rev. 2017, 11, 326. [Google Scholar] [CrossRef]
- Huang, Y.; Hong, W.; Wei, X. The Molecular Mechanisms and Therapeutic Strategies of EMT in Tumor Progression and Metastasis. J. Hematol. Oncol.J Hematol Oncol 2022, 15, 129. [Google Scholar] [CrossRef]
- Falcone, I.; Conciatori, F.; Bazzichetto, C.; Ferretti, G.; Cognetti, F.; Ciuffreda, L.; Milella, M. Tumor Microenvironment: Implications in Melanoma Resistance to Targeted Therapy and Immunotherapy. Cancers 2020, 12, 2870. [Google Scholar] [CrossRef] [PubMed]
- Soekmadji, C.; Nelson, C.C. The Emerging Role of Extracellular Vesicle-Mediated Drug Resistance in Cancers: Implications in Advanced Prostate Cancer. BioMed Res. Int. 2015, 2015, 454837. [Google Scholar] [CrossRef]
- Mastronikolis, N.S.; Kyrodimos, E.; Spyropoulou, D.; Delides, A.; Giotakis, E.; Piperigkou, Z.; Karamanos, N.K. The Role of Exosomes in Epithelial–to-Mesenchymal Transition and Cell Functional Properties in Head and Neck Cancer. Cancers 2023, 15, 2156. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xing, J.; Xu, K.; Liu, D.; Zhuo, Y. Exosomes in the Tumor Microenvironment: Promoting Cancer Progression. Front. Immunol. 2022, 13, 1025218. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular Vesicles: Exosomes, Microvesicles, and Friends. J Cell Biol 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New Insights into the Mechanisms of Epithelial–Mesenchymal Transition and Implications for Cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Xu, Z.; Zhang, Y.; Dai, H.; Han, B. Epithelial–Mesenchymal Transition-Mediated Tumor Therapeutic Resistance. Molecules 2022, 27, 4750. [Google Scholar] [CrossRef]
- Said, S.S.; Ibrahim, W.N. Cancer Resistance to Immunotherapy: Comprehensive Insights with Future Perspectives. Pharmaceutics 2023, 15, 1143. [Google Scholar] [CrossRef] [PubMed]
- Weadick, B.; Nayak, D.; Persaud, A.K.; Hung, S.W.; Raj, R.; Campbell, M.J.; Chen, W.; Li, J.; Williams, T.M.; Govindarajan, R. EMT-Induced Gemcitabine Resistance in Pancreatic Cancer Involves the Functional Loss of Equilibrative Nucleoside Transporter 1. Mol. Cancer Ther. 2021, 20, 410–422. [Google Scholar] [CrossRef]
- Huang, M.; Zhang, D.; Wu, J.Y.; Xing, K.; Yeo, E.; Li, C.; Zhang, L.; Holland, E.; Yao, L.; Qin, L.; et al. Wnt-Mediated Endothelial Transformation into Mesenchymal Stem Cell–like Cells Induces Chemoresistance in Glioblastoma. Sci. Transl. Med. 2020, 12, eaay7522. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Zhang, R.; An, X.; Li, Z.; Fang, C.; Pan, B.; Chen, W.; Xu, G.; Han, W. LncRNA HOXA-AS3 Confers Cisplatin Resistance by Interacting with HOXA3 in Non-Small-Cell Lung Carcinoma Cells. Oncogenesis 2019, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kirave, P.; Gondaliya, P.; Kulkarni, B.; Rawal, R.; Garg, R.; Jain, A.; Kalia, K. Exosome Mediated miR-155 Delivery Confers Cisplatin Chemoresistance in Oral Cancer Cells via Epithelial-Mesenchymal Transition. Oncotarget 2020, 11, 1157–1171. [Google Scholar] [CrossRef]
- Bebawy, M.; Combes, V.; Lee, E.; Jaiswal, R.; Gong, J.; Bonhoure, A.; Grau, G.E.R. Membrane Microparticles Mediate Transfer of P-Glycoprotein to Drug Sensitive Cancer Cells. Leukemia 2009, 23, 1643–1649. [Google Scholar] [CrossRef] [PubMed]
- Levchenko, A.; Mehta, B.M.; Niu, X.; Kang, G.; Villafania, L.; Way, D.; Polycarpe, D.; Sadelain, M.; Larson, S.M. Intercellular Transfer of P-Glycoprotein Mediates Acquired Multidrug Resistance in Tumor Cells. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 1933–1938. [Google Scholar] [CrossRef]
- Wang, B.; Tan, Z.; Guan, F. Tumor-Derived Exosomes Mediate the Instability of Cadherins and Promote Tumor Progression. Int. J. Mol. Sci. 2019, 20, 3652. [Google Scholar] [CrossRef] [PubMed]
- Hardin, H.; Helein, H.; Meyer, K.; Robertson, S.; Zhang, R.; Zhong, W.; Lloyd, R. Thyroid Cancer Stem-Like Cell Exosomes: Regulation of EMT via Transfer of LncRNAs. Lab. Investig. J. Tech. Methods Pathol. 2018, 98, 1133–1142. [Google Scholar] [CrossRef]
- Olarescu, N.C.; Gunawardane, K.; Hansen, T.K.; Møller, N.; Jørgensen, J.O.L. Normal Physiology of Growth Hormone in Adults. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., Dungan, K., Grossman, A., Hershman, J.M., Kaltsas, G., Koch, C., Kopp, P., Korbonits, M., McLachlan, R., Morley, J.E., New, M., Perreault, L., Purnell, J., Rebar, R., Singer, F., Trence, D.L., Vinik, A., Wilson, D.P., Eds.; MDText.com, Inc.: South Dartmouth (MA), 2000. [Google Scholar]
- Dal, J.; Leisner, M.Z.; Hermansen, K.; Farkas, D.K.; Bengtsen, M.; Kistorp, C.; Nielsen, E.H.; Andersen, M.; Feldt-Rasmussen, U.; Dekkers, O.M.; et al. Cancer Incidence in Patients With Acromegaly: A Cohort Study and Meta-Analysis of the Literature. J. Clin. Endocrinol. Metab. 2018, 103, 2182–2188. [Google Scholar] [CrossRef]
- Laron, Z.; Kauli, R.; Lapkina, L.; Werner, H. IGF-I Deficiency, Longevity and Cancer Protection of Patients with Laron Syndrome. Mutat. Res. Mutat. Res. 2017, 772, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Guevara-Aguirre, J.; Balasubramanian, P.; Guevara-Aguirre, M.; Wei, M.; Madia, F.; Cheng, C.-W.; Hwang, D.; Martin-Montalvo, A.; Saavedra, J.; Ingles, S.; et al. Growth Hormone Receptor Deficiency Is Associated With a Major Reduction in Pro-Aging Signaling, Cancer and Diabetes in Humans. Sci. Transl. Med. 2011, 3, 70ra13. [Google Scholar] [CrossRef] [PubMed]
- Harvey, S. Extrapituitary Growth Hormone. Endocrine 2010, 38, 335–359. [Google Scholar] [CrossRef]
- Kopchick, J.J.; Basu, R.; Berryman, D.E.; Jorgensen, J.O.L.; Johannsson, G.; Puri, V. Covert Actions of Growth Hormone: Fibrosis, Cardiovascular Diseases and Cancer. Nat. Rev. Endocrinol. 2022, 18, 558–573. [Google Scholar] [CrossRef]
- Non-Pituitary GH Regulation of the Tissue Microenvironment in: Endocrine-Related Cancer Volume 30 Issue 7 (2023). Available online: https://erc.bioscientifica.com/view/journals/erc/30/7/ERC-23-0028.xml (accessed on 9 March 2024).
- Sustarsic, E.G.; Junnila, R.K.; Kopchick, J.J. Human Metastatic Melanoma Cell Lines Express High Levels of Growth Hormone Receptor and Respond to GH Treatment. Biochem. Biophys. Res. Commun. 2013, 441. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.; Baumgaertel, N.; Wu, S.; Kopchick, J.J. Growth Hormone Receptor Knockdown Sensitizes Human Melanoma Cells to Chemotherapy by Attenuating Expression of ABC Drug Efflux Pumps. Horm. Cancer 2017, 8, 143–156. [Google Scholar] [CrossRef]
- Basu, R.; Kopchick, J.J. The Effects of Growth Hormone on Therapy Resistance in Cancer. Cancer Drug Resist. 2019. [Google Scholar] [CrossRef]
- Arumugam, A.; Subramani, R.; Nandy, S.B.; Terreros, D.; Dwivedi, A.K.; Saltzstein, E.; Lakshmanaswamy, R. Silencing Growth Hormone Receptor Inhibits Estrogen Receptor Negative Breast Cancer through ATP-Binding Cassette Sub-Family G Member 2. Exp. Mol. Med. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Qian, Y.; Basu, R.; Mathes, S.C.; Arnett, N.A.; Duran-Ortiz, S.; Funk, K.R.; Brittain, A.L.; Kulkarni, P.; Terry, J.C.; Davis, E. Growth Hormone Upregulates Mediators of Melanoma Drug Efflux and Epithelial-to-Mesenchymal Transition in Vitro and in Vivo. Cancers 2020, 12, 3640. [Google Scholar] [CrossRef]
- Wu, Z.-S.; Yang, K.; Wan, Y.; Qian, P.-X.; Perry, J.K.; Chiesa, J.; Mertani, H.C.; Zhu, T.; Lobie, P.E. Tumor Expression of Human Growth Hormone and Human Prolactin Predict a Worse Survival Outcome in Patients with Mammary or Endometrial Carcinoma. J. Clin. Endocrinol. Metab. 2011, 96, E1619–1629. [Google Scholar] [CrossRef]
- Waters, M.J.; Conway-Campbell, B.L. The Oncogenic Potential of Autocrine Human Growth Hormone in Breast Cancer. Proc. Natl. Acad. Sci. 2004, 101, 14992–14993. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Lopez-Valdez, R.; Salcido, A.; Boopalan, T.; Arumugam, A.; Nandy, S.; Lakshmanaswamy, R. Growth Hormone Receptor Inhibition Decreases the Growth and Metastasis of Pancreatic Ductal Adenocarcinoma. Exp. Mol. Med. 2014, 46, e117. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Wu, W.; Yuan, Y.; Pandey, V.; Wu, Z.; Lu, X.; Zhang, W.; Chen, Y.; Wu, M.; Zhang, M.; et al. Human Growth Hormone and Human Prolactin Function as Autocrine/Paracrine Promoters of Progression of Hepatocellular Carcinoma. Oncotarget 2016, 7, 29465–29479. [Google Scholar] [CrossRef]
- Brittain, A.L.; Basu, R.; Qian, Y.; Kopchick, J.J. Growth Hormone and the Epithelial-to-Mesenchymal Transition. J. Clin. Endocrinol. Metab. 2017, 102, 3662–3673. [Google Scholar] [CrossRef]
- Basu, R.; Wu, S.; Kopchick, J.J. Targeting Growth Hormone Receptor in Human Melanoma Cells Attenuates Tumor Progression and Epithelial Mesenchymal Transition via Suppression of Multiple Oncogenic Pathways. Oncotarget 2017, 8, 21579–21598. [Google Scholar] [CrossRef]
- Steinbichler, T.B.; Dudás, J.; Skvortsov, S.; Ganswindt, U.; Riechelmann, H.; Skvortsova, I.-I. Therapy Resistance Mediated by Exosomes. Mol. Cancer 2019, 18, 58. [Google Scholar] [CrossRef] [PubMed]
- Skarkova, V.; Vitovcova, B.; Matouskova, P.; Manethova, M.; Kazimirova, P.; Skarka, A.; Brynychova, V.; Soucek, P.; Vosmikova, H.; Rudolf, E. Role of N-Cadherin in Epithelial-to-Mesenchymal Transition and Chemosensitivity of Colon Carcinoma Cells. Cancers 2022, 14, 5146. [Google Scholar] [CrossRef]
- Tune, B.X.J.; Sim, M.S.; Poh, C.L.; Guad, R.M.; Woon, C.K.; Hazarika, I.; Das, A.; Gopinath, S.C.B.; Rajan, M.; Sekar, M.; et al. Matrix Metalloproteinases in Chemoresistance: Regulatory Roles, Molecular Interactions, and Potential Inhibitors. J. Oncol. 2022, 2022, 3249766. [Google Scholar] [CrossRef]
- Dart, A. EMT in Chemoresistance. Nat. Rev. Cancer 2023, 23, 349–349. [Google Scholar] [CrossRef]
- Basu, R.; Kulkarni, P.; Swegan, D.; Duran-Ortiz, S.; Ahmad, A.; Caggiano, L.J.; Davis, E.; Walsh, C.; Brenya, E.; Koshal, A.; et al. Growth Hormone Receptor Antagonist Markedly Improves Gemcitabine Response in a Mouse Xenograft Model of Human Pancreatic Cancer 2024.
- Wang, X.; Qiao, D.; Chen, L.; Xu, M.; Chen, S.; Huang, L.; Wang, F.; Chen, Z.; Cai, J.; Fu, L. Chemotherapeutic Drugs Stimulate the Release and Recycling of Extracellular Vesicles to Assist Cancer Cells in Developing an Urgent Chemoresistance. Mol. Cancer 2019, 18, 182. [Google Scholar] [CrossRef]
- Qin, X.; Yu, S.; Zhou, L.; Shi, M.; Hu, Y.; Xu, X.; Shen, B.; Liu, S.; Yan, D.; Feng, J. Cisplatin-Resistant Lung Cancer Cell–Derived Exosomes Increase Cisplatin Resistance of Recipient Cells in Exosomal miR-100–5p-Dependent Manner. Int. J. Nanomedicine 2017, 12, 3721–3733. [Google Scholar] [CrossRef]
- Wei, F.; Ma, C.; Zhou, T.; Dong, X.; Luo, Q.; Geng, L.; Ding, L.; Zhang, Y.; Zhang, L.; Li, N.; et al. Exosomes Derived from Gemcitabine-Resistant Cells Transfer Malignant Phenotypic Traits via Delivery of miRNA-222-3p. Mol. Cancer 2017, 16, 132. [Google Scholar] [CrossRef] [PubMed]
- Tritos, N.A.; Biller, B.M.K. Pegvisomant: A Growth Hormone Receptor Antagonist Used in the Treatment of Acromegaly. Pituitary 2017, 20, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Kopchick, J.J. Discovery and Mechanism of Action of Pegvisomant. Eur. J. Endocrinol. 2003, 148 Suppl 2, S21–25. [Google Scholar] [CrossRef]
- Kaseb, A.O.; Haque, A.; Vishwamitra, D.; Hassan, M.M.; Xiao, L.; George, B.; Sahu, V.; Mohamed, Y.I.; Carmagnani Pestana, R.; Lombardo, J.L.; et al. Blockade of Growth Hormone Receptor Signaling by Using Pegvisomant: A Functional Therapeutic Strategy in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 986305. [Google Scholar] [CrossRef] [PubMed]
- Kaulsay, K.K.; Mertani, H.C.; Törnell, J.; Morel, G.; Lee, K.-O.; Lobie, P.E. Autocrine Stimulation of Human Mammary Carcinoma Cell Proliferation by Human Growth Hormone. Exp. Cell Res. 1999, 250, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Mukhina, S.; Mertani, H.C.; Guo, K.; Lee, K.-O.; Gluckman, P.D.; Lobie, P.E. Phenotypic Conversion of Human Mammary Carcinoma Cells by Autocrine Human Growth Hormone. Proc. Natl. Acad. Sci. 2004, 101, 15166–15171. [Google Scholar] [CrossRef] [PubMed]
- Bougen, N.M.; Yang, T.; Chen, H.; Lobie, P.E.; Perry, J.K. Autocrine Human Growth Hormone Reduces Mammary and Endometrial Carcinoma Cell Sensitivity to Mitomycin C. Oncol. Rep. 2011, 26, 487–493. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.; Kulkarni, P.; Qian, Y.; Walsh, C.; Arora, P.; Davis, E.; Duran-Ortiz, S.; Funk, K.; Ibarra, D.; Kruse, C.; et al. Growth Hormone Upregulates Melanocyte-Inducing Transcription Factor Expression and Activity via JAK2-STAT5 and SRC Signaling in GH Receptor-Positive Human Melanoma. Cancers 2019, 11, 1352. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-J.; You, M.-L.; Chong, Q.-Y.; Pandey, V.; Zhuang, Q.-S.; Liu, D.-X.; Ma, L.; Zhu, T.; Lobie, P.E. Autocrine Human Growth Hormone Promotes Invasive and Cancer Stem Cell-Like Behavior of Hepatocellular Carcinoma Cells by STAT3 Dependent Inhibition of CLAUDIN-1 Expression. Int. J. Mol. Sci. 2017, 18, 1274. [Google Scholar] [CrossRef]
- Vouyovitch, C.M.; Perry, J.K.; Liu, D.X.; Bezin, L.; Vilain, E.; Diaz, J.-J.; Lobie, P.E.; Mertani, H.C. WNT4 Mediates the Autocrine Effects of Growth Hormone in Mammary Carcinoma Cells. Endocr. Relat. Cancer 2016, 23, 571–585. [Google Scholar] [CrossRef]
- Wang, J.-J.; Chong, Q.-Y.; Sun, X.-B.; You, M.-L.; Pandey, V.; Chen, Y.-J.; Zhuang, Q.-S.; Liu, D.-X.; Ma, L.; Wu, Z.-S.; et al. Autocrine hGH Stimulates Oncogenicity, Epithelial-Mesenchymal Transition and Cancer Stem Cell-like Behavior in Human Colorectal Carcinoma. Oncotarget 2017, 8, 103900–103918. [Google Scholar] [CrossRef] [PubMed]
- Osaki, M.; Okada, F. Exosomes and Their Role in Cancer Progression. Yonago Acta Med. 2019, 62, 182–190. [Google Scholar] [CrossRef] [PubMed]
- McCready, J.; Sims, J.D.; Chan, D.; Jay, D.G. Secretion of Extracellular Hsp90α via Exosomes Increases Cancer Cell Motility: A Role for Plasminogen Activation. BMC Cancer 2010, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Freitas, J.T.; Jozic, I.; Bedogni, B. Wound Healing Assay for Melanoma Cell Migration. Methods Mol. Biol. Clifton NJ 2021, 2265, 65–71. [Google Scholar] [CrossRef]
- Hwang, S.T.; Yang, M.H.; Kumar, A.P.; Sethi, G.; Ahn, K.S. Corilagin Represses Epithelial to Mesenchymal Transition Process Through Modulating Wnt/β-Catenin Signaling Cascade. Biomolecules 2020, 10, 1406. [Google Scholar] [CrossRef] [PubMed]
- Niland, S.; Riscanevo, A.X.; Eble, J.A. Matrix Metalloproteinases Shape the Tumor Microenvironment in Cancer Progression. Int. J. Mol. Sci. 2021, 23, 146. [Google Scholar] [CrossRef] [PubMed]
- Mukhina, S.; Mertani, H.C.; Guo, K.; Lee, K.-O.; Gluckman, P.D.; Lobie, P.E. Phenotypic Conversion of Human Mammary Carcinoma Cells by Autocrine Human Growth Hormone. Proc. Natl. Acad. Sci. 2004, 101, 15166–15171. [Google Scholar] [CrossRef] [PubMed]
- Pandey, V.; Perry, J.K.; Mohankumar, K.M.; Kong, X.-J.; Liu, S.-M.; Wu, Z.-S.; Mitchell, M.D.; Zhu, T.; Lobie, P.E. Autocrine Human Growth Hormone Stimulates Oncogenicity of Endometrial Carcinoma Cells. Endocrinology 2008, 149, 3909–3919. [Google Scholar] [CrossRef]
- Dolo, V.; Ginestra, A.; Cassarà, D.; Violini, S.; Lucania, G.; Torrisi, M.R.; Nagase, H.; Canevari, S.; Pavan, A.; Vittorelli, M.L. Selective Localization of Matrix Metalloproteinase 9, Β1 Integrins, and Human Lymphocyte Antigen Class I Molecules on Membrane Vesicles Shed by 8701-BC Breast Carcinoma Cells. Cancer Res. 1998, 58, 4468–4474. [Google Scholar]
- Ginestra, A.; La Placa, M.D.; Saladino, F.; Cassarà, D.; Nagase, H.; Vittorelli, M.L. The Amount and Proteolytic Content of Vesicles Shed by Human Cancer Cell Lines Correlates with Their in Vitro Invasiveness. Anticancer Res. 1998, 18, 3433–3437. [Google Scholar] [PubMed]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10. [Google Scholar] [CrossRef] [PubMed]
- Ginestra, A.; Monea, S.; Seghezzi, G.; Dolo, V.; Nagase, H.; Mignatti, P.; Vittorelli, M.L. Urokinase Plasminogen Activator and Gelatinases Are Associated with Membrane Vesicles Shed by Human HT1080 Fibrosarcoma Cells. J. Biol. Chem. 1997, 272, 17216–17222. [Google Scholar] [CrossRef]
- Alfarouk, K.O.; Stock, C.-M.; Taylor, S.; Walsh, M.; Muddathir, A.K.; Verduzco, D.; Bashir, A.H.H.; Mohammed, O.Y.; Elhassan, G.O.; Harguindey, S.; et al. Resistance to Cancer Chemotherapy: Failure in Drug Response from ADME to P-Gp. Cancer Cell Int. 2015, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.-W.; Fojo, A.; Chin, J.E.; Roninson, I.B.; Richert, N.; Pastan, I.; Gottesman, M.M. Human Multidrug-Resistant Cell Lines: Increased Mdr1 Expression Can Precede Gene Amplification. Science 1986, 232, 643–645. [Google Scholar] [CrossRef] [PubMed]
- Basu, R.; Qian, Y.; Mathes, S.; Terry, J.; Arnett, N.; Riddell, T.; Stevens, A.; Funk, K.; Bell, S.; Bokal, Z.; et al. Growth Hormone Receptor Antagonism Downregulates ATP-Binding Cassette Transporters Contributing to Improved Drug Efficacy against Melanoma and Hepatocarcinoma in Vivo. Front. Oncol. 2022, 12, 936145. [Google Scholar] [CrossRef]
- Basu, R.; Qian, Y.; Kopchick, J.J. MECHANISMS IN ENDOCRINOLOGY: Lessons from Growth Hormone Receptor Gene-Disrupted Mice: Are There Benefits of Endocrine Defects? Eur. J. Endocrinol. 2018, 178, R155–R181. [Google Scholar] [CrossRef]
- Raji, G.R.; Sruthi, T.V.; Edatt, L.; Haritha, K.; Sharath Shankar, S.; Sameer Kumar, V.B. Horizontal Transfer of miR-106a/b from Cisplatin Resistant Hepatocarcinoma Cells Can Alter the Sensitivity of Cervical Cancer Cells to Cisplatin. Cell. Signal. 2017, 38, 146–158. [Google Scholar] [CrossRef]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-Resistance in Prostate Cancer: Evaluating Associated Phenotypic Changes and Potential for Resistance Transfer via Exosomes. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Lv, M.; Zhu, X.; Chen, W.; Zhong, S.; Hu, Q.; Ma, T.; Zhang, J.; Chen, L.; Tang, J.; Zhao, J. Exosomes Mediate Drug Resistance Transfer in MCF-7 Breast Cancer Cells and a Probable Mechanism Is Delivery of P-Glycoprotein. Tumor Biol. 2014, 35, 10773–10779. [Google Scholar] [CrossRef]
- Bebawy, M.; Combes, V.; Lee, E.; Jaiswal, R.; Gong, J.; Bonhoure, A.; Grau, G.E.R. Membrane Microparticles Mediate Transfer of P-Glycoprotein to Drug Sensitive Cancer Cells. Leukemia 2009, 23, 1643–1649. [Google Scholar] [CrossRef]
- Levchenko, A.; Mehta, B.M.; Niu, X.; Kang, G.; Villafania, L.; Way, D.; Polycarpe, D.; Sadelain, M.; Larson, S.M. Intercellular Transfer of P-Glycoprotein Mediates Acquired Multidrug Resistance in Tumor Cells. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 1933–1938. [Google Scholar] [CrossRef]
- Xu, X.; Zheng, Y.; Luo, L.; You, Z.; Chen, H.; Wang, J.; Zhang, F.; Liu, Y.; Ke, Y. Glioblastoma Stem Cells Deliver ABCB4 Transcribed by ATF3 via Exosomes Conferring Glioblastoma Resistance to Temozolomide. Cell Death Dis. 2024, 15, 318. [Google Scholar] [CrossRef]
- Zhu, T.; Starling-Emerald, B.; Zhang, X.; Lee, K.-O.; Gluckman, P.D.; Mertani, H.C.; Lobie, P.E. Oncogenic Transformation of Human Mammary Epithelial Cells by Autocrine Human Growth Hormone. Cancer Res. 2005, 65, 317–324. [Google Scholar] [CrossRef]
- Bougen, N.M.; Steiner, M.; Pertziger, M.; Banerjee, A.; Brunet-Dunand, S.E.; Zhu, T.; Lobie, P.E.; Perry, J.K. Autocrine Human GH Promotes Radioresistance in Mammary and Endometrial Carcinoma Cells. Endocr. Relat. Cancer 2012, 19, 625–644. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.K.; Wu, Z.-S.; Mertani, H.C.; Zhu, T.; Lobie, P.E. Tumour-Derived Human Growth Hormone As a Therapeutic Target in Oncology. Trends Endocrinol. Metab. 2017, 28, 587–596. [Google Scholar] [CrossRef]
- Zhu, Z.; Mukhina, S.; Zhu, T.; Mertani, H.C.; Lee, K.-O.; Lobie, P.E. P44/42 MAP Kinase-Dependent Regulation of Catalase by Autocrine Human Growth Hormone Protects Human Mammary Carcinoma Cells from Oxidative Stress-Induced Apoptosis. Oncogene 2005, 24, 3774–3785. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Guo, P.; He, Y.; Chen, Z.; Chen, L.; Luo, Y.; Qi, L.; Liu, Y.; Wu, Q.; Cui, Y.; et al. HCC-Derived Exosomes Elicit HCC Progression and Recurrence by Epithelial-Mesenchymal Transition through MAPK/ERK Signalling Pathway. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, H.; Sun, X.; Wang, X.; Ren, T.; Huang, Y.; Zhang, R.; Zheng, B.; Guo, W. Exosomal PD-L1 and N-Cadherin Predict Pulmonary Metastasis Progression for Osteosarcoma Patients. J. Nanobiotechnology 2020, 18, 151. [Google Scholar] [CrossRef]
- Rahman, M.A.; Barger, J.F.; Lovat, F.; Gao, M.; Otterson, G.A.; Nana-Sinkam, P. Lung Cancer Exosomes as Drivers of Epithelial Mesenchymal Transition. Oncotarget 2016, 7, 54852–54866. [Google Scholar] [CrossRef]
- Wang, L.; Yang, G.; Zhao, D.; Wang, J.; Bai, Y.; Peng, Q.; Wang, H.; Fang, R.; Chen, G.; Wang, Z.; et al. CD103-Positive CSC Exosome Promotes EMT of Clear Cell Renal Cell Carcinoma: Role of Remote MiR-19b-3p. Mol. Cancer 2019, 18, 86. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Liang, W.; Fu, M.; Huang, Z.-H.; Li, X.; Zhang, W.; Zhang, P.; Qian, H.; Jiang, P.-C.; Xu, W.-R.; et al. Exosomes-Mediated Transfer of Long Noncoding RNA ZFAS1 Promotes Gastric Cancer Progression. J. Cancer Res. Clin. Oncol. 2017, 143, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Han, K.-Y.; Dugas-Ford, J.; Seiki, M.; Chang, J.-H.; Azar, D.T. Evidence for the Involvement of MMP14 in MMP2 Processing and Recruitment in Exosomes of Corneal Fibroblasts. Invest. Ophthalmol. Vis. Sci. 2015, 56, 5323–5329. [Google Scholar] [CrossRef] [PubMed]
| ABC transporter | Cell lines | Exosomes | ||||
|---|---|---|---|---|---|---|
| Control | GH | Doxo | Doxo+GH | Doxo+GH+Peg | ||
| ABCC1 | Malme-3M | 1.0 | 5.0 | 5.4 | 4.9 | 1.4 |
| SK-MEL-28 | 1.0 | 2.4 | 3.3 | 3.3 | 1.7 | |
| SK-MEL-30 | 1.0 | 7.2 | 6.0 | 6.1 | 1.4 | |
| ABCC2 | Malme-3M | 1.0 | 2.8 | 2.4 | 2.5 | 0.8 |
| SK-MEL-28 | 1.0 | 2.2 | 2.5 | 3.4 | 2.3 | |
| ABCB1 | SK-MEL-30 | 1.0 | 3.3 | 3.9 | 5.6 | 1.9 |
| Malme-3M | 1.0 | 3.2 | 3.9 | 3.5 | 1.2 | |
| ABCG2 | SK-MEL-28 | 1.0 | 1.4 | 1.5 | 1.6 | 1.0 |
| SK-MEL-30 | 1.0 | 2.7 | 3.2 | 2.9 | 1.9 | |
| Recipient cells | ||||||
| ExoControl | ExoGH | ExoDoxo | ExoDoxo+GH | ExoDoxo+GH+Peg | ||
| ABCC1 | Malme-3M | 1.0 | 2.0 | 2.0 | 1.6 | 0.5 |
| SK-MEL-28 | 1.0 | 1.0 | 1.6 | 1.4 | 0.8 | |
| ABCB1 | Malme-3M | 1.0 | 1.5 | 1.8 | 1.5 | 0.7 |
| SK-MEL-28 | 1.0 | 1.0 | 1.3 | 1.1 | 0.5 | |
| SK-MEL-30 | 1.0 | 1.5 | 2.1 | 1.5 | 0.9 | |
| ABCG2 | Malme-3M | 1.0 | 3.1 | 3.7 | 5.0 | 2.7 |
| SK-MEL-28 | 1.0 | 1.6 | 2.9 | 3.2 | 0.9 | |
| SK-MEL-30 | 1.0 | 1.7 | 1.3 | 1.5 | 1.8 | |
| ABC transporter | Cell lines | Exosomes | ||||
|---|---|---|---|---|---|---|
| Control | GH | Doxo | Doxo+GH | Doxo+GH+Peg | ||
| MMP2 | Malme-3M | 1.0 | 1.6 | 1.6 | 2.0 | 1.0 |
| SK-MEL-28 | 1.0 | 2.0 | 2.1 | 2.3 | 1.4 | |
| SK-MEL-30 | 1.0 | 1.9 | 1.4 | 2.0 | 0.7 | |
| N-cadherin | Malme-3M | 1.0 | 1.0 | 2.0 | 3.5 | 1.6 |
| SK-MEL-28 | 1.0 | 0.8 | 0.8 | 0.8 | 1.9 | |
| SK-MEL-30 | 1.0 | 4.5 | 1.7 | 4.0 | 2.9 | |
| Recipient cells | ||||||
| ExoControl | ExoGH | ExoDoxo | ExoDoxo+GH | ExoDoxo+GH+Peg | ||
| MMP2 | Malme-3M | 1.0 | 1.2 | 1.1 | 1.0 | 0.8 |
| SK-MEL-28 | 1.0 | 0.8 | 1.9 | 0.8 | 0.8 | |
| SK-MEL-30 | 1.0 | 1.6 | 1.8 | 2.0 | 1.4 | |
| N-cadherin | Malme-3M | 1.0 | 2.5 | 1.9 | 3.1 | 1.5 |
| SK-MEL-28 | 1.0 | 2.0 | 1.4 | 1.9 | 1.2 | |
| SK-MEL-30 | 1.0 | 1.0 | 1.2 | 1.0 | 0.7 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





