Submitted:
28 June 2024
Posted:
01 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
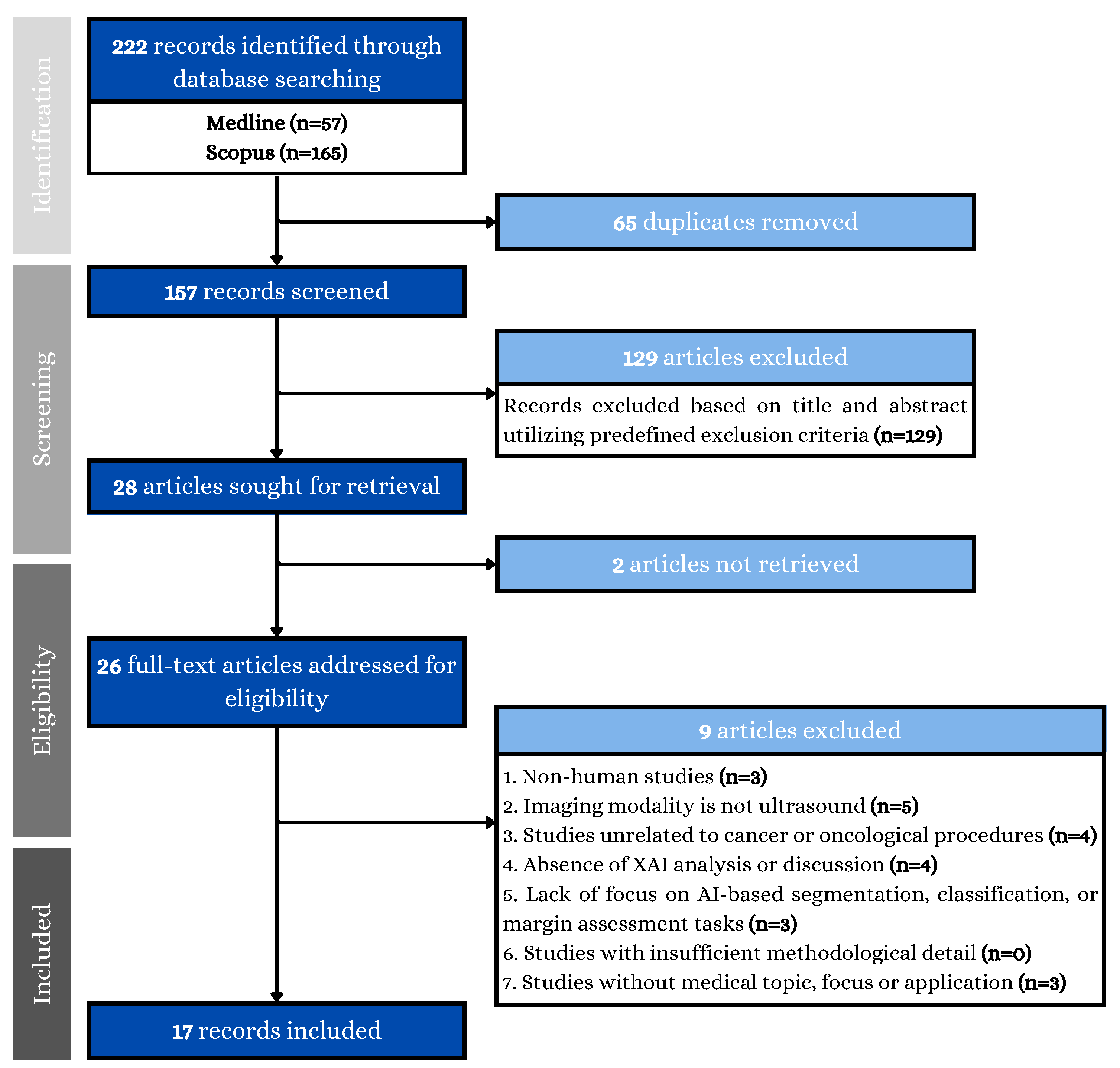
2.1. Search Strategy and Data Collection
2.2. Study Eligibility Criteria
2.3. Data Extraction
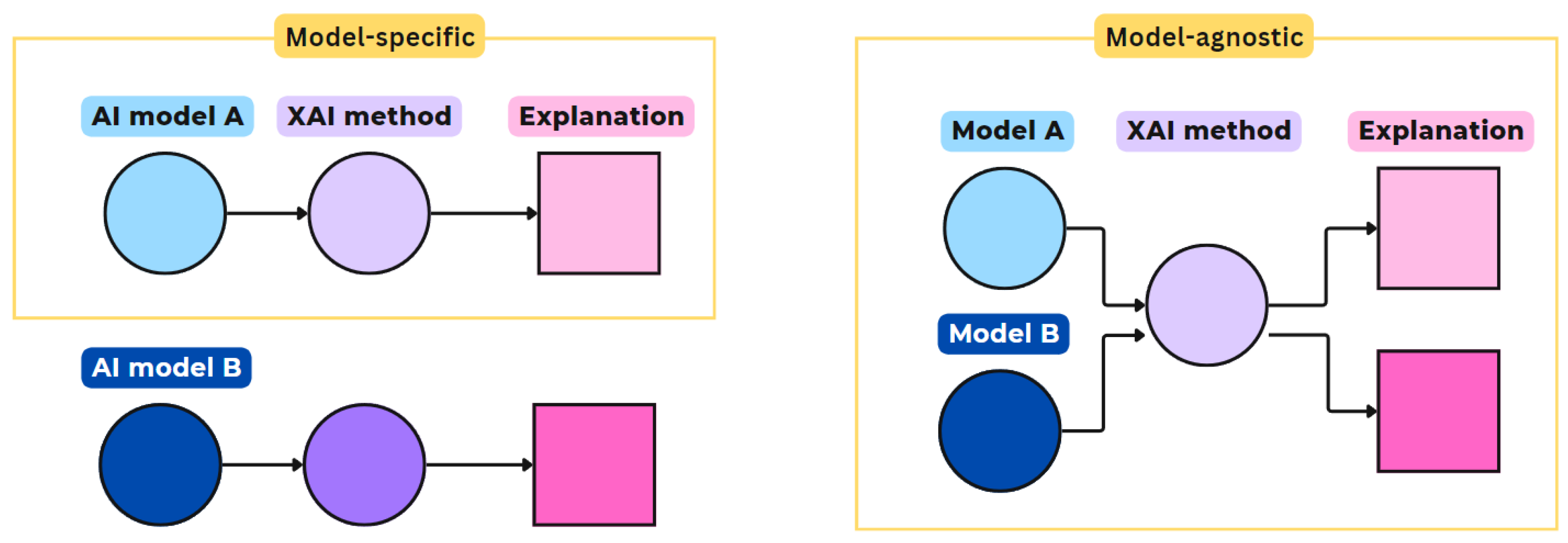
- Model-Specific vs. Model-Agnostic: Model-specific explanation methods are limited to application on particular AI model architectures, e.g. a specific convolutional network model (CNN), see Figure 1. These methods leverage the underlying network’s internal characteristics and use reverse engineering to generate their explanations. Model-agnostic explanation methods only operate on the model input and output, thus, independent of the model architecture. They aim to clarify the model’s underlying function, for example, by approximating with another, simpler model that should be explainable. Other agnostic methods attribute weights to each model variable, depending on its influence on the , to decompose the importance between variables [36].
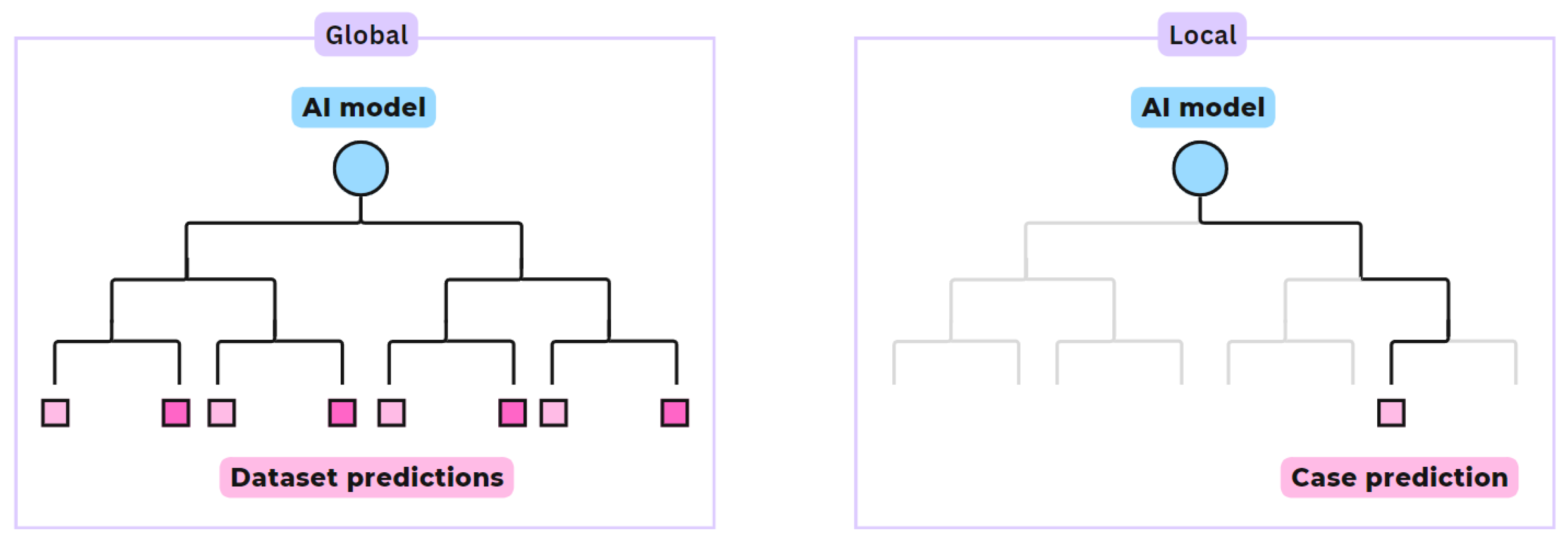
- Global vs. Local scope: Global scope explanations provide general relationships learned by the model, by assessing common patterns in the overall dataset that drive the model’s predictions, see Figure 2. Local scope methods offer explanations of the model’s specific prediction for a given input or single case.
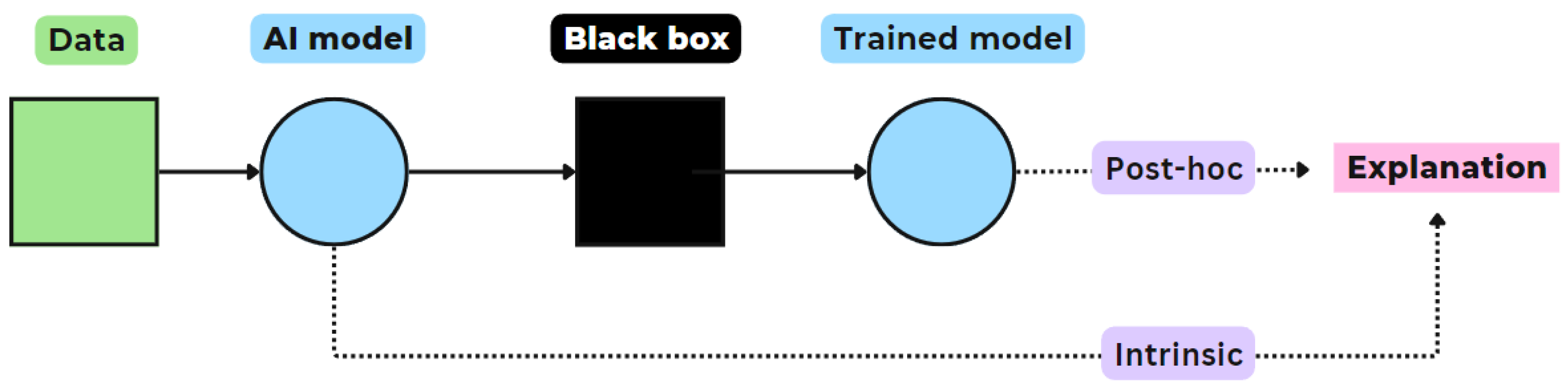
- Intrinsic vs. Post-hoc explanation: Intrinsic explanation models can construct general, internal relationships between input and output made during predictions, due to their simple structure (e.g. decision trees, linear regression model, or support vector machine), see Figure 3. Post-hoc explanations are applied to analyze models after these have completed training, providing insight into the learned relationships. The important difference is that post-hoc explanations train a neural network and attempt to explain the behavior of the black box network after that. In contrast, intrinsic explanations force the neural network to be explainable itself [32].
2.4. Main Outcomes
3. Results
3.1. Data Collection
3.2. General Study Characteristics
3.3. Imaging Modality
3.4. AI Models
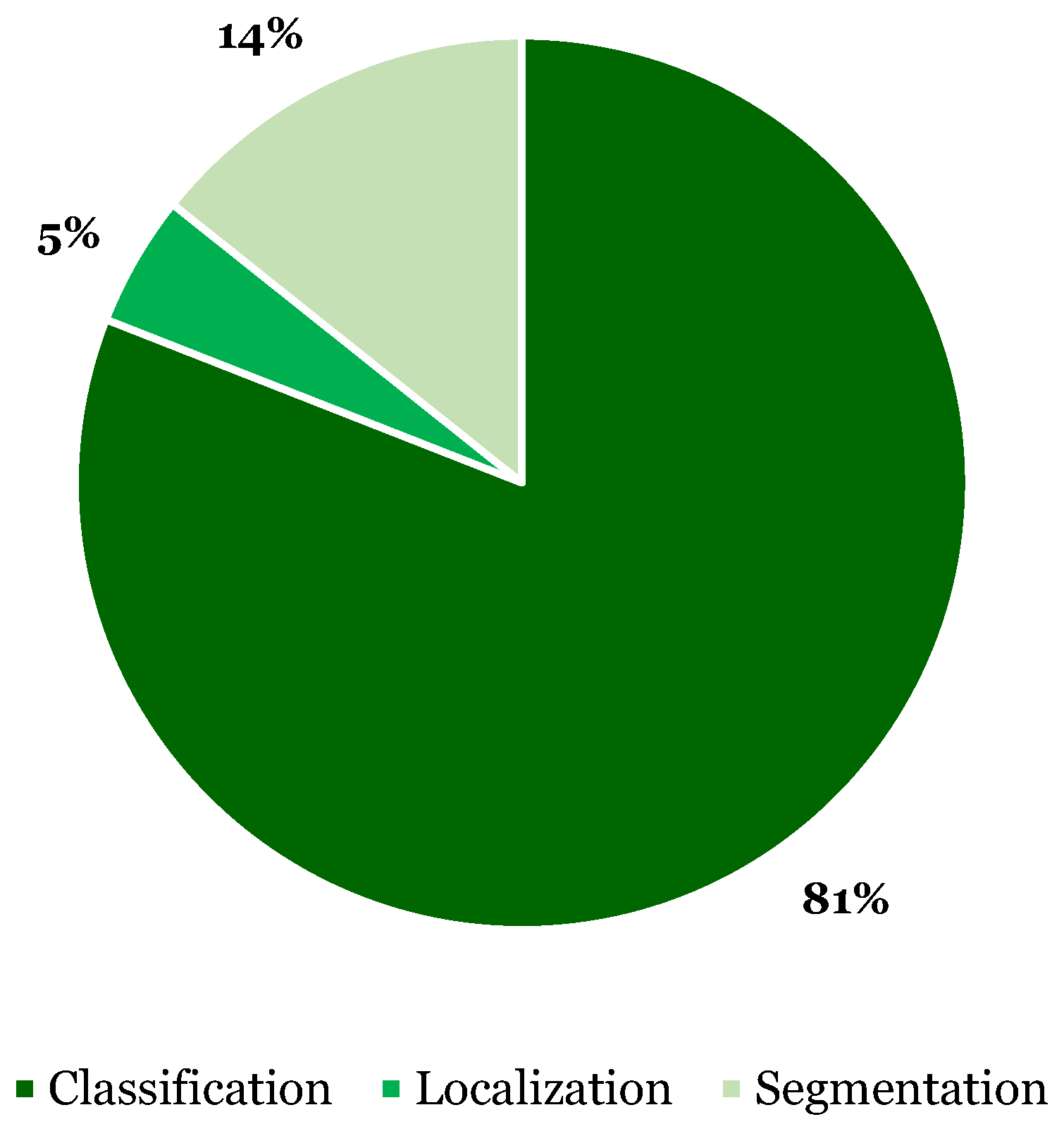
3.5. XAI Methods
- Model: Specific (71.4%) vs. Agnostic (28.6%)
- Scope: Local (85.7%) vs. Global scope (14.3%)
- Explanation: Intrinsic (42.9%) vs. Post-hoc (57.1%)
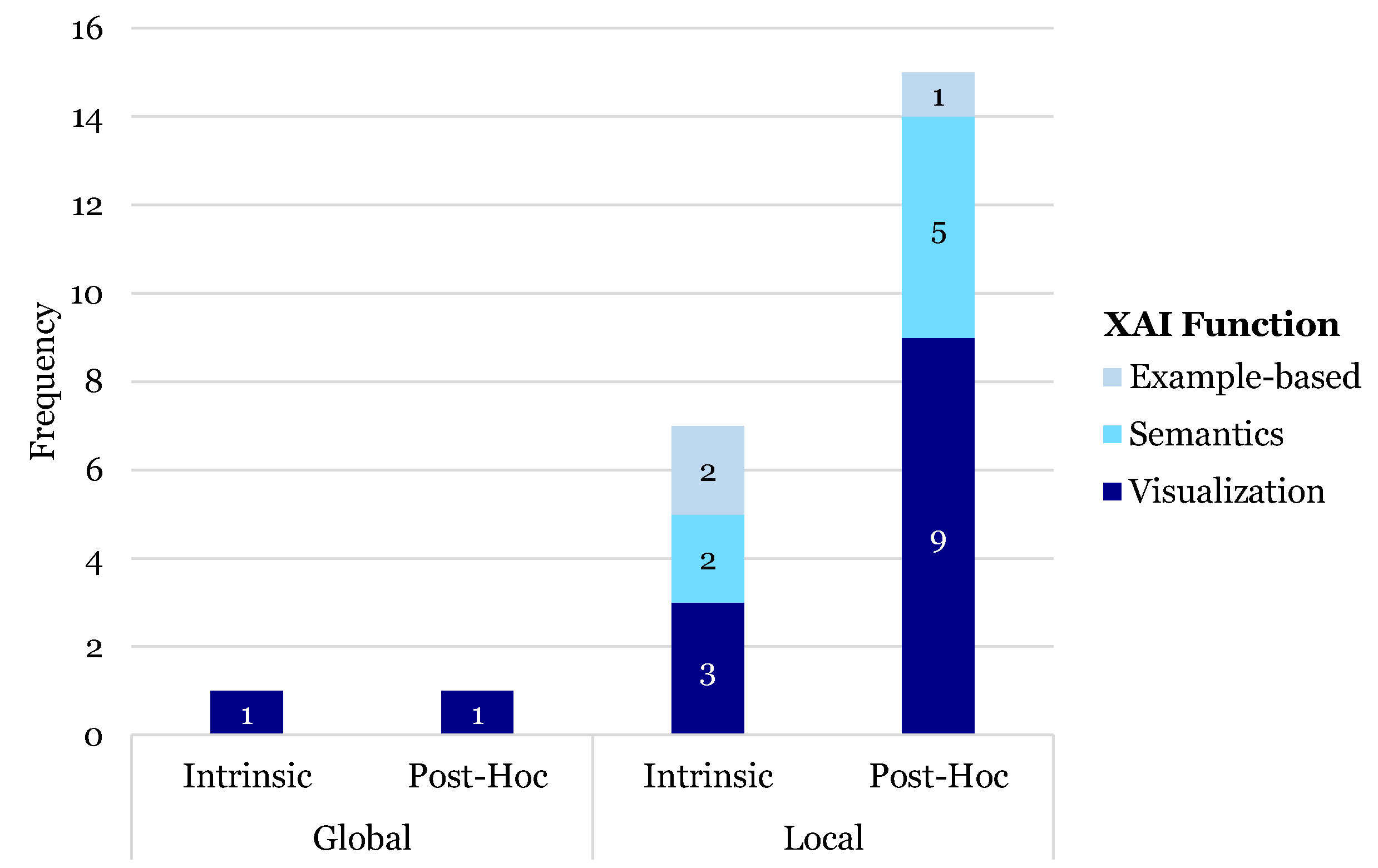
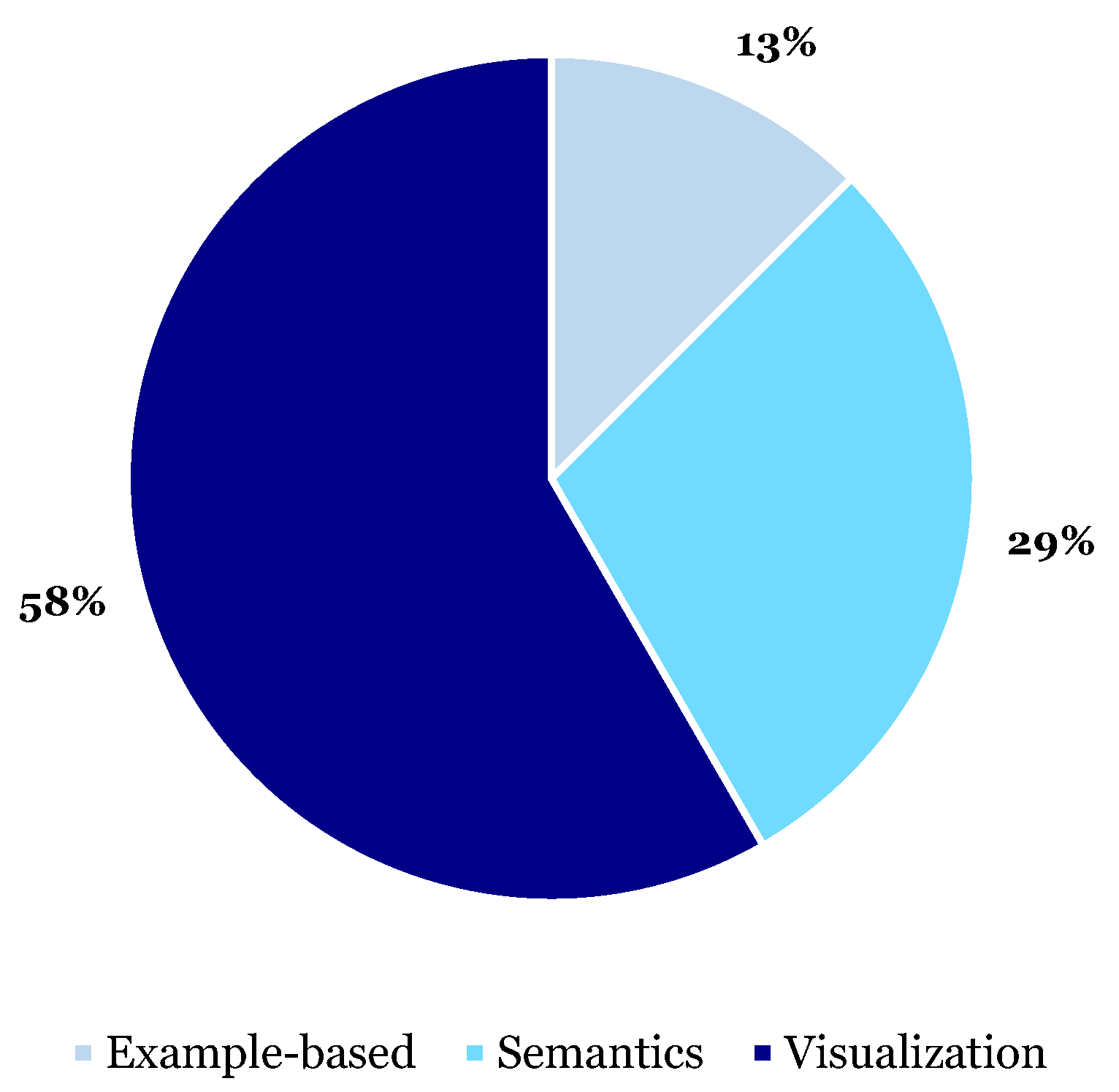
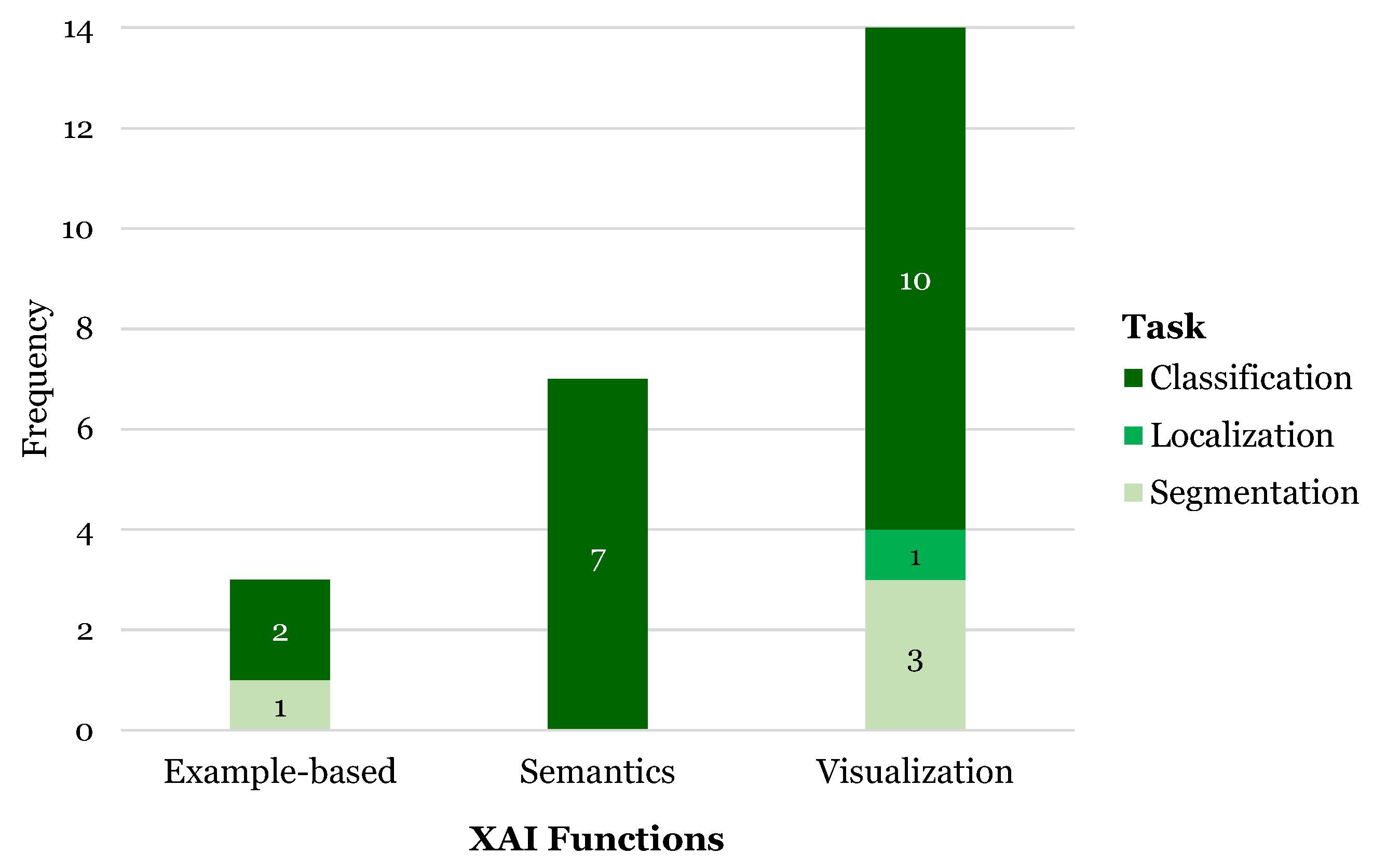
3.6. XAI Functions
3.6.1. Visualization
3.6.2. Semantics
3.6.3. Example-Based
3.7. XAI Advantages and Disadvantages
3.8. XAI Evaluation
3.8.1. Qualitative
3.8.2. Quantitative
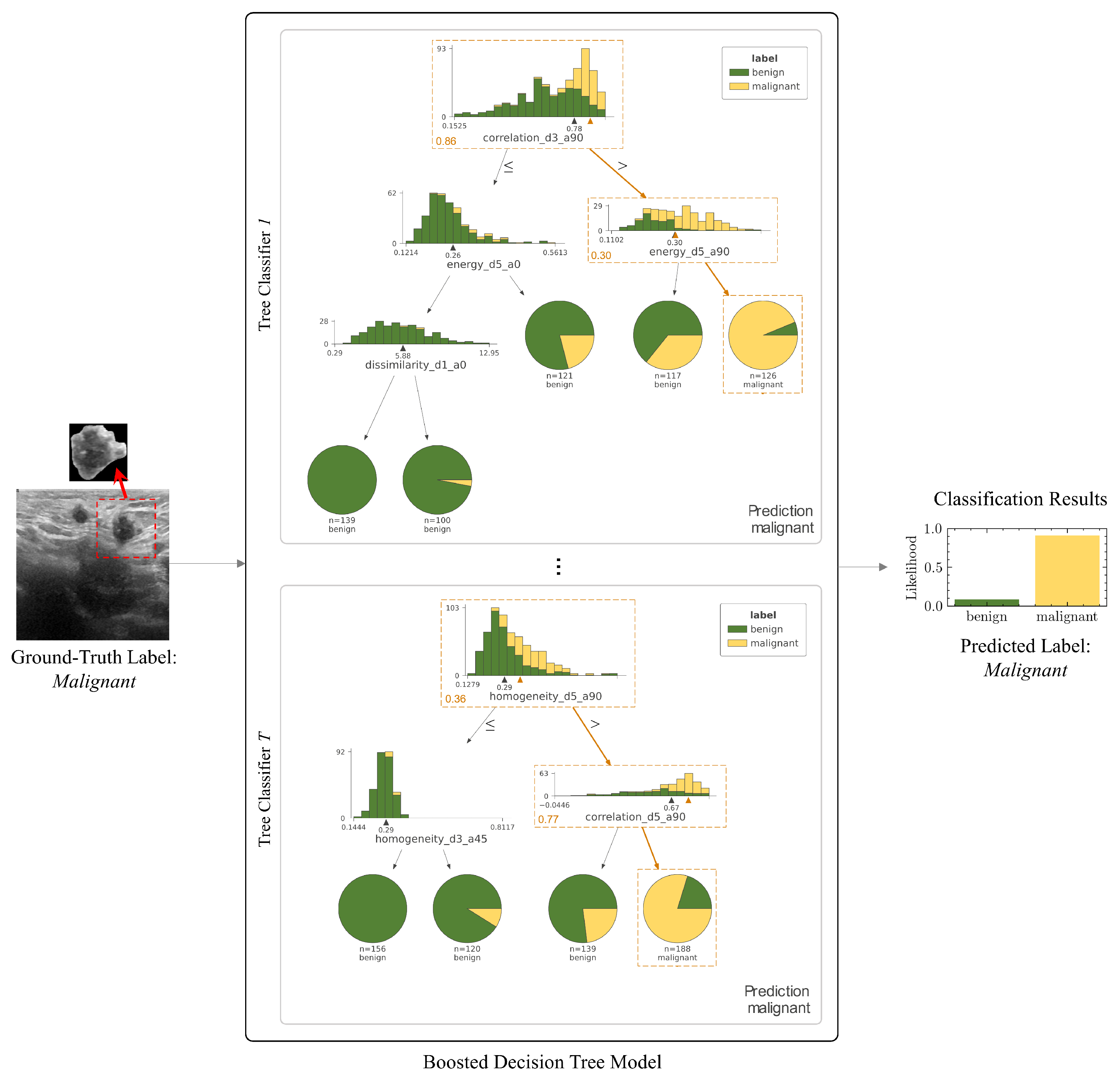
- Shapley values are derived from game theory and provide a method to distribute the impact among contributors (features) in a cooperative game (prediction model). Each feature value’s contribution is determined by assessing the change in prediction when a feature is added or removed for all possible combinations of features. The aim is to fairly attribute the model’s output to its input features, providing insights into which features are most important for predictions. In three studies, SHAP values are used to determine which clinical features (e.g. tumor size, shape, or texture) most significantly impact the models’ classification of a tumor as benign or malignant [43,44,48].
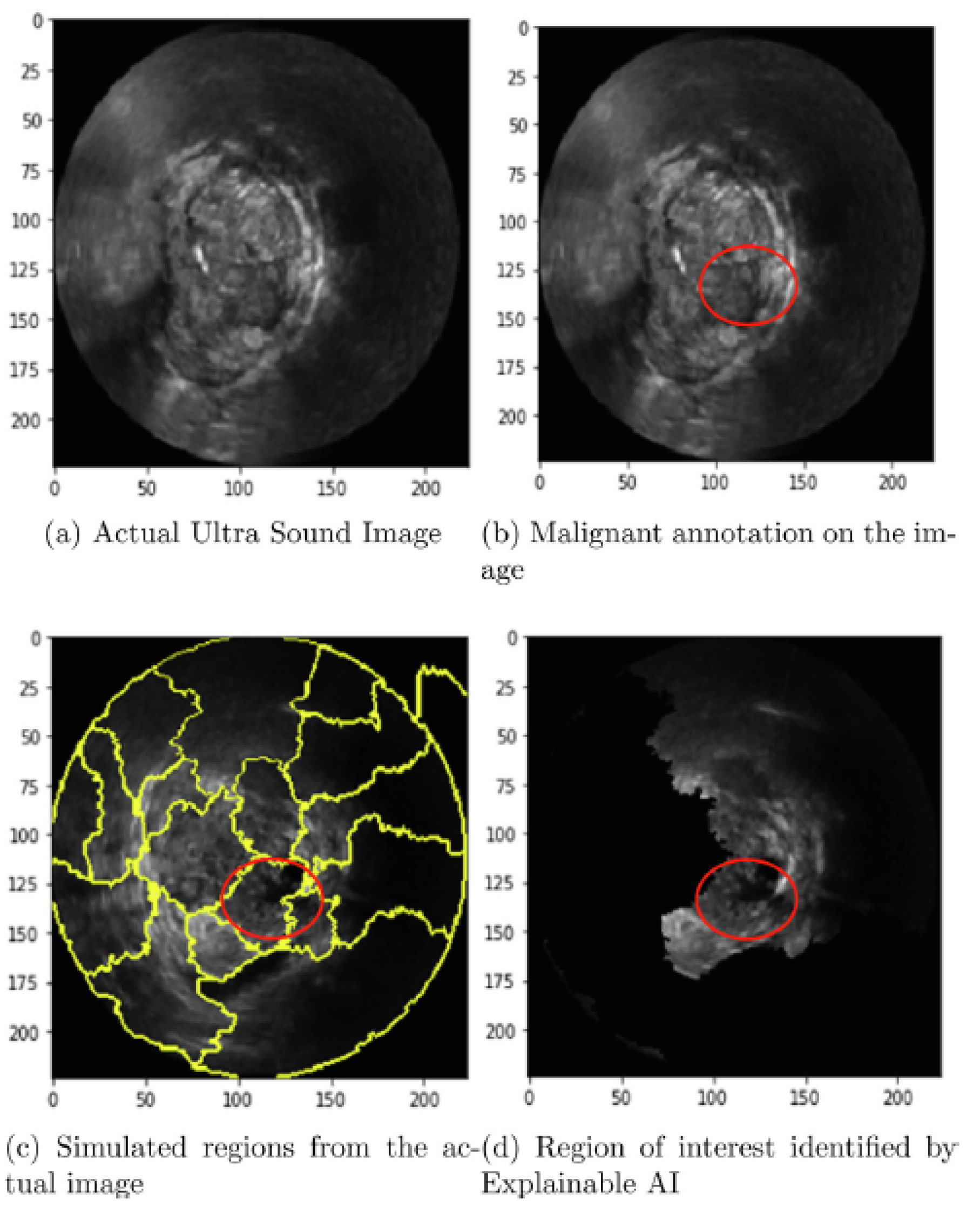
- The Zero-mean Normalized Cross-Correlation (ZNCC) score presents a statistical measure to assess the similarity between two images. It calculates the degree of similarity between two images through a normalized cross-correlation measurement formula, subtracting their mean and dividing by their standard deviation. The ZNCC score ranges from -1 to 1, where 1 indicates perfect correlation, 0 indicates no correlation, and -1 indicates perfect inverse correlation. In Tasnim et al. (2024) the ZNCC score quantitatively assessed the feature separation ability of the Activation Maximization generated images in a benign-malignant (i.e. binary) classification problem [50].
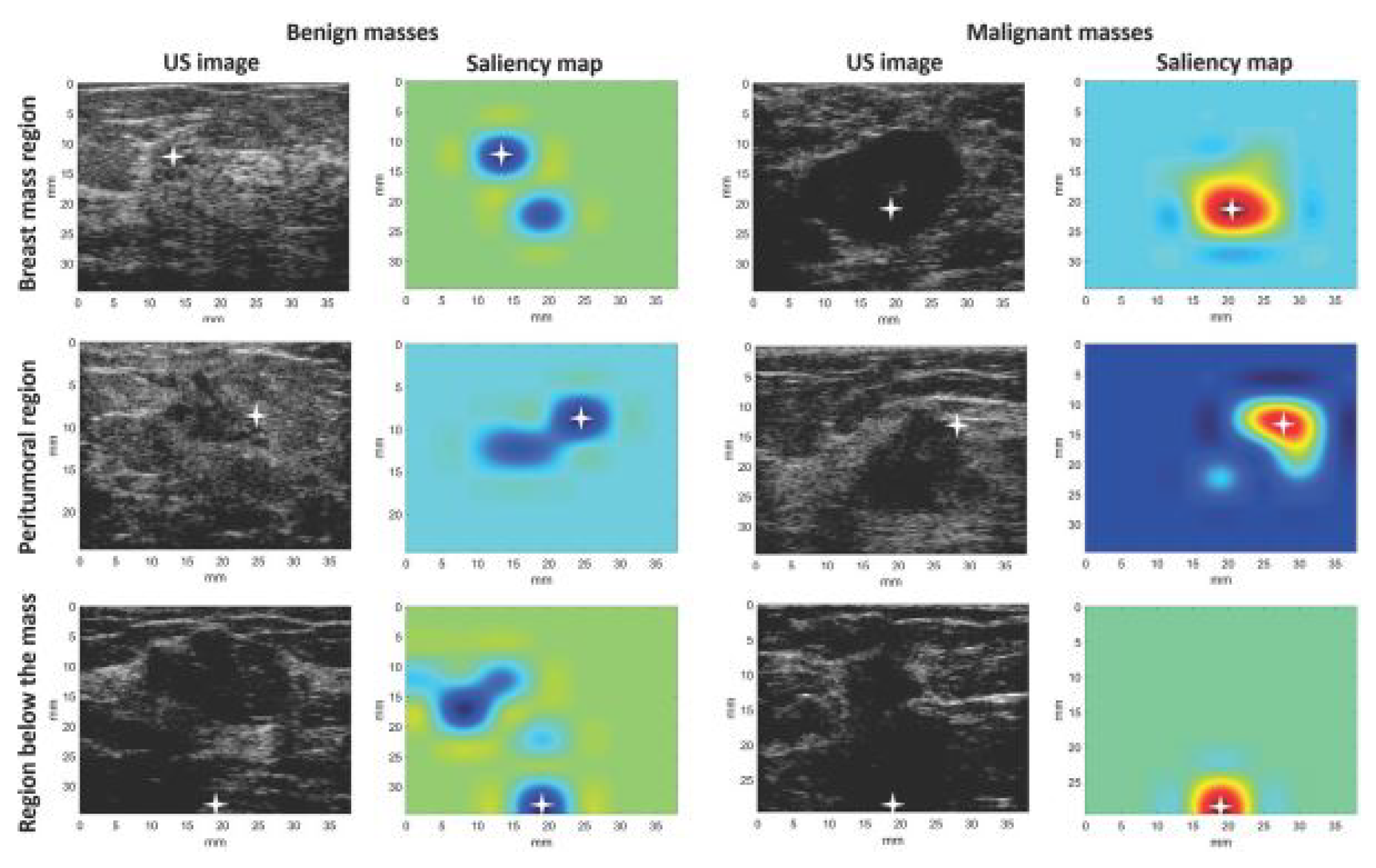
- The Pointing game metric is a QT evaluation method used to assess how well the areas identified by saliency maps align with relevant regions in medical images. It evaluates whether the most significant activation points in the saliency map correspond to specific anatomical or pathological features in the analyzed images. Byra et al. (2022) used the pointing game to verify if the CAM saliency maps highlighted significant regions for accurate diagnosis i.e. breast mass region, peritumoral region, or region below the breast mass [39], see Figure 13.
- The Resemblance votes metric is used in Dong et al. (2021) for QT evaluation of how well the ROE identified by the AI aligns with the regions considered important by physicians for making diagnostic decisions [40]. The metric categorizes the ROE into three resemblance levels as perceived by clinicians: High Resemblance (HR), where the ROE closely matches the features used by physicians; Medium Resemblance (MR), where the ROE partially matches; and Low Resemblance (LR), where there is little to no match. Considering the perceived resemblance of AI predictions actively included clinicians and can aid in validating and improving the model’s explainability and utility in clinical practice.
4. Discussion
4.1. Key Findings
4.2. Comparison with Existing Literature
4.3. Limitations
4.4. Strengths
4.5. Implications & Future Research
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B
| Author (year) | US Modality | System (Manufacturer) | Probe / Transducer | Frequency | Acquisition |
|---|---|---|---|---|---|
| Al-Jebrni, A.H. (2023) [37] | US | EPIQ 7 (Philips) + DU8 (Technos) | NA | NA | Continuous |
| lightgrayBasu, S. (2023) [38] | Transabdominal US | Logic S8 (GE Healthcare) | Convex low-frequency | 1.0–5.0 MHz | Multiple shot, static B-mode |
| Byra, M. (2022) [39] | Breast US | SonixTouch Research (Ultrasonix) | L14-5/38 linear probe | 10 MHz | Single shot, static B-mode |
| Dong, F. (2021) [40] | US | Resona 7 (Mindray) | L11-3U linear array | 2.0–11 MHz | Continuous |
| Han, X. (2022) [41] | US | NA (Siemens), NA (Philips) | NA | NA | Continuous |
| Hassan, M. R. (2022) [42] | Transrectal US | Hi-Vision 5500 (Hitachi) | NA | 7.5 MHz | Continuous |
| C41V end-fire probe (Nobulus) | NA | 2.0–10 MHz | Continuous | ||
| Karimzadeh, M. (2023) [43] | Breast US | LOGIQ E9 + LOGIQ E9 Agile (GE Healthcare) | ML6-15-D Matrix linear probe | NA | Continuous |
| VIVID 7 (GE Healthcare), LOGIQ E9 (GE Healthcare), EUB-6500 (Hitachi), iU22 (Philips), ACUSON S2000 (Siemens) | NA | NA | Continuous | ||
| Lombardi, A. (2023) [44] | Transvaginal or transabdominal US & 3D US | NA | NA | 5.0–9.0 MHz (transvaginal), 3.5–5.0 MHz (transabdominal) | Continuous or 3D volume scan |
| Martizzi, D. (2021) [45] | Quantitative Transmission US | NA | NA | NA | 3D volume scan |
| Morris, J. (2023) [46] | US | NA | NA | NA | Continuous |
| Qian, X. (2021) [47] | US | Aixplorer (SuperSonic) | NA | NA | Continuous |
| Rezazadeh, A. (2022) [48] | Breast US | LOGIQ E9 (GE Healthcare) + LOGIQ E9 Agile (GE Healthcare) | ML6-15-D Matrix linear probe | 1.0–5.0 MHz | Continuous |
| Song, D. (2023) [49] | US | NA | NA | NA | Single shot |
| Tasnim, J. (2024) [50] | Quantitative US | LOGIQ E9 + LOGIQ E9 Agile (GE Healthcare) | ML6-15-D Matrix linear probe | 1.0–5.0 MHz | Continuous |
| Voluson730 scanner (GE Healthcare) | S-VNW5-10 small-part transducer | 5.0–10 MHz | Continuous | ||
| ACUSON Sequoia C512 (Siemens) | 17L5 HD linear array transducer | 8.5 MHz | Continuous | ||
| iU22 (Philips) | Linear probe | 7.0–15 MHz | Continuous | ||
| Sonix-Touch Research (Ultrasonix) | L14-5/38 linear transducer | 10 MHz | Continuous | ||
| Thomas, J. (2020) [51] | US | NA (GE Healthcare), NA (Philips), NA (Sonosite) | NA | 8.0–13 MHz | Continuous |
| Zhang, B. (2021) [52] | Breast US | LOGIQ E9 + LOGIQ E9 Agile (GE Healthcare) | ML6-15-D Matrix linear probe | 1.0–5.0 MHz | Continuous |
| VIVID 7 (GE Healthcare), LOGIQ E9 (GE Healthcare), EUB-6500 (Hitachi), iU22 (Philips), ACUSON S2000 (Siemens) | L11-3U linear array | 1.0–5.0 MHz | Continuous | ||
| Zheng, H. (2024) [53] | Endoscopic US | NA | NA | NA | Continuous |
References
- Grand View Research. AI In Healthcare Market Size, Share & Trends Analysis Report By Component (Hardware, Services), By Application, By End-use, By Technology, By Region, And Segment Forecasts, 2024 - 2030. https://www.grandviewresearch.com/industry-analysis/artificial-intelligence-ai-healthcare-market, 2024.
- Sanskrutisathe. AI in Healthcare Market Size and Growth. https://medium.com/@sanskrutisathe01/ai-in-healthcare-market-size-and-growth-2ae9b8463121, 2024.
- World Health Organization. Global strategy on human resources for health: Workforce 2030. https://apps.who.int/iris/bitstream/handle/10665/250368/9789241511131-eng.pdf, 2016.
- Choi, M.; Sempungu, J.K.; Lee, E.H.; Lee, Y.H. Living longer but in poor health: healthcare system responses to ageing populations in industrialised countries based on the Findings from the Global Burden of Disease Study 2019. BMC Public Health 2024, 24, 576. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, S.; Jackson, C. Three in five globally say their healthcare system is overstretched. https://www.ipsos.com/en/three-five-globally-say-their-healthcare-system-overstretched, 2022.
- Page, B.; Irving, D.; Amalberti, R.; Vincent, C. Health services under pressure: a scoping review and development of a taxonomy of adaptive strategies. BMJ Quality & Safety. [CrossRef]
- Bohr, A.; Memarzadeh, K. Chapter 2 - The rise of artificial intelligence in healthcare applications. In Artificial Intelligence in Healthcare; Academic Press, 2020; pp. 25–60. [CrossRef]
- Zhang, B.; Shi, H.; Wang, H. Machine Learning and AI in Cancer Prognosis, Prediction, and Treatment Selection: A Critical Approach. Journal of Multidisciplinary Healthcare 2023, 16, 1779–1791. [Google Scholar] [CrossRef] [PubMed]
- Wolff, J.; Pauling, J.; Keck, A.; Baumbach, J. The Economic Impact of Artificial Intelligence in Health Care: Systematic Review. J Med Internet Res 2020, 22, e16866. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Coelho, L. How Artificial Intelligence Is Shaping Medical Imaging Technology: A Survey of Innovations and Applications. Bioengineering 2023, 10, 1435. [Google Scholar] [CrossRef] [PubMed]
- Borys, K.; Schmitt, Y.A.; Nauta, M.; Seifert, C.; Krämer, N.; Friedrich, C.M.; Nensa, F. Explainable AI in medical imaging: An overview for clinical practitioners – Saliency-based XAI approaches. European Journal of Radiology 2023, 162, 110787. [Google Scholar] [CrossRef] [PubMed]
- Abrantes, J.; Rouzrokh, P. Explaining explainability: The role of XAI in medical imaging. European Journal of Radiology 2024, 173, 111389. [Google Scholar] [CrossRef]
- Singh, A.; Sengupta, S.; Lakshminarayanan, V. Explainable Deep Learning Models in Medical Image Analysis. Journal of Imaging 2020, 6, 52. [Google Scholar] [CrossRef] [PubMed]
- Reyes, M.; Meier, R.; Pereira, S.; Silva, C.A.; Dahlweid, F.M.; Tengg-Kobligk, H.v.; Summers, R.M.; Wiest, R. On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiology: Artificial Intelligence 2020, 2, e190043. [Google Scholar] [CrossRef] [PubMed]
- Hacker, P.; Passoth, J.H. , Varieties of AI Explanations Under the Law. From the GDPR to the AIA, and Beyond. In xxAI - Beyond Explainable AI, Lecture Notes in Artificial Intelligence; Springer International Publishing, 2022; pp. 343–373. [CrossRef]
- Antoniadi, A.M.; Du, Y.; Guendouz, Y.; Wei, L.; Mazo, C.; Becker, B.A.; Mooney, C. Current Challenges and Future Opportunities for XAI in Machine Learning-Based Clinical Decision Support Systems: A Systematic Review. Applied Sciences 2021, 11, 5088. [Google Scholar] [CrossRef]
- Barredo Arrieta, A.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; Garcia, S.; Gil-Lopez, S.; Molina, D.; Benjamins, R.; et al. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Information Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef]
- Linardatos, P.; Papastefanopoulos, V.; Kotsiantis, S. Explainable AI: A Review of Machine Learning Interpretability Methods. Entropy 2020, 23, 18. [Google Scholar] [CrossRef] [PubMed]
- Thormundsson, B. Global explainable AI market revenues 2022. https://www.statista.com/statistics/1256246/worldwide-explainable-ai-market-revenues/, 2024.
- Jacovi, A. Trends in explainable AI (XAI) literature. https://medium.com/@alonjacovi/trends-in-explainable-ai-xai-literature-a1db485e871, 2023.
- Longo, L.; Brcic, M.; Cabitza, F.; Choi, J.; Confalonieri, R.; Ser, J.D.; Guidotti, R.; Hayashi, Y.; Herrera, F.; Holzinger, A.; et al. Explainable Artificial Intelligence (XAI) 2.0: A manifesto of open challenges and interdisciplinary research directions. Information Fusion 2024, 106, 102301. [Google Scholar] [CrossRef]
- Ali, S.; Abuhmed, T.; El-Sappagh, S.; Muhammad, K.; Alonso-Moral, J.M.; Confalonieri, R.; Guidotti, R.; Del Ser, J.; Díaz-Rodríguez, N.; Herrera, F. Explainable Artificial Intelligence (XAI): What we know and what is left to attain Trustworthy Artificial Intelligence. Information Fusion 2023, 99, 101805. [Google Scholar] [CrossRef]
- World Health Organization. Global cancer burden growing, amidst mounting need for services. https://www.who.int/news/item/01-02-2024-global-cancer-burden-growing–amidst-mounting-need-for-services, 2024.
- Weerarathna, I.N.; Kamble, A.R.; Luharia, A. Artificial Intelligence Applications for Biomedical Cancer Research: A Review. Cureus 2023, 15, e48307. [Google Scholar] [CrossRef] [PubMed]
- Jaber, N. Can Artificial Intelligence Help See Cancer in New, and Better, Ways? National Cancer Institute 2022. [Google Scholar]
- Wijkhuizen, M.; van Karnenbeek, L.; Geldof, F.; Ruers, T.J.; Dashtbozorg, B. Ultrasound tumor detection using an adapted Mask-RCNN with a continuous objectness score. In Proceedings of the Medical Imaging with Deep Learning; 2024. [Google Scholar]
- Natali, T.; Wijkhuizen, M.; Kurucz, L.; Fusaglia, M.; van Leeuwen, P.J.; Ruers, T.J.; Dashtbozorg, B. Automatic real-time prostate detection in transabdominal ultrasound images. In Proceedings of the Medical Imaging with Deep Learning; 2024. [Google Scholar]
- Hoogteijling, N.; Veluponnar, D.; de Boer, L.; Dashtbozorg, B.; Peeters, M.J.V.; van Duijnhoven, F.; Ruers, T. Toward automatic surgical margin assessment using ultrasound imaging during breast cancer surgery. European Journal of Surgical Oncology 2023, 49, e108–e109. [Google Scholar] [CrossRef]
- Veluponnar, D.; de Boer, L.L.; Geldof, F.; Jong, L.J.S.; Da Silva Guimaraes, M.; Vrancken Peeters, M.J.T.; van Duijnhoven, F.; Ruers, T.; Dashtbozorg, B. Toward intraoperative margin assessment using a deep learning-based approach for automatic tumor segmentation in breast lumpectomy ultrasound images. Cancers 2023, 15, 1652. [Google Scholar] [CrossRef]
- Geldof, F.; Pruijssers, C.W.; Jong, L.J.S.; Veluponnar, D.; Ruers, T.J.; Dashtbozorg, B. Tumor Segmentation in Colorectal Ultrasound Images Using an Ensemble Transfer Learning Model: Towards Intra-Operative Margin Assessment. Diagnostics 2023, 13, 3595. [Google Scholar] [CrossRef] [PubMed]
- Weld, A.; Dixon, L.; Anichini, G.; Dyck, M.; Ranne, A.; Camp, S.; Giannarou, S. Identifying Visible Tissue in Intraoperative Ultrasound Images during Brain Surgery: A Method and Application. arXiv preprint arXiv:2306.01190, arXiv:2306.01190 2023. [CrossRef]
- van der Velden, B.H.; Kuijf, H.J.; Gilhuijs, K.G.; Viergever, M.A. Explainable artificial intelligence (XAI) in deep learning-based medical image analysis. Medical Image Analysis 2022, 79, 102470. [Google Scholar] [CrossRef]
- Medline Embase Database. https://www.embase.com.
- Scopus Bibliographic Database. https://www.scopus.com.
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Systematic Reviews 2021, 10, 89. [Google Scholar] [CrossRef]
- Visani, G. Explainable Machine Learning, XAI Review: Model Agnostic Tools. https://towardsdatascience.com/explainable-machine-learning-9d1ca0547ae0, 2020.
- Al-Jebrni, A.H.; Ali, S.G.; Li, H.; Lin, X.; Li, P.; Jung, Y.; Kim, J.; Feng, D.D.; Sheng, B.; Jiang, L.; et al. SThy-Net: a feature fusion-enhanced dense-branched modules network for small thyroid nodule classification from ultrasound images. The Visual Computer 2023, 39, 3675–3689. [Google Scholar] [CrossRef]
- Basu, S.; Gupta, M.; Rana, P.; Gupta, P.; Arora, C. RadFormer: Transformers with global–local attention for interpretable and accurate Gallbladder Cancer detection. Medical Image Analysis 2023, 83, 102676. [Google Scholar] [CrossRef] [PubMed]
- Byra, M.; Dobruch-Sobczak, K.; Piotrzkowska-Wroblewska, H.; Klimonda, Z.; Litniewski, J. Explaining a Deep Learning Based Breast Ultrasound Image Classifier with Saliency Maps. Journal of Ultrasonography 2022, 22, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; She, R.; Cui, C.; Shi, S.; Hu, X.; Zeng, J.; Wu, H.; Xu, J.; Zhang, Y. One step further into the blackbox: a pilot study of how to build more confidence around an AI-based decision system of breast nodule assessment in 2D ultrasound. European Radiology 2021, 31, 4991–5000. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Chang, L.; Song, K.; Cheng, L.; Li, M.; Wei, X. Multitask network for thyroid nodule diagnosis based on TI-RADS. Medical Physics 2022, 49, 5064–5080. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.R.; Islam, M.F.; Uddin, M.Z.; Ghoshal, G.; Hassan, M.M.; Huda, S.; Fortino, G. Prostate cancer classification from ultrasound and MRI images using deep learning based Explainable Artificial Intelligence. Future Generation Computer Systems 2022, 127, 462–472. [Google Scholar] [CrossRef]
- Karimzadeh, M.; Vakanski, A.; Xian, M.; Zhang, B. Post-Hoc Explainability of BI-RADS Descriptors in a Multi-Task Framework for Breast Cancer Detection and Segmentation. In Proceedings of the 2023 IEEE 33rd International Workshop on Machine Learning for Signal Processing (MLSP). IEEE Computer Society; 2023. [Google Scholar] [CrossRef]
- Lombardi, A.; Arezzo, F.; Sciascio, E.D.; Ardito, C.; Mongelli, M.; Lillo, N.D.; Fascilla, F.D.; Silvestris, E.; Kardhashi, A.; Putino, C.; et al. A human-interpretable machine learning pipeline based on ultrasound to support leiomyosarcoma diagnosis. Artificial Intelligence in Medicine 2023, 146, 102697. [Google Scholar] [CrossRef]
- Martizzi, D.; Huang, Y.; Malik, B.; Ray, P.D. Breast mass detection and classification using PRISM™ eXplainable Network based Machine Learning (XNML™) platform for Quantitative Transmission (QT) ultrasound tomography. In Proceedings of the Proc. SPIE 11602, Medical Imaging 2021: Ultrasonic Imaging and Tomography, 2021, Vol. 11602. [CrossRef]
- Morris, J.; Liu, Z.; Liang, H.; Nagala, S.; Hong, X. ThyExp: An explainable AI-assisted Decision Making Toolkit for Thyroid Nodule Diagnosis based on Ultra-sound Images. In Proceedings of the Proceedings of the 32nd ACM International Conference on Information and Knowledge Management.; pp. 20235371–5375. [CrossRef]
- Qian, X.; Pei, J.; Zheng, H.; Xie, X.; Yan, L.; Zhang, H.; Han, C.; Gao, X.; Zhang, H.; Zheng, W.; et al. Prospective assessment of breast cancer risk from multimodal multiview ultrasound images via clinically applicable deep learning. Nature Biomedical Engineering 2021, 5, 522–532. [Google Scholar] [CrossRef]
- Rezazadeh, A.; Jafarian, Y.; Kord, A. Explainable Ensemble Machine Learning for Breast Cancer Diagnosis Based on Ultrasound Image Texture Features. Forecasting 2022, 4, 262–274. [Google Scholar] [CrossRef]
- Song, D.; Yao, J.; Jiang, Y.; Shi, S.; Cui, C.; Wang, L.; Wang, L.; Wu, H.; Tian, H.; Ye, X.; et al. A new xAI framework with feature explainability for tumors decision-making in Ultrasound data: comparing with Grad-CAM. Computer Methods and Programs in Biomedicine 2023, 235, 107527. [Google Scholar] [CrossRef]
- Tasnim, J.; Hasan, M.K. CAM-QUS guided self-tuning modular CNNs with multi-loss functions for fully automated breast lesion classification in ultrasound images. Physics in Medicine and Biology 2024, 69, 015018. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Haertling, T. AIBx, Artificial Intelligence Model to Risk Stratify Thyroid Nodules. Thyroid 2020, 30, 878–884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Vakanski, A.; Xian, M. Bi-Rads-Net: An Explainable Multitask Learning Approach for Cancer Diagnosis in Breast Ultrasound Images. In Proceedings of the 2021 IEEE 31st International Workshop on Machine Learning for Signal Processing (MLSP); 2021. [Google Scholar] [CrossRef]
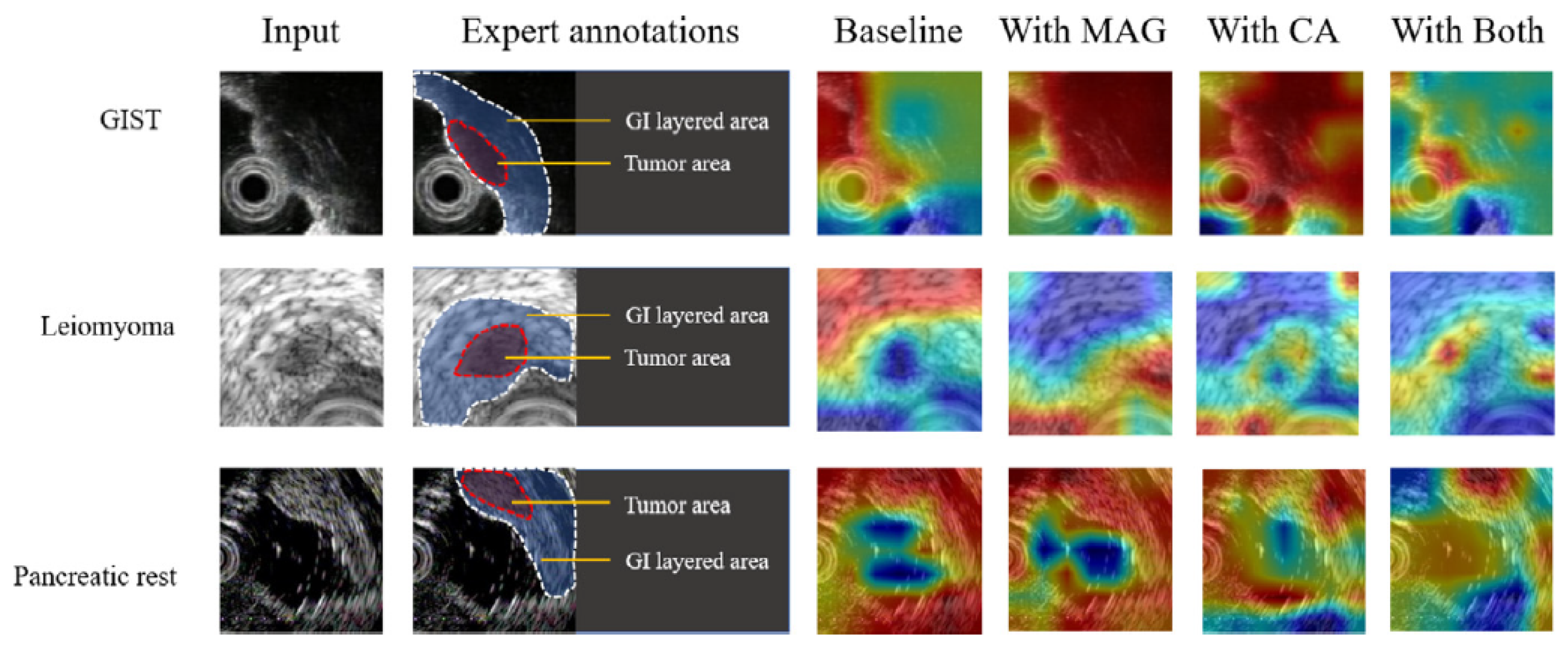
- Zheng, H.; Dong, Z.; Liu, T.; Zheng, H.; Wan, X.; Bao, J. Enhancing gastrointestinal submucosal tumor recognition in endoscopic ultrasonography: A novel multi-attribute guided contextual attention network. Expert Systems with Applications 2024, 242, 122725. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, S.; Song, J.; Pan, B.; Bai, G.; Zhao, L. XAI Benchmark for Visual Explanation. arXiv preprint arXiv:2310.08537, arXiv:2310.08537 2023. [CrossRef]
- Amann, J.; Blasimme, A.; Vayena, E.; Frey, D.; Madai, V.I. Explainability for artificial intelligence in healthcare: a multidisciplinary perspective. BMC Medical Informatics and Decision Making 2020, 20, 310. [Google Scholar] [CrossRef] [PubMed]
| Author (year) | Cancer Type | Clinical Application | Study Type | Imaging | Acquisition Type | Dataset | Total Patients (Lesions) | Total Images | Healthy Images (%) | Benign Images (%) | Malignant Images (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Al-Jebrni A.H. (2023) [37] | Thyroid | Diagnosis | R | US | Continuous | STNU | 1810 (2068) | 4136 | - | 1884 (45.6%) | 2252 (54.4%) |
| lightgrayBasu S. (2023) [38] | Gallbladder | Diagnosis | R | US | Single shot | GBUS | 218 (147) | 1255 | 432 (34.4%) | 558 (44.5%) | 265 (21.1%) |
| Byra, M. (2022) [39] | Breast | Diagnosis | R | US | Single shot | Clinical | NA (272) | 272 | - | 149 (54.8%) | 123 (45.2%) |
| Dong, F. (2021) [40] | Breast | Diagnosis | R | US | Continuous | Clinical | 367 (785) | 579 | - | 247 (42.7%) | 332 (57.3%) |
| Han, X. (2022) [41] | Thyroid | Diagnosis | P | US | Single Continuous | Clinical | 3906 (3906) | 3906 | - | 1696 (43.4%) | 2210 (56.6%) |
| Hassan, M. R. (2022) [42] | Prostate | Diagnosis | R | US + MRI | Continuous (US) + Static slice (MRI) | Cancer Imaging Archive | 1151 (NA) | 611119 | - | NA | NA |
| Karimzadeh, M. (2023) [43] | Breast | Screening | R | US | Continuous | BUSI, BUSIS, HMSS | NA (2917) | 2186 | - | NA | NA |
| Lombardi, A. (2023) [44] | Leiomyogenic | Surgery (pre-operative) | R | US | Continuous, 3D volume scan | Clinical | 68 (68) | 68 | - | 60 (88.2%) | 8 (11.8%) |
| Martizzi, D. (2021) [45] | Breast | Screening | P | US | 3D volume scan | Clinical | 70 (60) | 70 | 10 (14.3%) | 41 (58.6%) | 19 (27.1%) |
| Morris, J. (2023) [46] | Thyroid | Diagnosis | R | US | Continuous | Clinical | 307 (831) | 831 | - | NA | NA |
| Qian, X. (2021) [47] | Breast | Screening | P | US | Continuous | Clinical | 634 (721) | 10815 | - | NA | NA |
| Rezazadeh, A. (2022) [48] | Breast | Diagnosis | R | US | Continuous | Public | 600 (697) | 780 | 133 (17.1%) | 210 (26.9%) | 487 (62.4%) |
| Song, D. (2023) [49] | Thyroid | Screening | R | US | Single shot | Clinical | 7236 (19341) | 19341 | - | 12943 (66.9%) | 6398 (33.1%) |
| Tasnim, J. (2024) [50] | Breast | Diagnosis | R | US | Continuous | BUSI, Mendeley, UDIAT, OMI, BUET-BUSD | NA (1494) | 1494 | - | 901 (60.3%) | 593 (39.7%) |
| Thomas, J. (2020) [51] | Thyroid | Surgery (pre-operative) | R | US | Continuous | Clinical | 402 (482) | 2025 | - | NA | NA |
| Zhang, B. (2021) [52] | Breast | Diagnosis | R | US | Continuous | BUSI, BUSIS | NA (1192) | 1192 | - | 727 (61.0%) | 465 (39.0%) |
| Zheng, H. (2024) [53] | Gastrointestinal stromal tumors | Diagnosis | R | US | Continuous | Clinical | 261 (261) | 1900 | - | - | 1900 (100%) |
| Classification (n=16) | ||||||
|---|---|---|---|---|---|---|
| Author (year) | Architecture | Task | Ground Truth | Accuracy | AUROC | Other Performance |
| Al-Jebrni, A.H. (2023) [37] | Inception-V3 | Tumor class benign or malignant | Clinical diagnosis | 0.874 | 0.905 | Sens 0.895, Spec 0.858 |
| lightgrayBasu, S. (2023)* [38] | BagNets33 | Tumor class benign or malignant (local-level) | Histopathology + Radiologist assigned lexicons | 0.921 | 0.971 | Sens 0.923, Spec 0.961 |
| Byra, M. (2022) [39] | ResNet | Tumor class benign or malignant | Pathology | 0.887 | 0.835 | Sens 0.801, Spec 0.868 |
| Dong, F. (2021) [40] | DenseNet-121 | Tumor class benign or malignant | Histopathology | 0.884 | 0.899 | Sens 0.879, Spec 0.892 |
| Han, X. (2022) [41] | DenseNets + SGE attention module | TI-RADS risk level (multi-class) & | Clinical diagnosis | 0.780 (w) | - | MAE 1.30 |
| Tumor class benign or malignant | Histopathology | 0.954 | 0.981 | Sens 0.988, Spec 0.912, PPV 0.928, NPV 0.985 | ||
| Hassan, M. R. (2022) [42] | VGG-16 + Random Forest | Tumor class benign or malignant | Radiologist manual annotation | 0.875 | - | - |
| Karimzadeh, M. (2023)* [43] | VGG-16 | BI-RADS risk level (multi-class) & | Clinical diagnosis | 0.852 (w) | - | - |
| Tumor class benign or malignant | Clinical diagnosis | 0.913 | - | Sens 0.940, Spec 0.858 | ||
| Lombardi, A. (2023) [44] | XGBoost | Tumor class benign or malignant | Clinical diagnosis | - | 0.994 | Sens 0.875, Spec 0.983, F1-score 0.875, Brier score 0.0187 |
| Morris, J. (2023) [46] | Local texture quantization | TI-RADS risk level (multi-class) | Clinical diagnosis | >0.80 | - | - |
| Qian, X. (2021) [47] | ResNet-18 + SENet | BI-RADS risk level (multi-class) | Histopathology | - | 0.955 | - |
| Rezazadeh, A. (2022) [48] | LightGBM | Tumor class benign or malignant | Clinical diagnosis | 0.91 | 0.93 | Prec 0.94, Rec 0.93, F1-score 0.93 |
| Song, D. (2023) [49] | DenseNet-121 | Tumor class benign or malignant | Pathology / Clinical diagnosis | NA | - | - |
| Tasnim, J. (2024) [50] | ResNet-18 + InceptionV3 | Tumor class benign or malignant | Clinical diagnosis | 0.915 | 0.952 | Sens 0.894, Spec 0.929, F1-score 0.893, MCC 0.824 |
| Thomas, J. (2020) [51] | ResNet 34 | Tumor class benign or malignant | Histopathology | 0.777 | - | Sens 0.849, Spec 0.743, PPV 0.609, NPV 0.912 |
| Zhang, B. (2021) [52] | VGG-16 | BI-RADS risk level (multi-class) & | Clinical diagnosis | 0.843 (w) | - | - |
| Tumor class benign or malignant & | Clinical diagnosis | 0.889 | - | Sens 0.838, Spec 0.923 | ||
| Likelihood of malignancy | Clinical diagnosis | - | - | R2 0.671, MSE 0.153 | ||
| Zheng, H. (2024) [53] | VGG-16 | Tumor class benign or malignant | Pathology | 0.932 | - | Prec 0.932, Rec 0.932, F1-score 0.932 |
| Segmentation (n=2) | ||||
|---|---|---|---|---|
| Author (year) | Architecture | Task | Ground Truth | Performance |
| Karimzadeh, M. (2023)* [43] | U-Net | Lesion mask | NA | DSC 0.827 |
| lightgrayMartizzi, D. (2021) [45] | Gaussian Mixture Models + Blob detection | ROI mask | Radiologist manual annotation of ROI | Recall 0.83 (benign) Recall 0.95 (malignant) |
| Localization (n=1) | ||||
| Author (year) | Architecture | Task | Ground Truth | Performance |
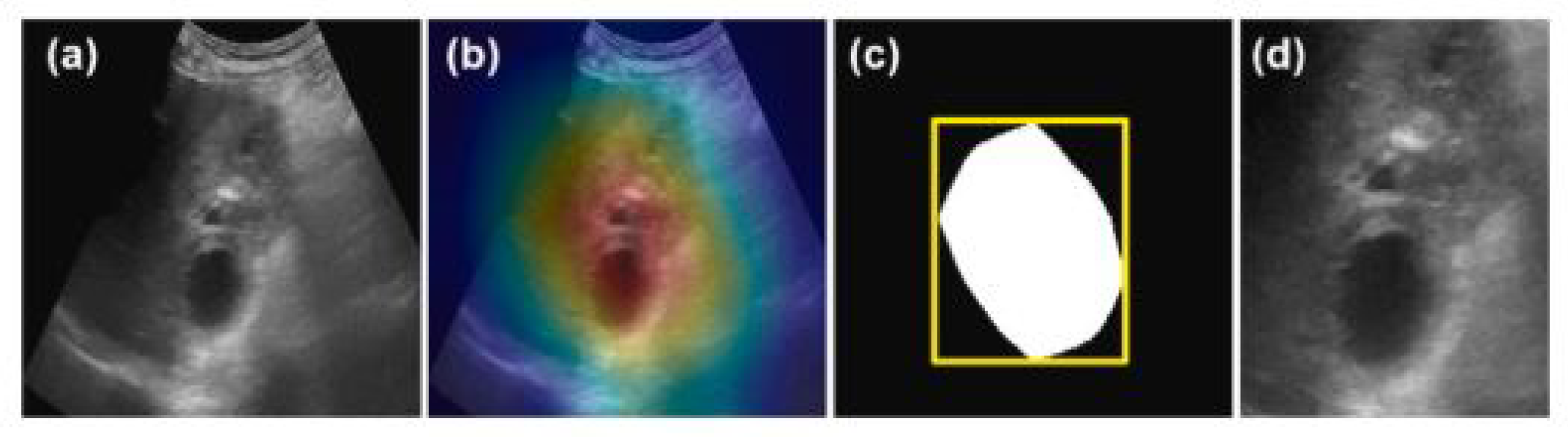
| Basu, S. (2023)* [38] | ResNet-50 | ROI identification (global-level) | Radiologist bounding box annotation | Mean IoU 0.484 Mean Intersection 0.934 |
| XAI Method | Model Specific | Scope | Explanation | AI Task | XAI Function | XAI Input | XAI Output | Analysis Type | Evaluation |
|---|---|---|---|---|---|---|---|---|---|
| Activation heat map [38] | No | Global | Intrinsic | Localization | Visualization | Images | Heat map with top activated image features | Visual inspection | QL |
| lightgrayActivation Maximization [50] | CNNs | Global | Post-hoc | Classification | Visualization | Feature maps | Activation maximizing output image patterns | Visual inspection, ZNCC score | QL, QT |
| Bag-of-Features [38] | Transformers | Local | Intrinsic | Classification | Semantics | ROI in US images | Most discriminative features for class prediction | Features mapped to radiological lexicon | QL |
| BI-RADS-Net [52] | MTL using CNNs | Local | Intrinsic | Classification | Semantics, Example based | Breast US images + feature maps | Predicted class probability + morphological feature explanations | Accuracy, Sensitivity and Specificity | QT |
| CAM [39,50] | CNNs | Local | Post-hoc | Classification | Visualization | Images | Saliency map | Visual inspection, Pointing game | QL, QT |
| Explainer [49] | CNNs | Local | Intrinsic | Classification | Visualization | Images + convoluted feature maps | Features the model uses to make the predictions | Visual inspection, Accuracy, Precision, Sensitivity, Specificity, F1-score, AUROC | QL, QT |
| Grad-CAM [37,41,47,49,53] | CNNs | Local | Post-hoc | Classification (n=6), Segmentation (n=1) | Visualization | Images | Saliency map | Visual inspection | QL |
| Image similarity AIBx [51]) | CNNs | Local | Post-hoc | Classification | Semantics, Example based | Images | Most similar images in database with known diagnoses | Confusion matrix | QT |
| LIME [42] | No | Local | Post-hoc | Classification | Visualization | Images | ROI identified | Visual inspection | QL |
| LTQ-E [46] | No | Local | Post-hoc | Classification | Semantics | US image + Embedded decision label model | Predicted class + morphological feature explanations | Accuracy | QT |
| MT-BI-RADS [43] | MTL using CNNs | Local | Intrinsic | Segmentation | Visualization | Breast US images | Tumor mask | Visual inspection, DSC | QL, QT |
| Region of Evidence (ROE) [40] | CNNs | Local | Post-hoc | Classification | Visualization | Images | Saliency map + Resemblance vote by expert physicians | Confusion matrix + breakdown in resemblance votes | QT |
| SHAP [43,44,48] | No | Local | Post-hoc | Classification | Semantics | Image + feature map | Significance (Shapley) value per feature | Shapley values, Confusion matrix | QL, QT |
| XNML [45] | PRISMTM platform (ML) | Local | Intrinsic | Segmentation | Visualization, Example based | 3D Quantitative Transmission US speed-of-sound maps | Color-coded segmentations | Visual inspection | QL |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





