1. Introduction
Shrimp is an economically important aquaculture species in China. However, efficient gene transfer and expression tool is lacking for both shrimp individuals and in vitro cultured shrimp cells. Many attempts had been tried to introduce foreign genes into the eggs or early embryos of penaeid shrimps but failed. This can be attributed to the inherent problems including the injection-induced burst especially for homolecithal eggs, the quick cleavage, and the low integration and survival rates associated with traditional gene delivery methods like microinjection, electroporation and particle bombardment [
1,
2,
3]. For the in vitro cultured shrimp cells, the gene transfer methods mediated by liposomes, mammalian viruses or insect baculoviruses had been tried but produced limited success with extremely low transfection or infection efficiencies in comparison with the results from mammalian tumor cells or insect cells due to the lacking of actively-dividing shrimp cells and extremely low tropism of the viruses used [
4,
5,
6,
7,
8,
9]. Thus, efficient gene transfer and expression tool is urgently needed for shrimps and shrimp cells.
It has been well recognized that a lot of great success in biological field can be attributed to a great degree to the use of mammalian virus-mediated gene transfer and expression systems such as retrovirus, lentivirus, adenovirus and adeno-associated viruses (AAV) because of their high infectivity and high gene delivery efficiency as well as low cytotoxicity [
10,
11,
12,
13]. However, all of the aforementioned mammalian viruses-mediated expression systems had extremely low tropism in shrimp cells even though they were improved to be pantropic by pseudo-typing with a foreign envelope glycoprotein of vesicular stomatitis virus (VSV-G) [
4,
5,
6,
7,
8,
14]. Pu et al. [
4] and Chen et al. [
5] further improved the tropism of the pantropic retrovirus and lentivirus expression systems in shrimp cells by introducing two envelope proteins of VP28 and VP19 of shrimp white spot syndrome virus (WSSV) into the envelope of the corresponding virions packaged via co-transfection method, respectively, however, their infection and expression efficiencies obtained in shrimp cells were far lower than what they behaved in mammalian tumor cells. Recently, Tao et al. [
8] modified the capsid of the packaged AAV-2 by introducing shrimp WSSV-sourced tegument protein of VP26 or shrimp infectious hypodermal and hematopoietic necrosis virus (IHHNV)-sourced capsid protein (IHCP) via co-transfection method and significantly improved the tropism of the modified AAV-2 in shrimp cells, but the infection and expression efficiencies obtained in shrimp cells were still much lower than it did in mammalian cells.
Attempts had also been tried on the improvement and applications of insect baculovirus expression system in shrimps and shrimp cells due to the fact that unmodified baculovirus could not efficiently infect shrimps and shrimp cell [
9,
15]. Puthumana et al. [
15] modified the baculovirus vector by insertion of shrimp virus-sourced promoters of Ie1 (WSSV) and P2 (IHHNV) and found that the modified baculovirus could successfully infect the shrimp adult tissues although the infection efficiency was still low (<20%). In contrast, Wu et al [
9] developed a shrimp WSSV envelope protein VP28-pseudo-typed baculovirus expression system and found that the improved baculovirus could infect the primarily cultured shrimp hemolymph cells at a very low efficiency (1.2%) but shrimp adult tissues at an extremely high efficiency of nearly 100%, suggesting a better performance of insect baculovirus in shrimps in contrast to mammalian viruses [
4,
5,
6,
7].
Compared with mammalian and insect viruses, shrimp virus has incomparable advantage in the tropism to shrimp cells. Thus, we believe that efficient infection and foreign gene expression in shrimp cells can be expected from the development of a shrimp virus-mediated gene transfer and expression system. Recently, based on a shrimp RNA virus of
Macrobrachium rosenbergii nodavirus (MrNV), Alenton et al. developed a shrimp viral vector, a replication-incompetent mutant MrNV(ΔRdRp)-GFP, for RNA delivery, paving the way for the oral delivery of antiviral therapeutics in farmed crustaceans [
16]. However, up to date, no shrimp DNA virus-mediated gene transfer and expression system have ever been established.
Infectious hypodermal and hematopoietic necrosis virus (IHHNV) belongs to the genus Penstylhamaparvovirus, family Parvoviridae and subfamily Hamaparvovirinae. It was also called PstDV because it was first found from
Penaeus stylirostris in Hawaii (USA) [
17,
18], and later found in other shrimp species, crabs and bivalves [
19,
20,
21,
22,
23]. IHHNV was found to be a unenveloped icosahedral DNA virus containing a linear single-stranded genomic DNA of no more than 4.1 kb in size, the smallest known penaeid shrimp virus [
24]. This makes it feasible for the genomic DNA of shrimp IHHNV to be engineered into an expression vector. The genomic DNA of IHHNV contains three open reading frames of ORF1, ORF2 and ORF3, driven by three promoters of P2, P11 and P61 in their upstream, respectively [
25,
26]. These three promoters were found to be active not only in shrimp cells but also in insect and fish cells. Moreover, the capsid protein (ORF3) of IHHNV had been over-expressed in bacterial or insect cells and then self-assembled in vitro into virus-like particles (VLPs) and successfully used as a nanocarrier to deliver foreign genes into shrimp cell [
27,
28,
29]. However, the use of VLPs as a gene delivery tool is limited due to the high cost and extensive labor in the production of VLPs and relatively low DNA-loading efficiency.
Suitable packaging cells are essential for a virus-mediated expression system. In consideration of the lacking of immortalized shrimp cell line and the accumulated evidences of the infectivity of several shrimp viruses in the insect cells [
30,
31,
32,
33,
34] and the successful propagation of IHHNV in a shrimp-insect hybrid cells of PmLyO-Sf9 35, in this study, the insect Sf9 cell line was chosen as a packaging cell line for the IHHNV-based expression vector although electron microscopy evidence on the formation of shrimp viral particles in the infected insect cells has not yet been reported.
This study aims to develop a shrimp virus (IHHNV)-mediated gene transfer and expression system using insect Sf9 cells as packaging cells. To achieve it, a near full-length genomic DNA of shrimp IHHNV had be isolated and then cyclized with a prokaryotic pUC19 backbone to obtain the cyclized plasmid of pUC19-IHHNV. After that, an IHHNV-based expression vector was constructed by inserting an expression cassette of PH-GUS-SV40 pA, the baculoviral polyhedron (PH) promoter-driven GUS (β-glucuronidase) gene, into pUC19-IHHNV immediately downstream of IHHNV and then packaged into IHHNV-like viral particles in the insect Sf9 cells by transfection. And then the gene transfer and expression efficiency of this shrimp IHHNV-mediated expression system was evaluated and further improved in three systems of insect Sf9 cells, shrimp hemolymph cells and adult tissues of infected shrimps via the expression of GUS reporter gene for proof-of-concept.
3. Conclusions
In this study, the genomic DNA of shrimp IHHNV had been successfully isolated and cyclized into a prokaryotic cloning plasmid of pUC19-IHHNV. This made it come true to multiply the genomic DNA of a shrimp virus in bacterial cells. Subsequently, a shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS was constructed by introducing a eukaryotic expression cassette of PH-GUS-MCS-SV40 pA into the aforementioned cyclized plasmid immediately after the IHHNV genome. It was also found that the insect Sf9 cells could be used as a packaging cell line for the shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS, confirmed by reinfection, transmission electron microscopy and electron microscopy negative staining assay. Shrimp IHHNV-like icosahedral virus particles with a similar size to wild-type IHHNV could be observed both in the Sf9 cells and the medium supernatant of pUC19-IHHNV-PH-GUS -infected Sf9 cells, however, as a late-stage promoter of insect baculovirus, the activation of PH promoter is dependent on the expression products of some early genes of baculovirus. Therefore, no GUS expression or blue signal could be detected in the single plasmid of pUC19-IHHNV-PH-GUS-transfected Sf9 cells, but many blue cells could be detected in the co-transfected Sf9 cells by pUC19-IHHNV-PH-GUS and Bacmid (or Bacmid-VP28). Moreover, a significant higher transfection and expression efficiency was obtained when baculoviral plasmid of Bacmid-VP28 encoding shrimp WSSV-sourced envelope protein VP28 was used instead of Bacmid.
In order to facilitate the expression efficiency of GUS reporter gene, the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid (or Bacmid-VP28) were used to infect the primarily cultured shrimp hemolymph cells and adult shrimps. However, in shrimp hemolymph cells, no GUS expression or blue cells could be detected after infection by the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid, possibly due to the extremely low tropism and infectivity of Bacmid in the shrimp hemolymph cells. Instead of this, obvious blue cells were observed in the shrimp hemolymph cells infected by the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28 possibly due to the higher infectivity and cell entry of Bacmid-VP28 in shrimp hemolymph cells. In a word, the infection and GUS expression capability of the mixed viruses could be greatly improved when Bacmid-VP28 was used to replace Bacmid. Unlike shrimp hemolymph cells, both of these two kinds of mixed viruses could infect the adult tissues of most of the injected shrimps with an infection efficiency of 70%-80% and in tissue-specific and dose-dependent manners, and this was confirmed by semi-quantitative RT-PCR analysis. Noteworthy, hearts had so strong a background GUS expression level as to interrupt the detection of infection and over-expression of foreign GUS gene.
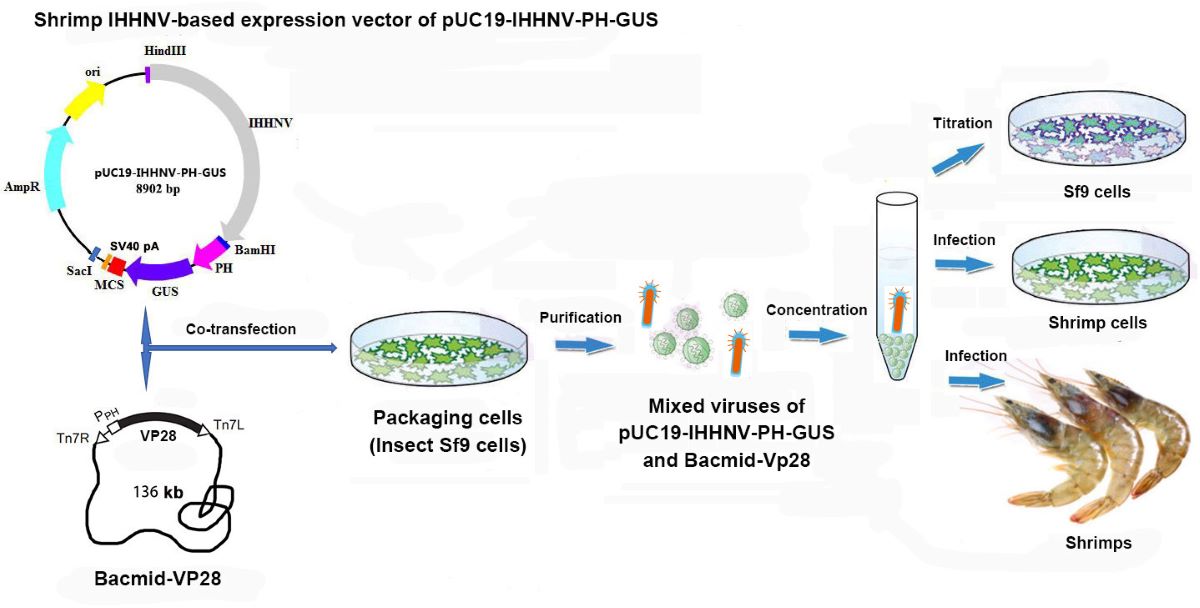
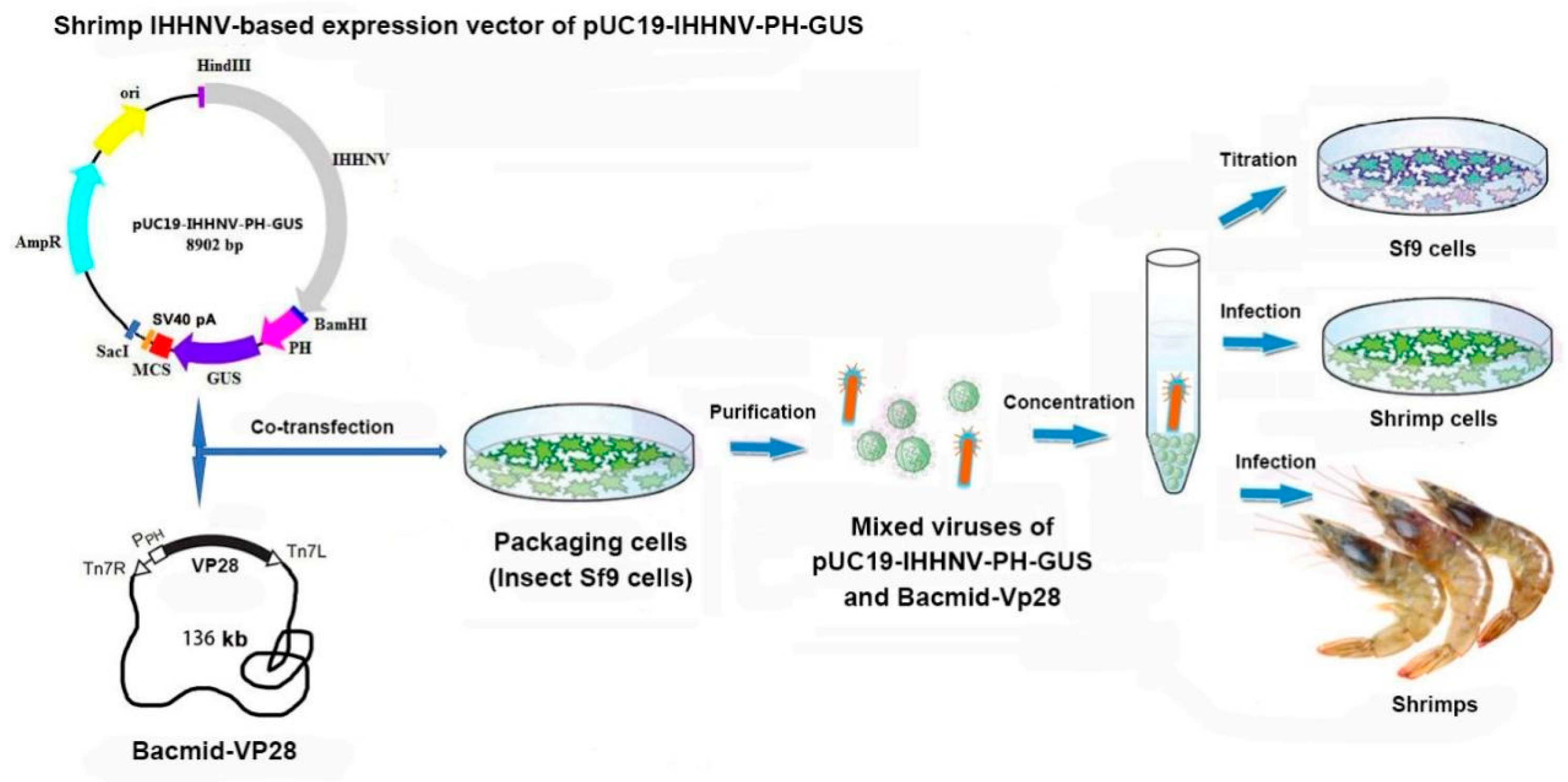
Taken together, a shrimp virus (IHHNV)-mediated gene transfer and expression system had been successfully developed in this study, which consisted of a shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS, a helper baculovirus of Bacmid-VP28 and one packaging cell line of insect Sf9 (
Figure 10). This useful research tool for efficient gene transfer and expression in shrimps will greatly promote the future works on the molecular breeding of transgenic shrimps with merit traits as well as immortalization of the in vitro cultured shrimp cells, although it still needs more improvements like gene delivery efficiency and biosafety assessment, for example, the use of IHHNV with full-length 5’ and 3’ ITR, the production of replication-defective IHHNV by splitting up of the capsid gene (ORF3) from the IHHNV genomic DNA.
4. Materials and Methods
4.1. Shrimps
Actively swimming shrimps (Metapenaeus ensis), 12 ± 3 cm in length and 15 ± 5 g in weight, were purchased from a local seafood market of Nanshan, Qingdao, China. They were acclimatized in an aerated seawater at ambient temperature of 18-25 °C for at least two days before used for viral infection and primary cell culture. The infectious hypodermic and hematopoietic necrosis virus (IHHNV)-infected shrimps were sampled from the local shrimp farm of Qingdao, China, immediately frozen and transported in liquid nitrogen to laboratory, and then stored at -80°C until used for the isolation of the genomic DNA of IHHNV. The care and use of shrimps were under supervision and approved by the scientific ethics committee of Ocean University of China.
4.2. Cells and Cell Culture
A continuous insect cell line of Sf9, derived from the ovarian tissues of the pupa of fall army worm, Spondoptera frugiperda, was used for the packaging and titration of shrimp IHHNV-based recombinant viruses, the baculoviruses of Bacmid (i.e. bMON14272, from Bac-to-Bac baculovirus expression system, Cat. No. 10360-014, Invitrogen), Bacmid-GUS encoding β-glucuronidase (GUS), and Bacmid-VP28 encoding the envelope protein (VP28) of shrimp WSSV. The Sf9 cells were maintained in SIM medium (Sino biology, China) supplemented with 10% fetal bovine serum (FBS, BI) at 28℃ in a 3% CO2 incubator.
Primarily cultured shrimp hemocytes were isolated from the circulating hemolymph of
M. ensis and then cultured in a 1.5× L-15-based or gelatin-containing 1.0× L-15-based growth medium as described previously by Han et al. [
36] and Zhao et al. [
37], respectively, until used for the infectivity analysis of the shrimp IHHNV-based recombinant viruses, or the baculoviruses of Bacmid, Bacmid-GUS and Bacmid-VP28. In brief, the shrimps were pre-treated overnight in aerated boiling-disinfected seawater containing 1 200 IU/mL penicillin and 1 200 IU/mL streptomycin. Then the shrimps were individually anesthetized by immersion in 75% ethanol for 3-5 min. Next, after sequential disinfection of the body surface of the shrimps by iodophor (Lircon, China) and 75% ethanol, the circulating hemolymph were drawn out from the thoracic sinus of the shrimp using 1-mL aseptic injection syringe preloaded with 200 μL 1.5× L-15-based shrimp growth medium, then mixed and seeded into a 96-well culture plate (100 μL/well) and cultured at 28 ℃ in a 3% CO
2 incubator for 4 hours. After that, the old medium in each well was replaced with fresh 1.5× L-15-based or 1× L-15 gelatin-containing growth medium and cultured until used.
4.3. Cloning and Cyclizing of the Genomic DNA of Shrimp IHHNV
The published full-length genomic DNA of shrimp infectious hypodermic and hematopoietic necrosis virus (IHHNV) was 4.1 bp in size, which had two highly variable inverted terminal repeats (ITR) in both ends. After many attempts, only 3833 bp in length of the genomic DNA of IHHNV, with shortened ITR in both ends, had been cloned in this study. In detail, the genomic DNA of IHHNV was amplified by PCR in two overlapped fragments, 1-1910 bp and 1870-3833 bp, respectively. In order to link them together, these two fragments had a 40 bp overlap located in position 1870-1910 bp which served as a homology arm for recombinase-based assembly. The genomic DNA of IHHNV was cyclized by head to tail joining with a linearized prokaryotic cloning plasmid of pUC19 which was pre-digested with double restriction endonucleases of Hind III and BamH I. Thus, an IHHNV-containing shuttle plasmid of PUC19-IHHNV was constructed to be used to multiply the genomic DNA of IHHNV in bacteria.
To clone and cyclize the genomic DNA of IHHNV, the anterior fragment of 1-1910 bp was amplified by a forward primer of 5’-TCACACAGGAAACAGCTATGACCATGATTACGCCAAGCTTTCGGAGCGCTTCGCAGGAAACCGTTACAA-3’, which had a 40 bp homologous arm (underlined) in the 5’-end corresponding to the 40 bp sequence in the upstream of the Hind III restriction endonuclease site in the plasmid of pUC19, and a reverse primer of 5’-GCATATTGTCGTAGTCTGGT-3’. The posterior fragment of 1870-3833 bp was amplified by a forward primer of 5’-GTCACTAATTACAAACCTGCAG-3’ and reverse primer of 5’-AACGACGGCCAGTGAATTCGAGCTCGGTACCCGGGGATCCCTTCGCAGAAACCGTTAAC-3’, which had a 40 bp homologous arm (underlined) in the 5’-end, too, but this homologous arm corresponded to the 40 bp complement sequence in the downstream of the BamH I restriction endonuclease site in the plasmid of pUC19. Finally, the linearized plasmid of pUC19 and the homology arm-carrying IHHNV fragments of 1-1910 bp and 1870-3833 bp were connected head to tail into one cyclized recombinant plasmid of pUC19-IHHNV using a large fragment homologous recombination kit (Gibson Assembly® Ultra Kit) according to the manual instruction. In brief, 40 ng anterior fragment, forty ng posterior fragment and 50 ng linearized pUC19 were first mixed in a 200-μL PCR tube and then 5 μL GA Ultra Master Mix A (2 ×) was added and mixed again. After that, the mixture was incubated and run under the following PCR program: 37°C for 5 min, 75°C for 20 min, and then decreased to 60°C at a rate of 0.1°C per second, then 60°C for 30 min, and finally decreased to 4°C at a rate of 0.1°C per second. Next, 10 μL GA Ultra Master Mix B (2 ×) was added and run at 45°C for 15 min. Finally, the ligation product was immediately transformed into the T1 competent bacteria cells for screening and sequencing of positive clones.
4.4. Construction of Shrimp IHHNV-Based Expression Vector of pUC19-IHHNV-PH-GUS
First, an expression cassette of PH-GUS-MCS-SV40 pA, 2383 bp in size, consisting of an insect baculovirus polyhedron (PH) promoter, GUS (β-glucuronidase) gene, a downstream MCS (multiple cloning site) and SV40 poly (A) signal (pA), was amplified by a forward primer of 5’-TCTGCGAAGGGATCCATCATGGAGATAATTAAAATGA-3’ and reverse primer of 5’-AGTGAATTCGAGCTCGATCCAGACATGATAAGAT-3’ using a donor plasmid of pFastBacTM1-GUS as template. The pFastBacTM1-GUS plasmid was purchased along with Bac-to-Bac baculovirus expression system (Invitrogen,catalog No. 10359-016 and 10360-014). For recombinase-based assembly, the above forward primer was added a 15 bp homologous arm in its 5’-end (underlined) corresponding to the 15 bp sequence in the upstream of the BamH I restriction endonuclease site in the plasmid of pUC19-IHHNV, and the above reverse primer was added a 15 bp homologous arm in its 5’-end (underlined) which corresponded to the 15 bp complement sequence in the downstream of the Sac I restriction endonuclease site in the plasmid of pUC19-IHHNV, respectively. Next, to construct the shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS, the homologous arm-containing fragment of PH-GUS-MCS-SV40 pA was inserted into the pUC19-IHHNV vector, which had been linearized by BamH I and Sac I, using a universal homologous recombination kit (Pro Ligation-Free Cloning Kit, abm Inc., Cat. No. E086) according to the manual instruction. In brief, the PH-GUS-MCS-SV40 pA fragment, linearized plasmid of pUC19-IHHNV and 2 × Pro Ligation-Free Mix were mixed and then incubated at 50℃ for 2 h. Then the ligation product was immediately transformed into DH5α competent bacterial cells for the screening and sequencing of positive clones.
4.5. Viral Packaging
The packaging of pUC19-IHHNV-PH-GUS, Bacmid and Bacmid-GUS were carried out as described previously by Wu et al [
9] with minor modification. In brief, Sf9 cells were seeded into a 48-well culture plate at a seeding density of 1 × 10
5 cells/well at about 16 h ahead of transfection, and when the cell confluency reached 70 %, the old medium in each well was replaced with a diluted SIM medium containing 13.5% SIM medium, 1.5 % FBS and 85% serum-free Grace medium (Gibco). After that, for each well, a total of 1.875 μL transfection reagent of Cellfectin II (Gibco) and 0.375 μg viral plasmid DNA were separately diluted in 25 μL serum-free Grace medium in two tubes and incubated at room temperature for 10 min. After that, they were combined and vortexed and incubated at room temperature for another 20-30 min. The next, the liposome -DNA transfection complex was evenly added into the well drop by drop and cultured at 28 °C for 4 h, and then the transfection reagent-containing medium was replaced with normal serum-containing SIM medium. 120 h later, the expression of
GUS reporter gene was assayed by X-Gluc staining (Solarbio, China) as described previously by Wu et al [
9]. The baculoviruses of Bacmid and Bacmid-GUS were chosen as negative and positive control, respectively, to verify the expression of the
GUS gene of the shrimp virus (IHHNV)-based expression vector of pUC19-IHHNV-PH-GUS in the Sf9 cells. Baculoviral plasmids of Bacmid and Bacmid-GUS were prepared according to the manual instruction of Bac-to-Bac baculovirus expression system (Invitrogen).
A total of two kinds of mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid (or pUC19-IHHNV-PH-GUS and Bacmid-VP28) were packaged in this study. Compared with the baculovirus of Bacmid, the recombinant baculovirus of Bacmid-VP28 encoded an important envelope protein of VP28 of shrimp WSSV and thus was reported to have better tropism to shrimp cells [
10]. Before packaging, firstly, the optimal co-transfection ratio of pUC19-IHHNV-PH-GUS to Bacmid was examined in a 48-well culture plate. The tested co-transfection ratios of pUC19-IHHNV-PH-GUS (0.375 μg) to Bacmid (μg) were 1: 1/3 (0.125), 1: 1/2 (0.1875), 1: 1 (0.375), 1: 2 (0.75) and 1: 3 (1.125) using a transfection reagent ratio of 5 μL Cellfectin II per μg DNA as reported by Wu et al [
9]. As shown in Fig.3, it was found that the optimal co-transfection ratios of pUC19-IHHNV-PH-GUS to Bacmid were 1: 1 (μg). Secondly, the optimal ratio of transfection reagent to the mixed plasmid DNAs was further examined in a 48-well culture plate with a co-transfection ratio of 1: 1 (pUC19-IHHNV-PH-GUS to Bacmid in μg). The tested ratios of the mixed plasmid DNAs (μg) to Cellfectin II (μL) were 1: 4, 1: 5 and 1: 6, respectively.
Finally, the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid (or pUC19-IHHNV-PH-GUS and Bacmid-VP28) were first packaged under the optimized co-transfection ratio and transfection reagent ratio in a 24-well culture plate, in detail, 1 μg pUC19-IHHNV-PH-GUS, 1 μg Bacmid (or Bacmid-VP28) and 12 μL transfection reagent of Cellfectin II were used in the co-transfection of Sf9 cells. Then, at 120 h post co-transfection, the medium containing detached Sf9 cells and cell debris was collected and then clarified twice by centrifugation at 12,000 ×g, 4 °C for 15 min and finally the medium supernatant of mixed viruses was saved and used to further infect the Sf9 cells cultured in 10-cm petri dishes by 400 μL viral supernatant per dish for the purpose of the multiplication of the mixed viruses.
4.6. Purification, Concentration and Titration of the Viruses
To purify the single virus of pUC19-IHHNV-PH-GUS, the transfected Sf9 cells were collected by trypsinization and centrifugation at 2,000 × g, 4 °C for 15 min and then frozen and thawed for three rounds. The next, the lysed cells were resuspended in sterile PBS (8.0 g NaCl, 0.2 g KCl, 0.2 g KH2PO4 and 3 g Na2HPO4·12H2O in 1 L dH2O, pH 7.0) and then clarified twice by centrifugation at 12,000 ×g, 4 °C for 15 min. Finally, the viral supernatant was collected and concentrated by ultracentrifugation at 140,000 ×g, 4 °C for 2 h. After ultracentrifugation, the supernatant was discarded and the viral precipitate was resuspended with PBS, aliquoted and stored at -80 °C.
To purify the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid (or pUC19-IHHNV-PH-GUS and Bacmid-VP28), the medium supernatant was first collected and saved, and then the co-transfected Sf9 cells were collected by trypsinization and centrifugation and then frozen and thawed for three rounds. The next, the lysed cells were resuspended in the saved medium supernatant and then clarified twice by centrifugation at 12,000 ×g, 4 °C for 15 min. Finally, the viral supernatant was collected and concentrated by ultracentrifugation at 140,000 ×g, 4 °C for 2 h. After ultracentrifugation, the virus precipitate was collected and resuspended with PBS, aliquoted and stored at -80 °C.
For viral titration, Sf9 cell monolayers were prepared in a 96-well culture plate at a seeding density of 4×10
4 cells per well at 16 h ahead of viral infection. Then the tested stock solutions of the mixed viruses were serially diluted tenfold ranging from 10 to 10
7 in serum- and antibiotic-free SIM medium. And then the old medium in each well was removed and 100 μL viral dilutions were added into each well. Five hours post-infection, the old medium was replaced with normal SIM medium. At 120 h post infection, the expression of GUS reporter gene was detected by X-gluc staining as described previously by Wu et al [
9]. The virus titers were calculated by the formula: TU/mL (transduction units per mL) = percentage of positive cell colonies × total cell numbers seeded / volume of viral stock (mL).
In consideration of the failure of PH promoter to drive the expression of GUS gene in the case of single viral vector transfection, the titration of the single virus of pUC19-IHHNV-PH-GUS was determined by the simultaneous co-transfection and titration of the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid. The purification, concentration and titration of the single baculoviruses of Bacmid-GUS and Bacmid were carried out as described previously by Wu et al [
9].
4.7. Verification of the Packaging Capacity of IHHNV-BASED expression Vector of pUC19-IHHNV-PH-GUS into Infective Virions in Sf9 Cells
The successful packaging of the shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS in Sf9 cells was verified by three methods of re-infection, transmission electron microscopy (TEM) and electron microscopy negative staining assay in this study.
For re-infection assay, the viral expression plasmid of pUC19-IHHNV-PH-GUS was transfected alone or co-transfected with Bacmid (or Bacmid-VP28) into the Sf9 cells as described previously under the optimized transfection conditions, at 120 h post transfection the medium supernatant was collected and clarified by centrifugation as described previously and then used to reinfect the Sf9 cells cultured in serum- and antibiotic- free SIM medium by an infection dose of 100 μL medium supernatant tested per well in a 48-well culture plate. The next day the old medium was replaced with normal SIM medium. At 120 h post infection, the expression of GUS gene was detected by X-gluc staining. The successful detection of GUS signal can verify the successful packaging of the mature virions of pUC19-IHHNV-PH-GUS in Sf9 cells.
For TEM and electron microscopy negative staining assay, Sf9 cells were inoculated into a 10-cm petri dish at a density of 6 × 106 cells per dish at 16 h ahead of transfection, and the old medium was replaced with 10 mL of diluted SIM medium containing 1.35 mL SIM, 0.15 mL FBS and 8.5 mL Grace medium when the confluency reached 70 %. Then, the plasmids of pUC19-IHHNV-PH-GUS and Bacmid were transfected individually or co-transfected into Sf9 cells using transfection reagent of Cellfectin II. In brief, in one tube 15 μg pUC19-IHHNV-PH-GUS plasmid, or 15 μg Bacmid, or 30 μg mixture of the above two plasmids, and in another tube 90 or 180 μL Cellfectin II, were diluted in 500 μL serum- and antibiotic-free Grace medium, respectively, and incubated at room temperature for 10 min. Then, the two tubes containing DNA or Cellfectin II were combined accordingly, mixed and then incubated at room temperature for another 20-30 min. The transfection complex was added drop by drop into the culture dish and then replaced to normal SIM medium at 4 h post transfection. At 120 h post transfection, the medium supernatant was collected and clarified by centrifugation and used to reinfect the Sf9 cells, which were freshly seeded in another 10-cm petri dish and the medium has been replaced with serum- and antibiotic- free SIM medium, with an infection dose of 400 μL medium supernatant per dish. At 120 h post infection, the medium supernatant of the pUC19-IHHNV-PH-GUS-transfected Sf9 cells was collected, clarified and saved at 4 °C for electron microscopy negative staining analysis by Savile Biotechnology (Shanghai, China). The next, two mL of 2.5 % glutaraldehyde was added into each dish after the medium has been removed, and the fixed Sf9 cells were removed by a cell scraper and collected by centrifugation. The supernatant was discarded and the cell pellet was resuspended in a new fixative solution carefully and incubated at room temperature for another 30 min and then stored at 4 °C until TEM assay and photographing by Saville Biotechnology (Shanghai, China).
4.8. Analysis of Infection and GUS Expression of Aforementioned Mixed Viruses in Primarily Cultured Shrimp Hemolymph Cells
The primarily cultured shrimp hemolymph cells in 96-well culture plate were infected with 100 μL viral solution per well containing 8 × 106 TU viruses of Bacmid (negative control) or pUC19-IHHNV-PH-GUS, or 8 × 106 TU mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid, or 8 × 105 TU mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28, all diluted in 1.5 × L-15 basic medium. At 4 h post infection, the virus-containing medium was replaced with 1.5 × L-15-based growth medium or gelatin-containing 1 × L-15-based growth medium and cultured at 28 °C for another 120 h. Then the expression of GUS gene was detected by X-gluc staining and observed under inverted phase contrast microscope (Nikon, Japan).
4.9. Analysis of Infection and GUS Expression of Aforementioned Mixed Viruses in the Adult Tissues of Shrimps
For the infection and GUS expression of mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid, firstly, the mixed-virus stock solution was serially diluted by 10, 10
2 and 10
3 folds in PBS, respectively, and then injected into the second abdominal segment of the shrimps by three different doses of 7 × 10
4, 7 × 10
5 and 7 × 10
6 TU per shrimp in an injection volume of 10 μL. The control shrimps were injected by the same volume of PBS instead. After injection, the injection site was immediately pressed with thumb for several seconds to prevent viruses and hemolymph from flowing out. After that, the injected shrimp was put back into the seawater and cultured for another 5 days. On the fifth day, various adult tissues of heart, gill, Oka organ, muscle and intestine of the injected shrimp were dissected out and the expression of GUS gene was detected by X-gluc staining as described previously by Wu et al [
9], respectively. In detail, the tested tissue was placed in 1.5-mL centrifuge tube after washed three times in 2 × PBS, and then incubated in the X-Gluc staining solution containing 1 M sodium phosphate (pH 7.0), 0.5 M Na
2EDTA (pH 8.0), 10% Triton X-100, 50 mM K
3Fe (CN)6 and 0.1 M 50 mg/ml X-Gluc overnight in a 28 °C incubator, and then the expression of GUS gene was detected by the intensity of blue color. After that, the stained tissues were further photographed under an inverted phase contrast microscope (Nikon).
For the infection and expression of mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28, only one infection dose of 4 × 106 TU per shrimp in the same injection volume of 10 μL was tested and the expression of GUS gene in the different tissues of the infected shrimps was detected by X-gluc staining as described previously and further confirmed by semi-quantitative RT-PCR analysis using GUS gene- specific primer pairs. For semi-quantitative RT-PCR analysis, first, the total RNAs of the tested adult tissues of hearts, gills, Oka organs, muscles and intestines of the infected and uninfected shrimps were isolated using TransZol Up Plus RNA Kit (TransGen Biotech, China) and then reversely transcribed into cDNAs using PrimeScript™ RT reagent Kit (TaKaRa), respectively, and used as the RT-PCR templates. Then, a 499 bp cDNA fragment of GUS gene was amplified using a forward primer of 5’-GCGTTACAAGAAAGCCGGGC-3’ and reverse primer of 5’-AGTCAACAGACGCGTGGTTA-3’. RT-PCR was performed in a 20-μL reaction volume including 10 μL of 2× Hieff PCR Master Mix, 1 μL forward primer (10 μM), 1 μL reverse primer (10 μM), 1 μL cDNA template and 7 μL H2O. RT-PCR reaction was run for 94 ◦C for 5 min, 30 cycles of 94 ◦C for 30 s, 58 ◦C for 30 s and 72 ◦C for 45 s, One cycle of 72 ◦C for 10 min. A 438 bp β-actin fragment was amplified using a forward primer of 5′-CCCAGAGCAAGCGAGGTA-3′ and reverse primer of 5′-CGGTGGTCGTGAAGGTGT-3′ and used as internal reference.
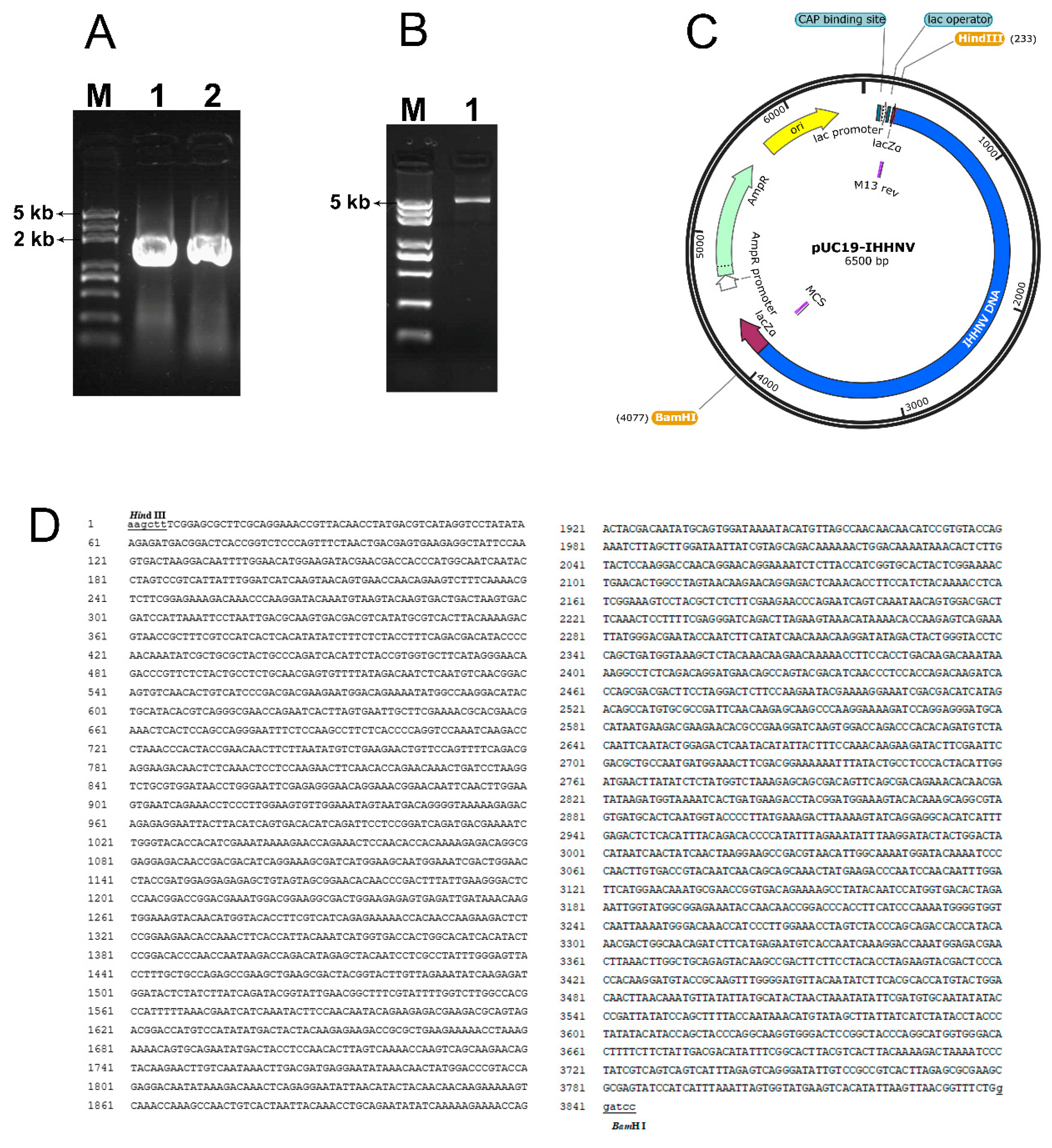
Figure 1.
Plasmid construction of pUC19-IHHNV by cloning and cyclizing the genomic DNA of the shrimp virus of IHHNV via a head-tail linkage with a prokaryotic cloning plasmid of pUC19. (A), the agarose gel electrophoresis result of the two genomic fragments amplified from the anterior and posterior part of the genomic DNA of shrimp IHHNV, respectively. Lane 1, the anterior fragment of 1 - 1910 bp. Lane 2, the posterior fragment of 1870 - 3833 bp. Lane M, DNA Marker. (B), the agarose gel electrophoresis result of the cyclized plasmid of pUC19-IHHNV. Lane 3, pUC19-IHHNV. (C), the cyclized plasmid map of pUC19-IHHNV (6.5 kb). (D), the sequencing result of the cyclized plasmid of pUC19-IHHNV. The restriction endonuclease sites are underlined.
Figure 1.
Plasmid construction of pUC19-IHHNV by cloning and cyclizing the genomic DNA of the shrimp virus of IHHNV via a head-tail linkage with a prokaryotic cloning plasmid of pUC19. (A), the agarose gel electrophoresis result of the two genomic fragments amplified from the anterior and posterior part of the genomic DNA of shrimp IHHNV, respectively. Lane 1, the anterior fragment of 1 - 1910 bp. Lane 2, the posterior fragment of 1870 - 3833 bp. Lane M, DNA Marker. (B), the agarose gel electrophoresis result of the cyclized plasmid of pUC19-IHHNV. Lane 3, pUC19-IHHNV. (C), the cyclized plasmid map of pUC19-IHHNV (6.5 kb). (D), the sequencing result of the cyclized plasmid of pUC19-IHHNV. The restriction endonuclease sites are underlined.
Figure 2.
Construction of the viral expression vector of pUC19-IHHNV-PH-GUS by inserting an exogenous concatemer of PH-GUS-MCS-SV40 pA into the cyclized plasmid of pUC19-IHHNV. (A), the agarose gel electrophoresis result of the PCR product of PPH-GUS-MCS-SV40 pA carrying a 30 bp pUC19-IHHNV-sourced homology arm in both ends. Lane1, PH-GUS-MCS-SV40 pA. Lane M, DNA Marker. (B), the agarose gel electrophoresis result of the viral expression plasmid of pUC19-IHHNV-PH-GUS. Lane 2, pUC19-IHHNV-PH-GUS (8.9 kb). (C), the plasmid map of pUC19-IHHNV-PH-GUS. (D), the sequencing result of the expression plasmid of pUC19-IHHNV-PH-GUS. The restriction endonuclease sites of BamH I and Sac I are lowercase and underlined. The sequence of PH promoter is shaded in gray color. The sequence of GUS gene is boxed in a larger size, and the start and stop codons are boxed in a smaller size. The terminator of SV40 pA is underlined in dotted line and the poly(A) signal is double underlined in dotted line. The multiple cloning site (MCS) is indicated in vertical lines.
Figure 2.
Construction of the viral expression vector of pUC19-IHHNV-PH-GUS by inserting an exogenous concatemer of PH-GUS-MCS-SV40 pA into the cyclized plasmid of pUC19-IHHNV. (A), the agarose gel electrophoresis result of the PCR product of PPH-GUS-MCS-SV40 pA carrying a 30 bp pUC19-IHHNV-sourced homology arm in both ends. Lane1, PH-GUS-MCS-SV40 pA. Lane M, DNA Marker. (B), the agarose gel electrophoresis result of the viral expression plasmid of pUC19-IHHNV-PH-GUS. Lane 2, pUC19-IHHNV-PH-GUS (8.9 kb). (C), the plasmid map of pUC19-IHHNV-PH-GUS. (D), the sequencing result of the expression plasmid of pUC19-IHHNV-PH-GUS. The restriction endonuclease sites of BamH I and Sac I are lowercase and underlined. The sequence of PH promoter is shaded in gray color. The sequence of GUS gene is boxed in a larger size, and the start and stop codons are boxed in a smaller size. The terminator of SV40 pA is underlined in dotted line and the poly(A) signal is double underlined in dotted line. The multiple cloning site (MCS) is indicated in vertical lines.

Figure 3.
GUS expression of shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS in Sf9 cells was dependent on the presence of insect baculovirus of Bacmid or Bacmid-VP28. The expression of GUS gene was detected by X-gluc staining at 120 h post transfection. (A1), (A2), (A3) and (A4), the un-transfected Sf9 cells (control). (B1), the Sf9 cells transfected by pUC19-IHHNV-PH-GUS. (C1), the Sf9 cells transfected by Bacmid. (B2), (C2), (D2), (E2) and (F2) are the Sf9 cells co-transfected by pUC19-IHHNV-PH-GUS and Bacmid at varied co-transfection ratio of 1:1/3 (B2), 1:1/2 (C2), 1:1 (D2), 1:2 (E2) and 1:3 (F2) but at the same transfection reagent ratio of 5 μL Cellfectin II per μg DNA. Panel B3, C3 and D3 are the Sf9 cells co-transfected by pUC19-IHHNV-PH-GUS and Bacmid at varied transfection reagent ratio of 4 (B3), 5 (C3) and 6 (D3) μL Cellfectin II per μg DNA but at the same co-transfection ratio of 1:1. (B4), the Sf9 cells co-transfected by pUC19-IHHNV-PH-GUS and Bacmid-VP28 at the optimized co-transfection ratio of 1:1 and transfection reagent ratio of 6 μL Cellfectin II per μg DNA. Scale bar, 100 μm.
Figure 3.
GUS expression of shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS in Sf9 cells was dependent on the presence of insect baculovirus of Bacmid or Bacmid-VP28. The expression of GUS gene was detected by X-gluc staining at 120 h post transfection. (A1), (A2), (A3) and (A4), the un-transfected Sf9 cells (control). (B1), the Sf9 cells transfected by pUC19-IHHNV-PH-GUS. (C1), the Sf9 cells transfected by Bacmid. (B2), (C2), (D2), (E2) and (F2) are the Sf9 cells co-transfected by pUC19-IHHNV-PH-GUS and Bacmid at varied co-transfection ratio of 1:1/3 (B2), 1:1/2 (C2), 1:1 (D2), 1:2 (E2) and 1:3 (F2) but at the same transfection reagent ratio of 5 μL Cellfectin II per μg DNA. Panel B3, C3 and D3 are the Sf9 cells co-transfected by pUC19-IHHNV-PH-GUS and Bacmid at varied transfection reagent ratio of 4 (B3), 5 (C3) and 6 (D3) μL Cellfectin II per μg DNA but at the same co-transfection ratio of 1:1. (B4), the Sf9 cells co-transfected by pUC19-IHHNV-PH-GUS and Bacmid-VP28 at the optimized co-transfection ratio of 1:1 and transfection reagent ratio of 6 μL Cellfectin II per μg DNA. Scale bar, 100 μm.
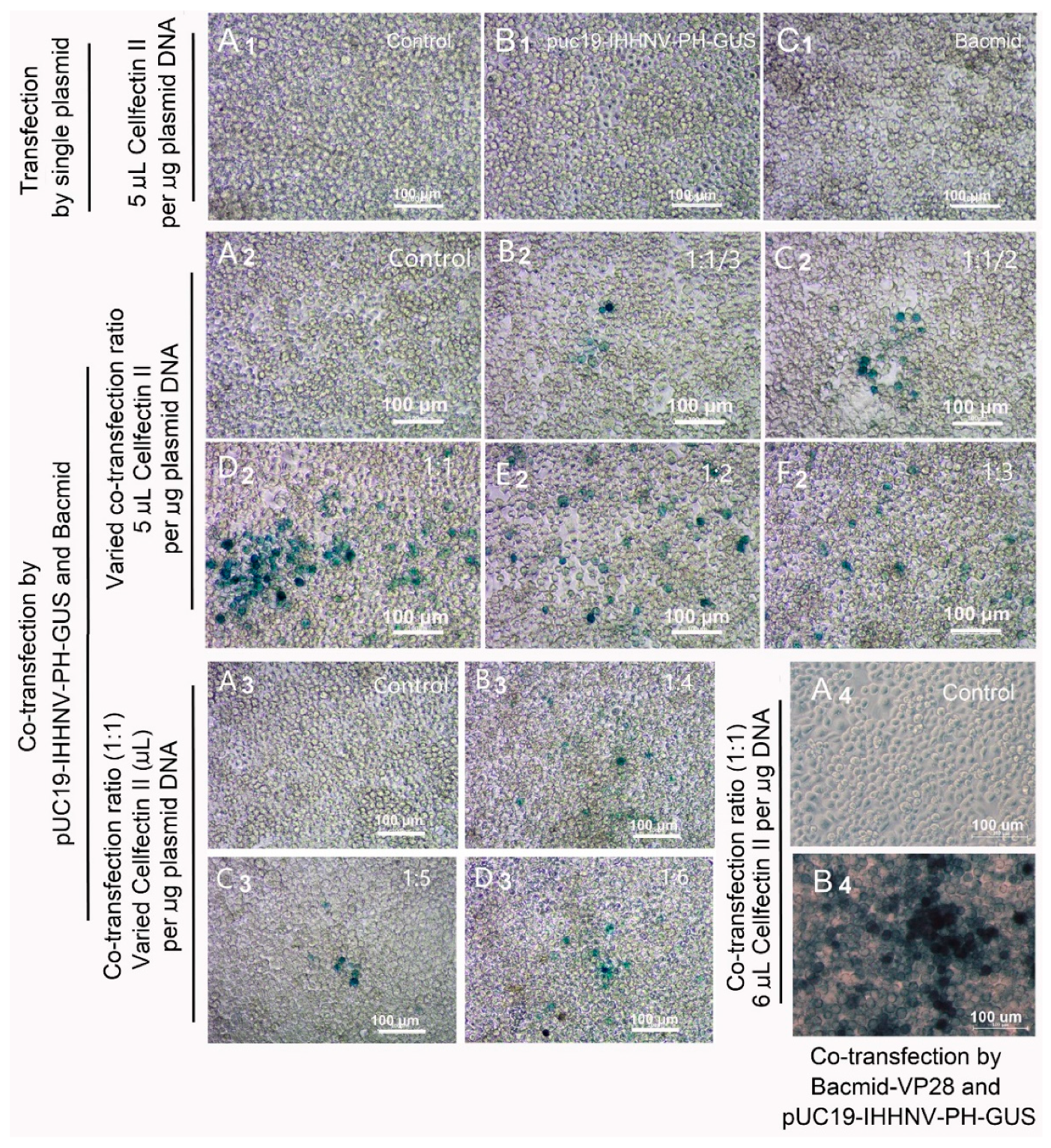
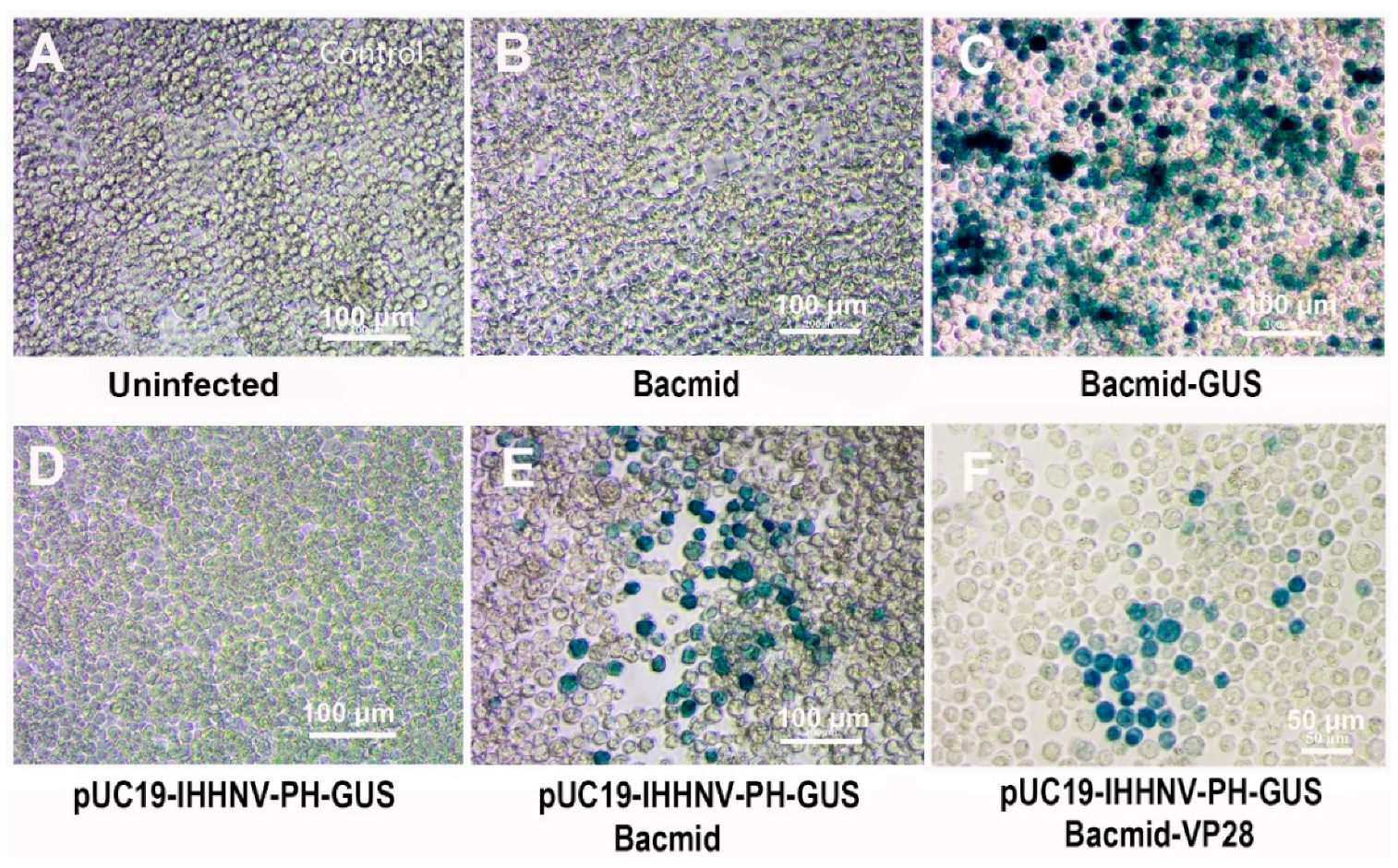
Figure 4.
Verification of the packaging capacity of shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS in Sf9 cells by reinfection assay. All the infected Sf9 cells were stained by X-gluc at 120 h post infection. (A), control Sf9 cells treated by the medium supernatant of un-transfected Sf9 cells. (B), Sf9 cells infected by the medium supernatant of Bacmid-transfected Sf9 cells. (C), Sf9 cells infected by the medium supernatant of Bacmid-GUS-transfected Sf9 cells. (D), Sf9 cells infected by the medium supernatant of pUC19-IHHNV-PH-GUS-transfected Sf9 cells. (E), Sf9 cells infected by the medium supernatant of co-transfected Sf9 cells by pUC19-IHHNV-PH-GUS and Bacmid. (F), Sf9 cells infected by the medium supernatant of co-transfected Sf9 cells by pUC19-IHHNV-PH-GUS and Bacmid-GUS. Scale bar, 100 μm in panels A-E and 50 μm in panel F.
Figure 4.
Verification of the packaging capacity of shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS in Sf9 cells by reinfection assay. All the infected Sf9 cells were stained by X-gluc at 120 h post infection. (A), control Sf9 cells treated by the medium supernatant of un-transfected Sf9 cells. (B), Sf9 cells infected by the medium supernatant of Bacmid-transfected Sf9 cells. (C), Sf9 cells infected by the medium supernatant of Bacmid-GUS-transfected Sf9 cells. (D), Sf9 cells infected by the medium supernatant of pUC19-IHHNV-PH-GUS-transfected Sf9 cells. (E), Sf9 cells infected by the medium supernatant of co-transfected Sf9 cells by pUC19-IHHNV-PH-GUS and Bacmid. (F), Sf9 cells infected by the medium supernatant of co-transfected Sf9 cells by pUC19-IHHNV-PH-GUS and Bacmid-GUS. Scale bar, 100 μm in panels A-E and 50 μm in panel F.
Figure 5.
Verification of the packaging capacity of shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS into infective virions in Sf9 cells by electron microscopy assay. Panels A-D were the results of transmission electron microscopy assay. Panel E was the result of electron microscopy negative staining assay. (A), un-infected Sf9 cells without any virions detected. (B), Sf9 cells reinfected by the medium supernatant of pUC19-IHHNV-PH-GUS-transfected Sf9 cells, only IHHNV-like virions could be observed. (C), Sf9 cells reinfected by the medium supernatant of Bacmid-transfected Sf9 cells, only Bacmid virions could be found. (D), Sf9 cells reinfected by the medium supernatant of the co-transfected Sf9 cells by pUC19-IHHNV-PH-GUS and Bacmid, and both IHHNV-like and Bacmid virions could be detected. (E), the packaged virions of pUC19-IHHNV-PH-GUS detected from the medium supernatant of pUC19-IHHNV-PH-GUS-transfected Sf9 cells. The insets in panels B-D are enlargement of the blue square box in dotted line. Scale bars were 2 μm in Panels A-D and 100 nm in Panel E.
Figure 5.
Verification of the packaging capacity of shrimp IHHNV-based expression vector of pUC19-IHHNV-PH-GUS into infective virions in Sf9 cells by electron microscopy assay. Panels A-D were the results of transmission electron microscopy assay. Panel E was the result of electron microscopy negative staining assay. (A), un-infected Sf9 cells without any virions detected. (B), Sf9 cells reinfected by the medium supernatant of pUC19-IHHNV-PH-GUS-transfected Sf9 cells, only IHHNV-like virions could be observed. (C), Sf9 cells reinfected by the medium supernatant of Bacmid-transfected Sf9 cells, only Bacmid virions could be found. (D), Sf9 cells reinfected by the medium supernatant of the co-transfected Sf9 cells by pUC19-IHHNV-PH-GUS and Bacmid, and both IHHNV-like and Bacmid virions could be detected. (E), the packaged virions of pUC19-IHHNV-PH-GUS detected from the medium supernatant of pUC19-IHHNV-PH-GUS-transfected Sf9 cells. The insets in panels B-D are enlargement of the blue square box in dotted line. Scale bars were 2 μm in Panels A-D and 100 nm in Panel E.

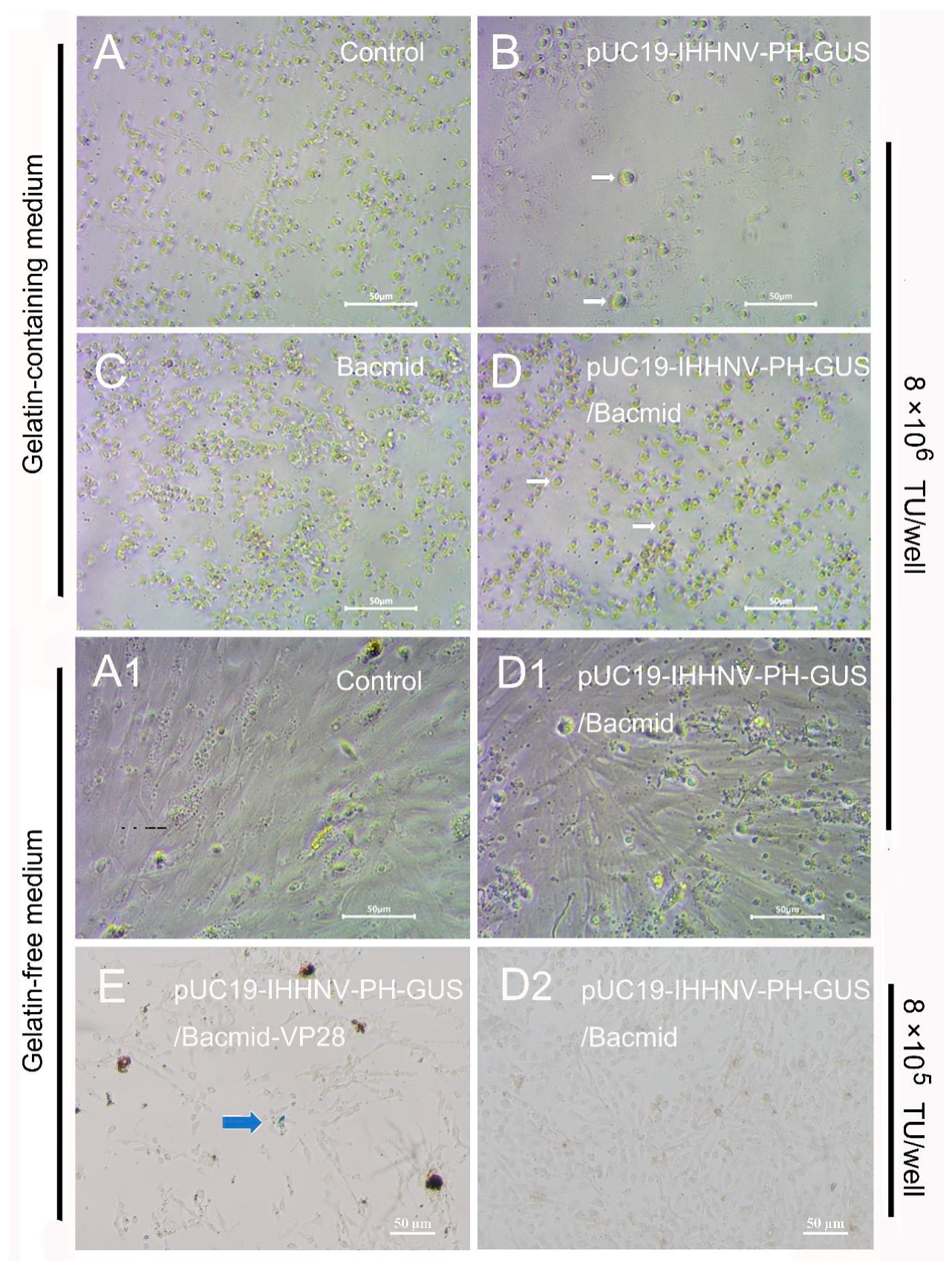
Figure 6.
Infection and GUS expression of the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid (or Bacmid-VP-28) in the primarily cultured shrimp hemolymph cells cultured in gelatin-containing 1× L-15-based medium or gelatin-free 1.5× L-15-based medium. GUS expression was detected by X-gluc staining at the 5th days post infection. (A) and (A1), uninfected cells. (B), cells infected by IHHNV-like viruses of pUC19-IHHNV-PH-GUS. (C), cells infected by baculovirus of Bacmid. (D), (D1) and (D2), cells infected by varied doses of mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid but cultured in gelatin-containing (D) or gelatin-free (D1 and D2) medium. (E), cells infected by mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28 and cultured in gelatin-free medium. The white arrows showed the cell hypertrophy. The blue arrow indicated the positive cells with GUS expression. Scale bar, 50 μm.
Figure 6.
Infection and GUS expression of the mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid (or Bacmid-VP-28) in the primarily cultured shrimp hemolymph cells cultured in gelatin-containing 1× L-15-based medium or gelatin-free 1.5× L-15-based medium. GUS expression was detected by X-gluc staining at the 5th days post infection. (A) and (A1), uninfected cells. (B), cells infected by IHHNV-like viruses of pUC19-IHHNV-PH-GUS. (C), cells infected by baculovirus of Bacmid. (D), (D1) and (D2), cells infected by varied doses of mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid but cultured in gelatin-containing (D) or gelatin-free (D1 and D2) medium. (E), cells infected by mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28 and cultured in gelatin-free medium. The white arrows showed the cell hypertrophy. The blue arrow indicated the positive cells with GUS expression. Scale bar, 50 μm.
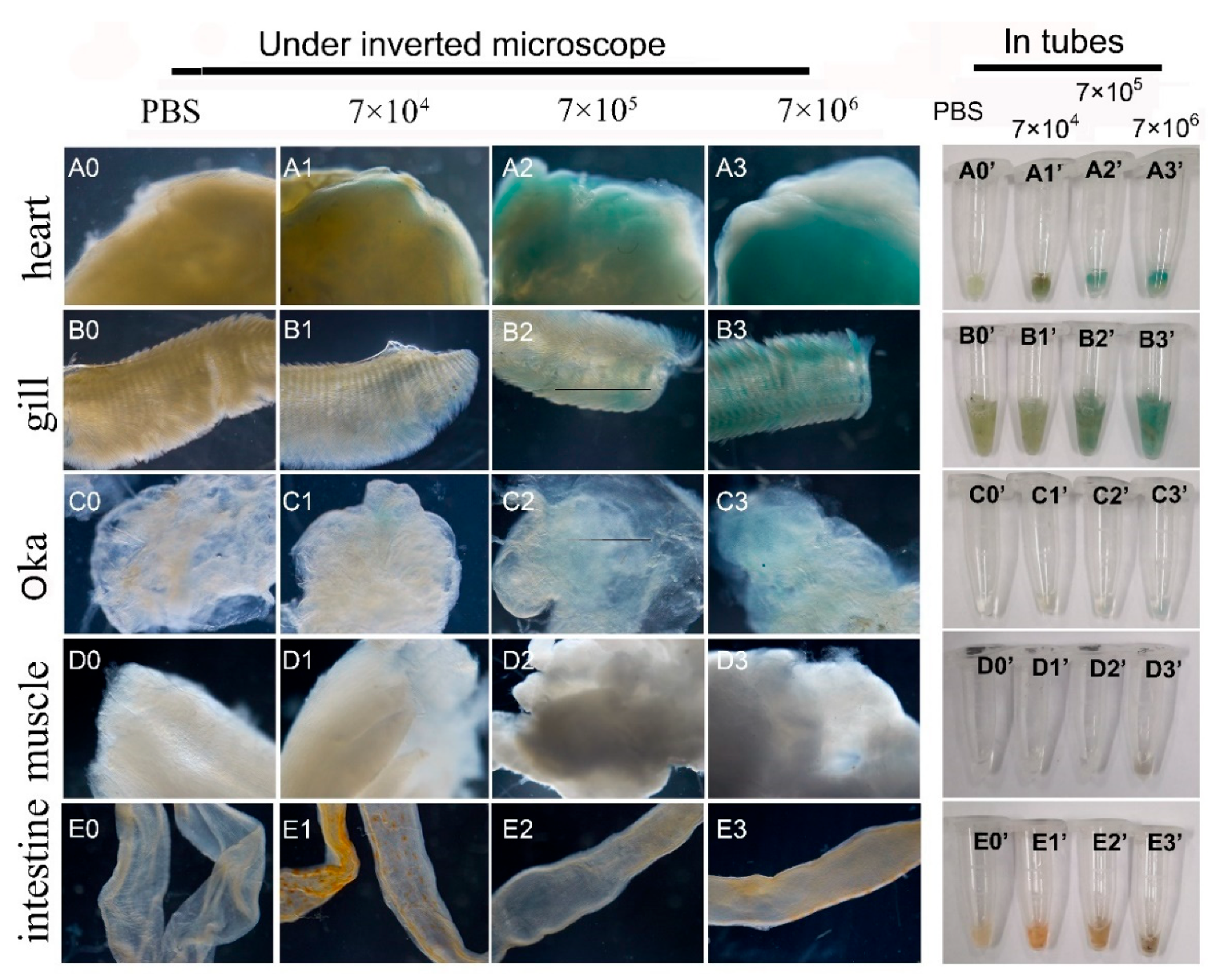
Figure 7.
Infection and GUS expression analysis in different tissues of adult shrimps infected by varied doses of mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid. The sampled adult tissues of the infected shrimps were first stained by X-gluc in a 1.5-mL centrifuge tube (right) and then observed under an inverted phase contrast microscope (left). (A0) - (E0) and (A0’) - (E0’), results of hearts, gills, Oka organs, muscles and intestines of the shrimps after injection of PBS, respectively. (A1) - (E1) and (A1’) - (E1’), results of the tested five tissues of the shrimps after injection of 7×104 TU mixed viruses, respectively. (A2) – (E2) and (A2’) – (E2’), results of tested five tissues of the shrimps after injection of 7×105 TU mixed viruses. (A3) – (E3) and (A3) – (E3), results of tested five tissues of the shrimp after injection of 7×106 TU mixed viruses.
Figure 7.
Infection and GUS expression analysis in different tissues of adult shrimps infected by varied doses of mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid. The sampled adult tissues of the infected shrimps were first stained by X-gluc in a 1.5-mL centrifuge tube (right) and then observed under an inverted phase contrast microscope (left). (A0) - (E0) and (A0’) - (E0’), results of hearts, gills, Oka organs, muscles and intestines of the shrimps after injection of PBS, respectively. (A1) - (E1) and (A1’) - (E1’), results of the tested five tissues of the shrimps after injection of 7×104 TU mixed viruses, respectively. (A2) – (E2) and (A2’) – (E2’), results of tested five tissues of the shrimps after injection of 7×105 TU mixed viruses. (A3) – (E3) and (A3) – (E3), results of tested five tissues of the shrimp after injection of 7×106 TU mixed viruses.
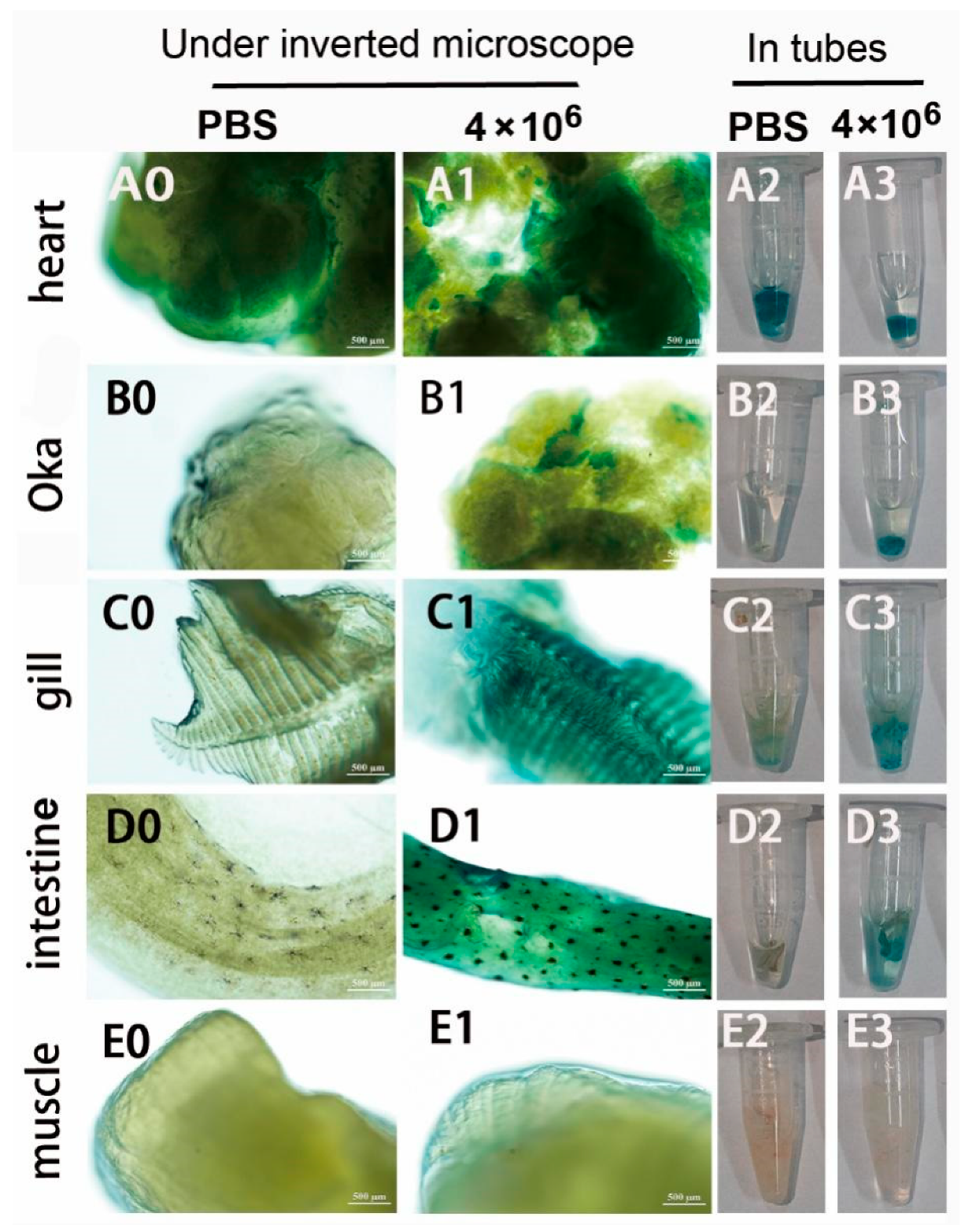
Figure 8.
Infection and GUS expression analysis in different tissues of adult shrimps infected by mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28. The sampled adult tissues of the infected shrimps were first stained by X-gluc in a 1.5-mL centrifuge tube (right) and then observed under an inverted phase contrast microscope (left). (A0) – (E0) and (A2) – (E2), results of hearts, Oka organs, gills, intestines and muscles of the shrimp after injection of PBS, respectively. (A1) – (E1) and (A3) – (E3), results of heart, Oka organ, gill, intestine and muscle of the shrimp after injection of 4×106 TU mixed viruses, respectively. Scale bar, 500 μm.
Figure 8.
Infection and GUS expression analysis in different tissues of adult shrimps infected by mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28. The sampled adult tissues of the infected shrimps were first stained by X-gluc in a 1.5-mL centrifuge tube (right) and then observed under an inverted phase contrast microscope (left). (A0) – (E0) and (A2) – (E2), results of hearts, Oka organs, gills, intestines and muscles of the shrimp after injection of PBS, respectively. (A1) – (E1) and (A3) – (E3), results of heart, Oka organ, gill, intestine and muscle of the shrimp after injection of 4×106 TU mixed viruses, respectively. Scale bar, 500 μm.
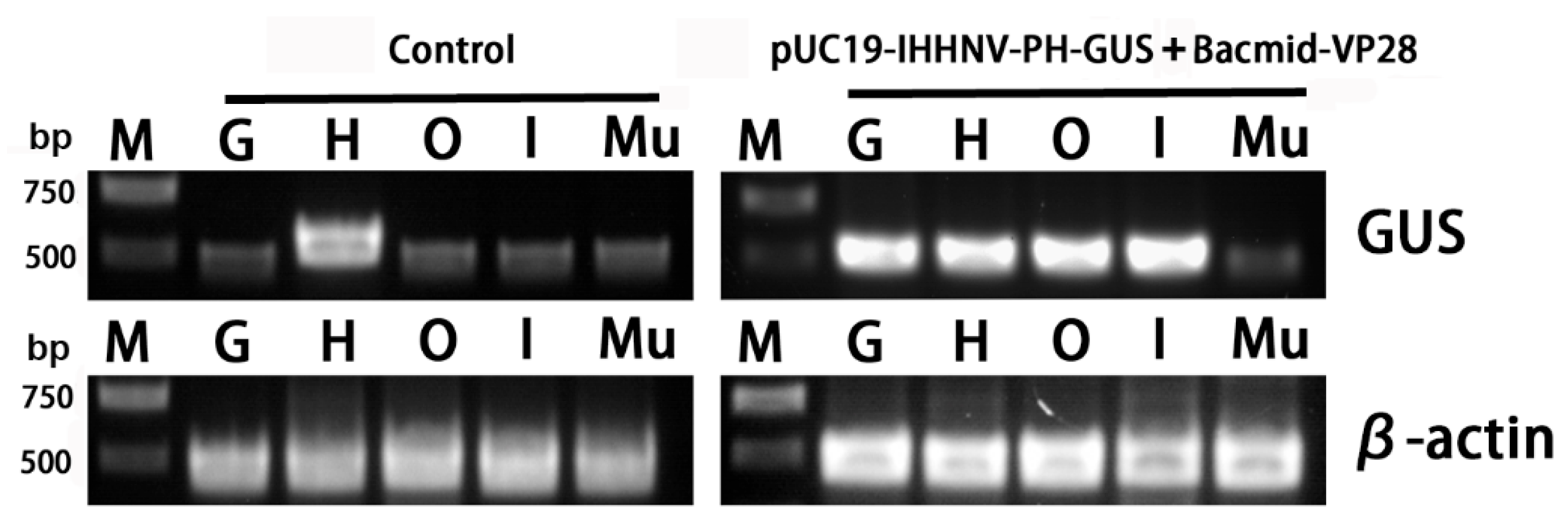
Figure 9.
Semi-quantitative RT-PCR analysis of the GUS gene expression in different tissues of the uninfected (control) and infected adult shrimps by mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28. M, Trans 2K plus DNA marker. G, gill. H, heart. O, Oka organ. I, intestine. Mu, muscle. GUS, β-glucuronidase gene. β-actin, internal reference gene.
Figure 9.
Semi-quantitative RT-PCR analysis of the GUS gene expression in different tissues of the uninfected (control) and infected adult shrimps by mixed viruses of pUC19-IHHNV-PH-GUS and Bacmid-VP28. M, Trans 2K plus DNA marker. G, gill. H, heart. O, Oka organ. I, intestine. Mu, muscle. GUS, β-glucuronidase gene. β-actin, internal reference gene.
Figure 10.
Schematic diagram of the components and technical process of shrimp IHHNV-mediated gene transfer and expression system.
Figure 10.
Schematic diagram of the components and technical process of shrimp IHHNV-mediated gene transfer and expression system.