Submitted:
25 June 2024
Posted:
26 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Site Descriptions and Sampling
2.2. Water Chemistry Analysis
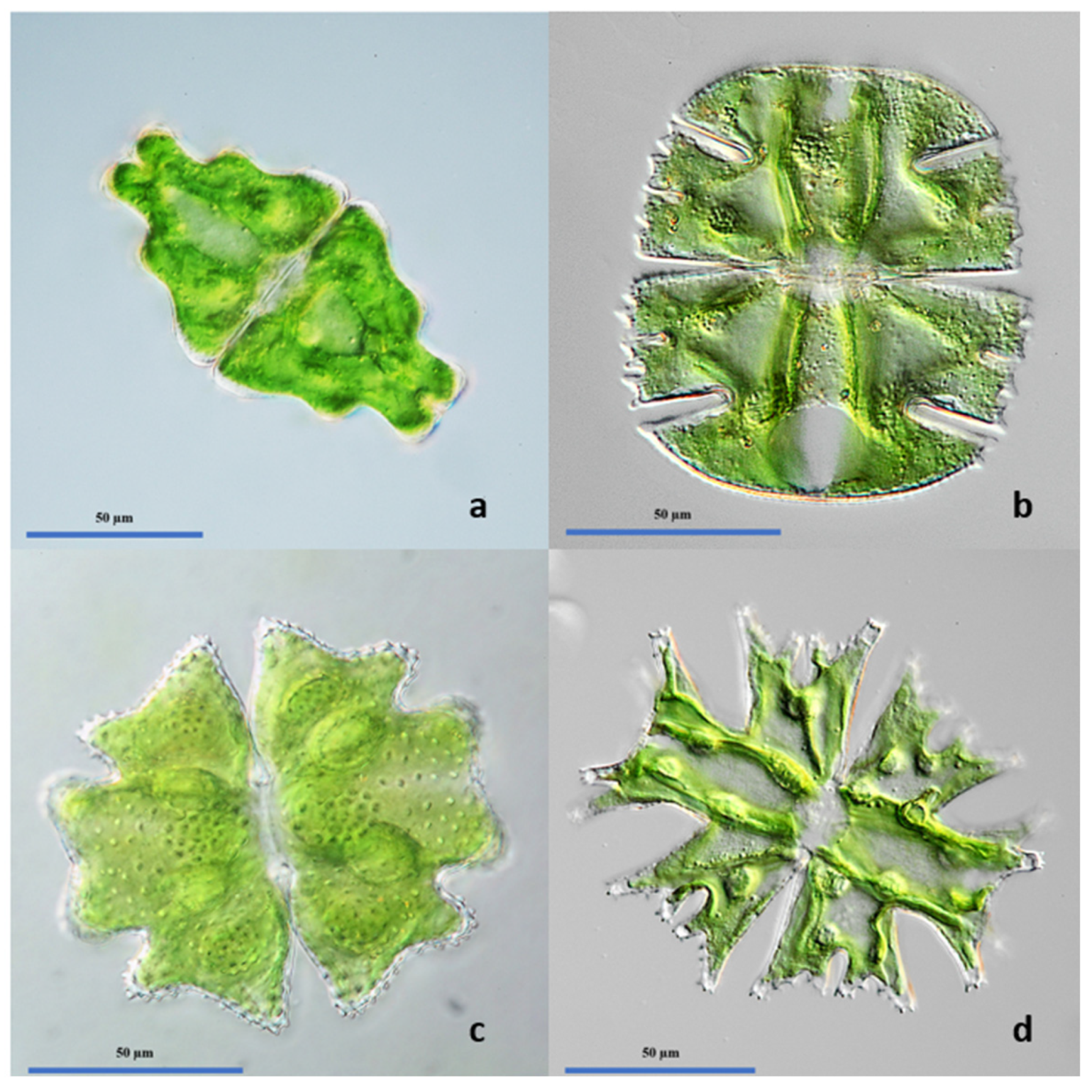
2.3. Light Microscopy and Species Identification
2.4. DNA Isolation and Metagenomic Sequencing
2.5. Bioinformatic and Statistical Analyses
3. Results
3.1. Water bodies in the Eifel National Park
3.2. Algal Biodiversity of the Investigated Ponds: Classical Approach
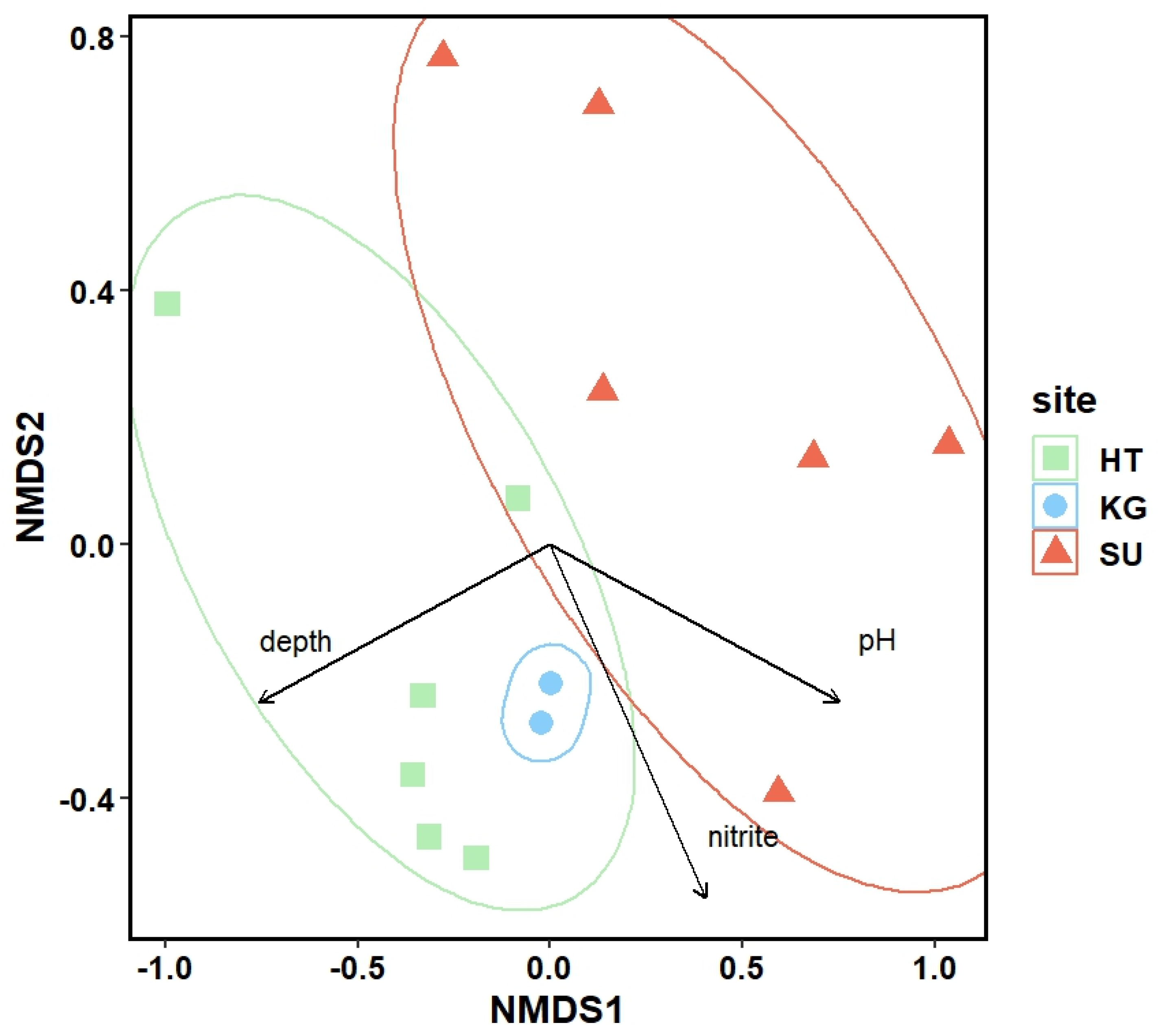
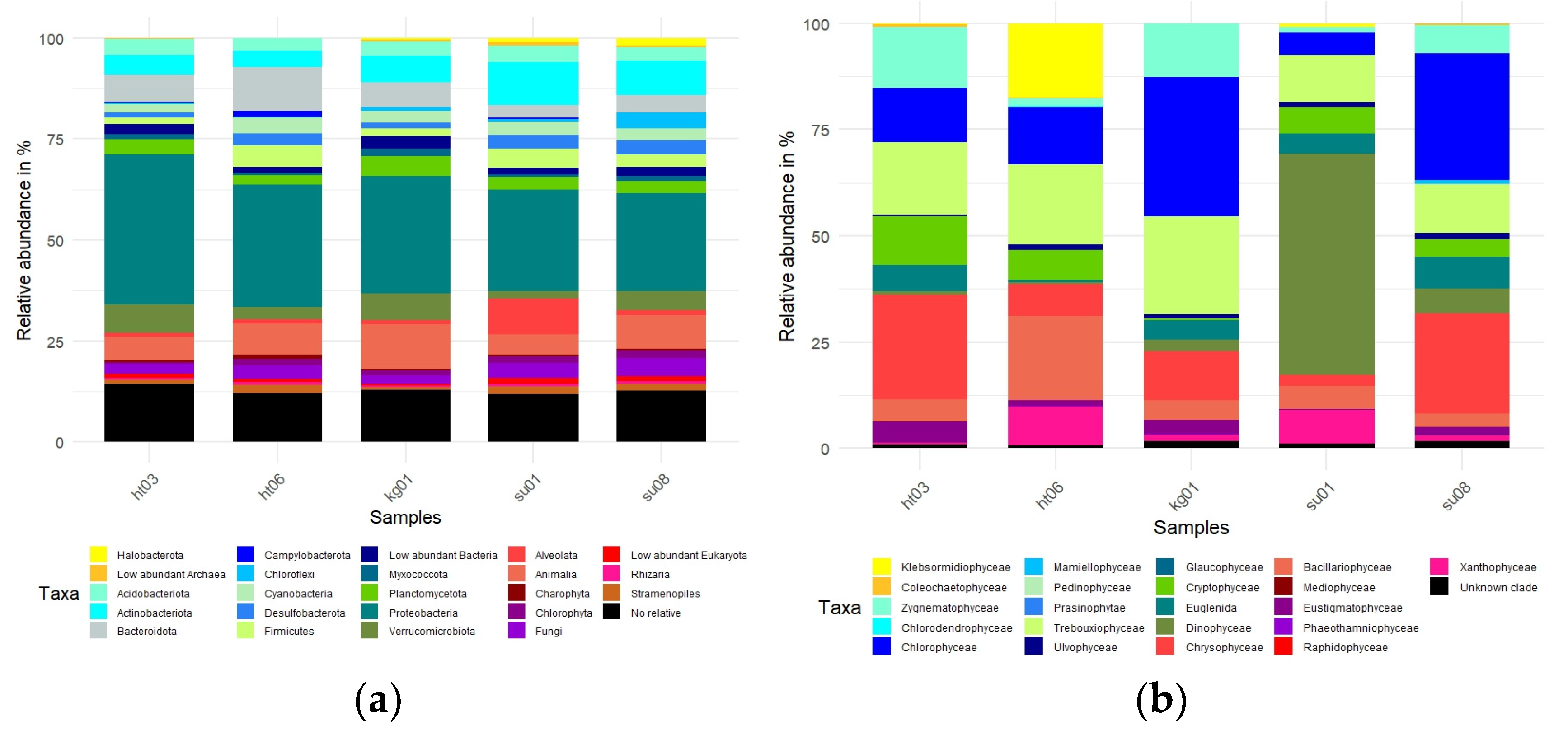
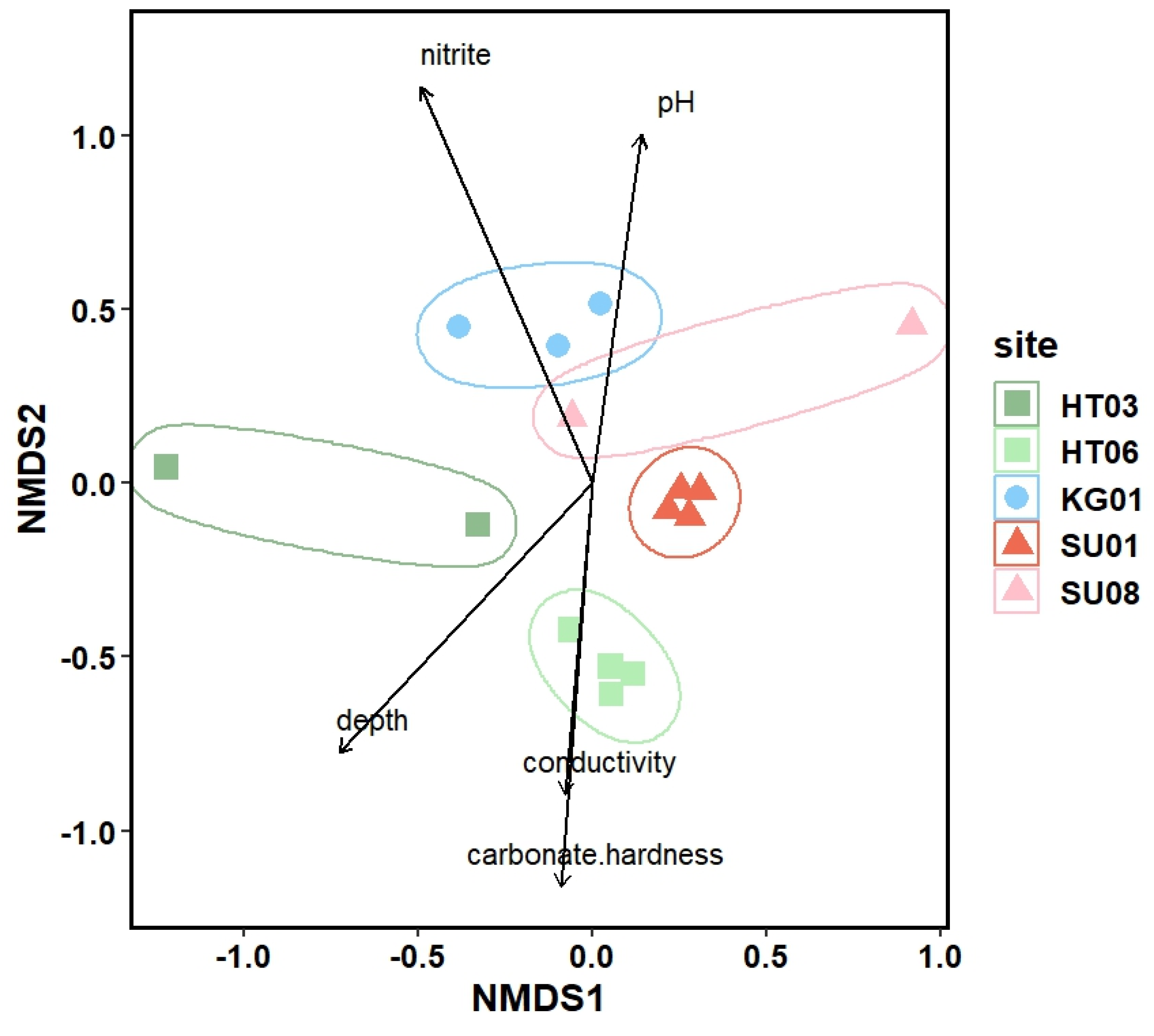
3.3. Microbial Diversity: Metagenomic Approach
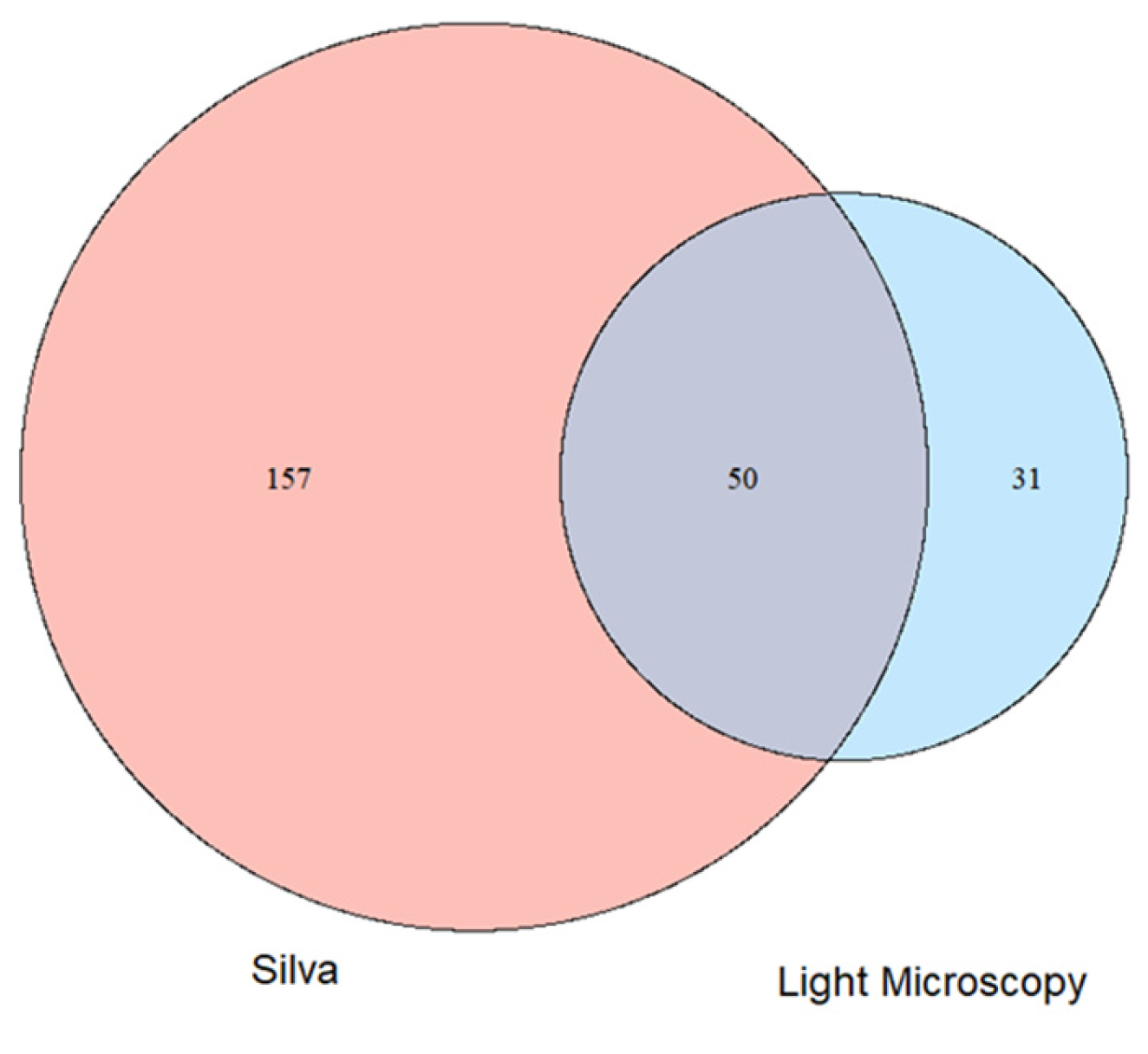
3.4. Comparison of the Alpha Biodiversity Found by Light Microscopic and Molecular Analysis of Photoautotrophs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Skinner CE (1932) Isolation in pure culture of green algae from soil by a simple technique. Plant Physiol 7:533–537. http s://doi.org/10.1104 /pp.7.3.533. [CrossRef]
- Bischoff H, Bold H (1963) Some soil algae from enchanted rock and related algal species. University of Texas Publications, Austin.
- Waterbury JB, Stanier RY (1978) Patterns of growth and development in pleurocapsalean Cyanobacteria. Microbiol Rev 42:2–44.
- Prescott GW (1964) How to know the fresh-water algae. Plenum Press, New York.
- Lind EM, Brook AJ (1980) A key to the commoner desmids of the English Lake District. Freshwater Biological Association.
- Cox EJ (1996) Identification of freshwater diatoms from live material. Chapman & Hall, London.
- John DM, Whitton BA, Brook AJ (2002) The freshwater algal flora of the British Isles: an identification guide to freshwater and terrestrial algae. Cambridge University Press, New York.
- Misawa S (1999) Rapid diagnosis of infectious diseases; features and limitations of the microscopic examination of clinical specimens. J Assoc Rapid Method Autom Microbiol 10:121–131.
- Manoylov KM (2014) Taxonomic identification of algae (Morphological and molecular): species concepts, methodologies, and their implications for ecological bioassessment. J Phycol 50:409–424. https://doi.org/10.1111 /jpy.1218 3. [CrossRef]
- Albrecht M, Pröschold T, Schumann R (2017) Identification of Cyanobacteria in a eutrophic coastal lagoon on the Southern Baltic coast. Front Microbiol 8:923. http s://doi.org/10.3389 /fmic b.2017.0092 3. [CrossRef]
- Vieira HH, Bagatini IL, Guinart CM, Vieira AAH (2016) tufA gene as molecular marker for freshwater Chlorophyceae. Algae 31:155–165. http s://doi.org/10.4490 /alga e.2016.31.4.14. [CrossRef]
- Buchheim MA, Chapman RL (1991) Phylogeny of the colonial green flagellates: a study of 18S and 26S rRNA sequence data. BioSystems 25:85–100. http s://doi.org/10.1016 /0303 -2647 (91)9001 5-D. [CrossRef]
- Wilmotte A (1994) Molecular evolution and taxonomy of the Cyanobacteria. In: Bryant D (ed) The molecular biology of cyanobacteria. Springer, Dordrecht, pp 1–25. 14. Doyle JJ, Doyle JL, Ballenger JA et al. (1997) A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am J Bot 84:541– 554. https://doi.org/10.2307 /2446 030. [CrossRef]
- Doyle JJ, Doyle JL, Ballenger JA et al. (1997) A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am J Bot 84:541– 554. https://doi.org/10.2307 /2446 030. [CrossRef]
- An SS, Friedel T, Hegewald E (1999) Phylogenetic relationships of Scenedesmus and Scenedesmus-like coccoid green algae as inferred from ITS-2 rDNA sequence comparison. Plant Biol 1:418–428. http s://doi.org/10.1111 /j.1438 -8677.1999.tb00 724.x. [CrossRef]
- Evans KM, Wortley AH, Mann DG (2007) An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 158:349–364. http s://doi.org/10.1016/j.prot is.2007.04.001:. [CrossRef]
- Sherwood AR, Vis ML, Entwisle TJ et al. (2008) Contrasting intra versus interspecies DNA sequence variation for representatives of the Batrachospermales (Rhodophyta): insights from a DNA barcoding approach. Phycol Res 56:269–279. http s://doi.org/10.1111 /j.1440 -1835.2008.0050 8.x. [CrossRef]
- Hall JD, Fucikova K, Lo C et al. (2010) An assessment of proposed DNA barcodes in freshwater green algae. Cryptogam Algol 31:529–555.
- Ward DM, Weller R, Bateson MM (1990) 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63–65. http s://doi.org/10.1038 /3461 83a0. [CrossRef]
- Schloss PD, Handelsman J (2005) Metagenomics for studying unculturable microorganisms: cutting the Gordian knot. Genome Biol 6:229. http s://doi.org/10.1186 /gb-2005 -6-8-229. [CrossRef]
- Shi XL, Marie D, Jardillier L et al. (2009) Groups without cultured representatives dominate eukaryotic picophytoplankton in the oligotrophic South East Pacific Ocean. PLoS ONE 4:e7657. https://doi.org/10.1371 /jour nal.pone.0007 657. [CrossRef]
- Massana R, del Campo J, Sieracki ME et al. (2014) Exploring the uncultured microeukaryote majority in the oceans: reevaluation of ribogroups within stramenopiles. ISME J 8:854–866. https://doi.org/10.1038 /isme j.2013.204. [CrossRef]
- Taberlet P, Coissac E, Pompanon F et al. (2012) Towards next-generation biodiversity assessment using DNA metabarcoding. Mol Ecol 21:2045–2050. http s://doi.org/10.1111 /j.1365 -294X.2012.0547 0.x. [CrossRef]
- Yoon T-H, Kang H-E, Kang C-K et al. (2016) Development of a costeffective metabarcoding strategy for analysis of the marine phytoplankton community. PeerJ 4:e2115. http s://doi.org/10.7717 /.
- Elferink S, Neuhaus S, Wohlrab S et al. (2017) Molecular diversity patterns among various phytoplankton size-fractions in West Greenland in late summer. Deep Res Part I Oceanogr Res Pap 121:54–69. http s://doi.org/10.1016 /j.dsr.2016.11.002peer j.2115. [CrossRef]
- Margulies M, Egholm M, Altman WE et al. (2005) Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–381. http s://doi.org/10.1038 /nature03 959. [CrossRef]
- Pawlowski J, Christen R, Lecroq B et al. (2011) Eukaryotic richness in the abyss: insights from pyrotag sequencing. PLoS ONE 6:e18169. http s://doi.org/10.1371 /jour nal.pone.0018 169:. [CrossRef]
- Urich T, Lanzén A, Qi J et al. (2008) Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE 3:e2527. http s://doi.org/10.1371 /jour nal.pone.0002 527. [CrossRef]
- Urich T, Lanzén A, Stokke R et al. (2014) Microbial community structure and functioning in marine sediments associated with diffuse hydrothermal venting assessed by integrated meta-omics. Environ Microbiol 16:2699–2710. http s://doi.org/10.1111 /1462-2920.1228 3. [CrossRef]
- Geisen S, Tveit AT, Clark IM et al. (2015) Metatranscriptomic census of active protists in soils. ISME J 9:2178–2190. http s://doi.org/10.1038 /isme j.2015. [CrossRef]
- Becker, B., & Pushkareva, E. (2023). Metagenomics provides a deeper assessment of the diversity of bacterial communities in polar soils than metabarcoding. Genes, 14(4), 812.
- Klimke W, O’Donovan C, White O et al. (2011) Solving the problem: genome annotation standards before the data deluge. Stand Genom Sci 5:168–193. http s://doi.org/10.4056 /sigs.2084 864. [CrossRef]
- Rippin, M., Lange, S., Sausen, N., Becker. B. (2018a) Biodiversity of biological soil crusts from the Polar Regions revealed by metabarcoding. FEMS Microbiology Ecology, Volume 94, Issue 4, fiy036, https://doi.org/10.1093/femsec/fiy036. [CrossRef]
- Rippin, M., Borchhardt, N., Williams, L., Colesie, C., Jung, P., Büdel, B., Karsten, U., Becker. B. (2018b) Genus richness of microalgae and Cyanobacteria in biological soil crusts from Svalbard and Livingston Island: morphological versus molecular approaches. Polar Biol 41: 909. https://doi.org/10.1007/s00300-018-2252-2. [CrossRef]
- Pushkareva, E., Elster, J., Holzinger, A., Niedzwiedz, S., & Becker, B. (2022). Biocrusts from Iceland and Svalbard: Does microbial community composition differ substantially?. Frontiers in Microbiology, 13, 1048522.
- Pushkareva, E., Elster, J., Kudoh, S., Imura, S., & Becker, B. (2024). Microbial community composition of terrestrial habitats in East Antarctica with a focus on microphototrophs. Frontiers in Microbiology, 14, 1323148.
- Pardey, A.; Twietmeyer, S. Artenvielfalt im Nationalpark Eifel.Natur in NRW 2018, 11-15.
- Nationalparkforstamt Eifel,Wald und Holz NRW. Leistungsbericht 2020.
- Coesel, Peter F.M., Meesters, Koos (J.): Desmids of the Lowlands, KNNV Publishing, the Netherlands, 2007.
- Hofmann, Gabriele; Werum, Marcus: Diatomeen im Süßwasser-Benthos von Mitteleuropa, Horst Lange Bertalot, A.R.G. Gantner Verlag K.G., Rugell, 2011.
- John, D.M.; Whitton, B.A.; Broo, A.J.: The freshwater Algal Flora of the British Isles, Cambridge University Press, United Kingdom, 2002.
- Lenzenweger, Rupert: Desmidiaceenflora von Österreich—Teil 1, Band 101, Gebrüder Borntraeger, Berlin, Stuttgart, 1996.
- Lenzenweger, Rupert: Desmidiaceenflora von Österreich—Teil 2, Band 102, Gebrüder Borntraeger, Berlin, Stuttgart, 1997.
- Lenzenweger, Rupert: Desmidiaceenflora von Österreich—Teil 3, Band 104, Gebrüder Borntraeger, Berlin, Stuttgart, 1997.
- Lenzenweger, Rupert: Desmidiaceenflora von Österreich—Teil 4, Band 111, Gebrüder Borntraeger, Berlin Stuttgart, 1996.
- Bolger, A.M., Lohse, M., and Usadel, B. (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120.
- Kopylova, E., Noé, L., and Touzet, H. (2012) SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217.
- Oksanen, J. (2013) Multivariate analysis of ecological communities in R:vegan tutorial. R documentation 43.
- The determination of ecological status in shallow lakes—a tested system (ECOFRAME) for implementation of the European Water Framework Directive. Aquatic Conservation: Marine and Freshwater Ecosystems, 13(6), 507-549.
- Moss, B., Stephen, D., Alvarez, C., Becares, E., Bund, W. V. D., Collings, S. E.,... & Wilson, D. (2003).
- Lindberg, K., Moestrup, Ø & Daugbjerg, N. (2005). Studies on woloszynskioid dinoflagellates I: Woloszynskia coronata re-examined using light and electron microscopy and partial LSU rDNA sequences, with description of Tovellia gen. nov. and Jadwigia gen. nov. (Tovelliaceae fam. nov.). Phycologia 44: 416-440.
- Moestrup, Ø. & Calado, A.J. (2018). Süßwasserflora von Mitteleuropa. Dinophyceae. Vol. 6 pp. [i]-xii, [1]-560, 421 figures. Berlin: Springer Spektrum.
- Paxinos, R.; Mitchell, J.G. A rapid Utermöhl method for estimating algal numbers. J.Plancton Res. 2000, 22, 2255–2262, https://doi.org/10.1093/plankt/22.12.2255. [CrossRef]
- Solden L, Lloyd K, Wrighton K (2016) The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr Opin Microbiol 31:217–226. http s://doi.org/10.1016 /j.mib.2016.04.020. [CrossRef]
| Site | Size (m²)1) | Depth (m) | Perennial | Soil composition | Vegetation cover (%)2) | Presence of Spaghnum |
|---|---|---|---|---|---|---|
| HT01 | 126 | 0.4 | Yes | Organic | 95 | Yes |
| HT02 | 314 | 0.5 | Yes | Organic | 30 | Yes |
| HT03 | 162 | 0.6 | Yes | Organic | 40 | Yes |
| HT04 | 195 | 0.4 | Yes | Organic | 50 | Yes |
| HT05 | 64 | 0.6 | Yes | Organic | 50 | Yes |
| HT06 | 24 | 0.6 | Yes | Organic | 100 | Yes |
| SU1 | 20 | 0.4 | No | Organic | 90 | Little |
| SU6 | 96 | 0.3 | No | Mineralic | 40 | No |
| SU7 | 64 | 0.3 | No | Mineralic | 30 | No |
| SU8 | 35 | 0.3 | No | Mineralic | 25 | No |
| SU9 | 40 | 0.5 | No | Organic | 100 | Little |
| SU10 | n.d. | 0.4 | No | Organic | 80 | No |
| KG1 | 84 | 0.5 | Yes | Organic | 90 | No |
| KG2 | 102 | 0.6 | Yes | Organic | 90 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





