Submitted:
22 June 2024
Posted:
24 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Tissue Collection
2.2. Cell Culture and Transfection
2.3. Pasmid Construction
2.4. Total RNA Was Extracted and Reverse-Transcribed
2.5. Real-Time Polymerase Chain Reaction
2.6. Western-Blot
2.7. Immunohistochemistry
2.8. Subcellular Localization
2.9. Cell Proliferation Analysis
2.10. Steroid Assay
2.11. Flow Cytometry Analysis
2.12. Statistics
3. Results
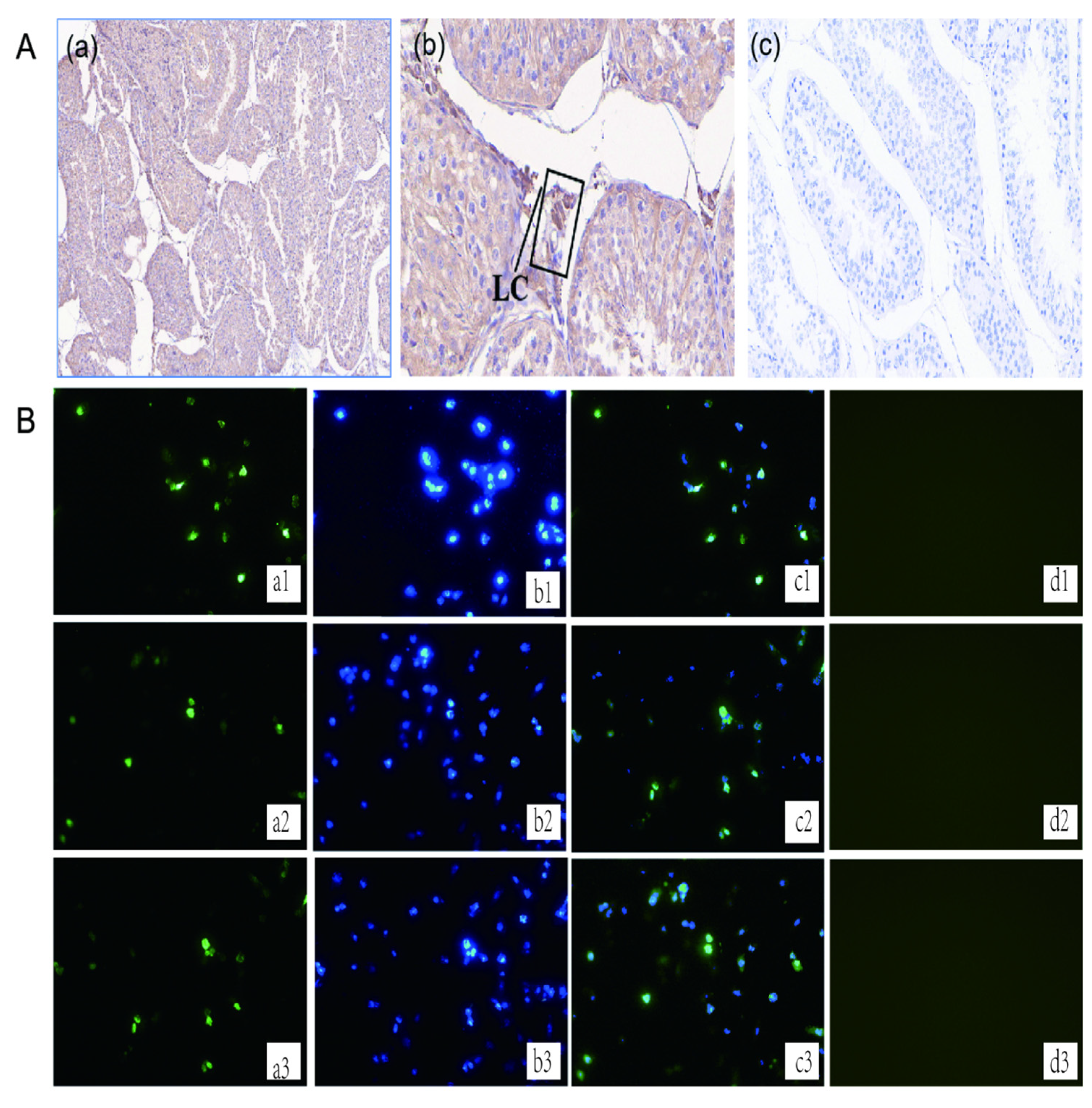
3.1. Localisation of GLOD4 in the Testis of Qianbei Ma Goats
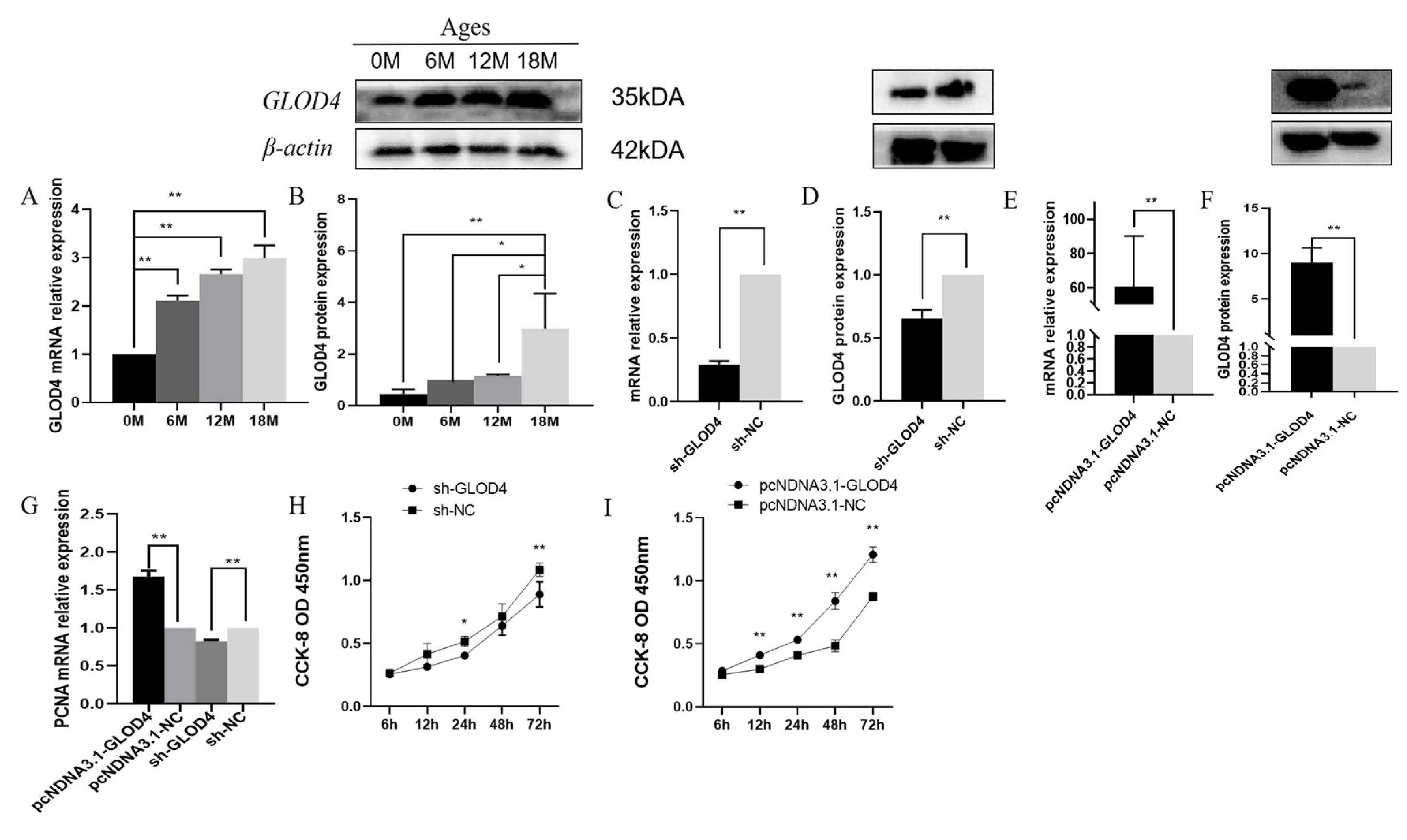
3.2. Expression Patterns of the GLOD4 Transcript at Different Developmental Stages of Qianbei Ma Goats Testes
3.3. GLOD4 Promotes Qianbei Ma Goat LCs Proliferation In Vitro
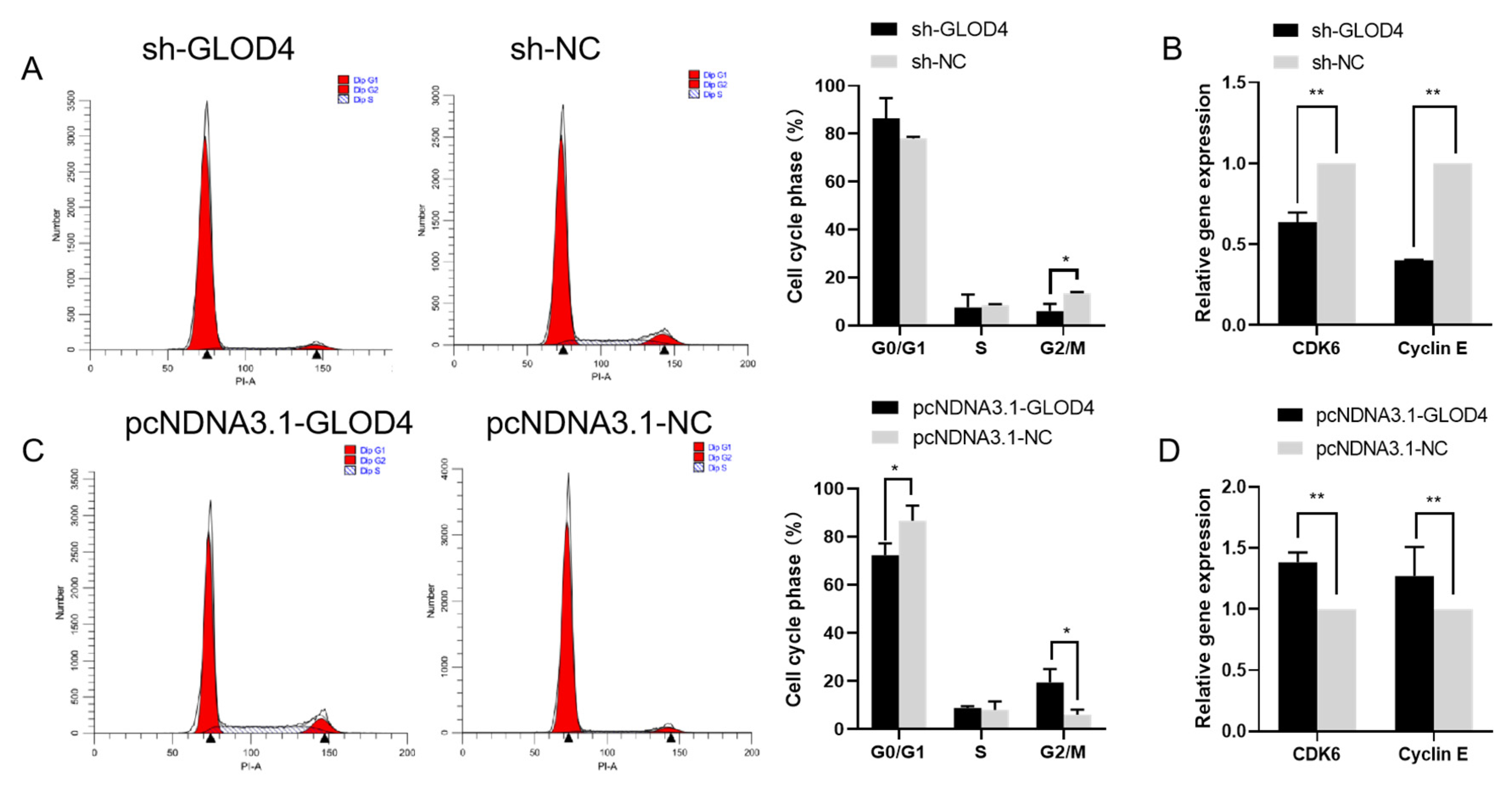
3.4. GLOD4 Promotes Cell Cycle Progress of Qianbei Ma Goat LCs In Vitro
3.5. Promotion of Testosterone Hormone Secretion by GLOD4
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix B
References
- Hess, R.A.; Renato, D.F.L. Spermatogenesis and cycle of the seminiferous epithelium. Asian J. Androl. 2008, 636, 1–15. [Google Scholar] [CrossRef]
- Staub, C.; Johnson, L. Review: Spermatogenesis in the bull. Animal 2018, 12, s27–s35. [Google Scholar] [CrossRef]
- Chalmel, F.; Rolland, A.D. Linking transcriptomics and proteomics in spermatogenesis. Reproduction 2015, 150, R149–R157. [Google Scholar] [CrossRef] [PubMed]
- Mruk, D.D.; Cheng, C.Y. The Mammalian Blood-Testis Barrier: Its Biology and Regulation. Endocr. Rev. 2015, 36, 564–591. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Chen, W.; Cheng, Z.; Wu, S.; He, J.; Han, L.; He, Z.; Qin, W. Novel Gene Regulation in Normal and Abnormal Spermatogenesis. Cells 2021, 10, 666. [Google Scholar] [CrossRef]
- Grande, G.; Barrachina, F.; Soler-Ventura, A.; Jodar, M.; Mancini, F.; Marana, R.; Chiloiro, S.; Pontecorvi, A.; Oliva, R.; Milardi, D. The Role of Testosterone in Spermatogenesis: Lessons From Proteome Profiling of Human Spermatozoa in Testosterone Deficiency. Front. Endocrinol. 2022, 13, 852661. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Chen, S.; Chen, Y.; Yuan, X.; Dong, H. Testicular fat deposition attenuates reproductive performance via decreased follicle-stimulating hormone level and sperm meiosis and testosterone synthesis in mouse. Anim. Biosci. 2024, 37, 50–60. [Google Scholar] [CrossRef]
- Schulster, M.; Bernie, A.M.; Ramasamy, R. The role of estradiol in male reproductive function. Asian J. Androl. 2016, 18, 435–440. [Google Scholar] [CrossRef]
- Rosati, L.; Di Fiore, M.M.; Prisco, M.; Di Giacomo, R.F.; Venditti, M.; Andreuccetti, P.; Chieffi, B.G.; Santillo, A. Seasonal expression and cellular distribution of star and steroidogenic enzymes in quail testis. Journal of Experimental Zoology. Part B, Molecular and Developmental Evolution 2019, 332, 198–209. [Google Scholar] [CrossRef]
- Qin, W.X.; Wan, F.; Sun, F.Y.; Zhang, P.P.; Han, L.W.; Huang, Y.; Jiang, H.Q.; Zhao, X.T.; He, M.; Ye, Y.; et al. Cloning and characterization of a novel gene (C17orf25) from the deletion region on chromosome 17p13.3 in hepatocelular carcinoma. Cell Res. 2001, 11, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Farrera, D.O.; Galligan, J.J. The Human Glyoxalase Gene Family in Health and Disease. Chem. Res. Toxicol. 2022, 35, 1766–1776. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.Y.; Xu, J.; Mao, Q.; Fu, L.L.; Gu, J.R.; De Zhu, J. The promoter analysis of the human C17orf25 gene, a novel chromosome 17p13.3 gene. Cell Res. 2002, 12, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic. Acids. Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Dihazi, G.H.; Bibi, A.; Jahn, O.; Nolte, J.; Mueller, G.A.; Engel, W.; Dihazi, H. Impact of the antiproliferative agent ciclopirox olamine treatment on stem cells proteome. World J. Stem Cells 2013, 5, 9–25. [Google Scholar] [CrossRef] [PubMed]
- Genini, S.; Zangerl, B.; Slavik, J.; Acland, G.M.; Beltran, W.A.; Aguirre, G.D. Transcriptional profile analysis of RPGRORF15 frameshift mutation identifies novel genes associated with retinal degeneration. Invest. Ophthalmol. Vis. Sci. 2010, 51, 6038–6050. [Google Scholar] [CrossRef] [PubMed]
- Suszynska-Zajczyk, J.; Wroblewski, J.; Utyro, O.; Luczak, M.; Marczak, L.; Jakubowski, H. Bleomycin hydrolase and hyperhomocysteinemia modulate the expression of mouse proteins involved in liver homeostasis. Amino Acids 2014, 46, 1471–1480. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, J.; Bose, N.; Gong, J.; Hall, D.; Rifkind, A.; Bhaumik, D.; Peiris, T.H.; Chamoli, M.; Le, CH.; Liu, J.; et al. A Caenorhabditis elegans Model Elucidates a Conserved Role for TRPA1-Nrf Signaling in Reactive alpha-Dicarbonyl Detoxification. Curr. Biol. 2016, 26, 3014–3025. [Google Scholar] [CrossRef]
- Heinrich, A.; Defalco, T. Essential roles of interstitial cells in testicular development and function. Andrology 2020, 8, 903–914. [Google Scholar] [CrossRef] [PubMed]
- Bittman, E.L. Timing in the Testis. J. Biol. Rhythms 2016, 31, 12–36. [Google Scholar] [CrossRef]
- Mendis-Handagama, S.M.; Ariyaratne, H.B. Effects of thyroid hormones on Leydig cells in the postnatal testis. Histol. Histopathol. 2004, 19, 985–997. [Google Scholar] [CrossRef] [PubMed]
- Mueller, J.P.; Getachew, T.; Rekik, M.; Rischkowsky, B.; Abate, Z.; Wondim, B.; Haile, A. Converting multi-trait breeding objectives into operative selection indexes to ensure genetic gains in low-input sheep and goat breeding programmes. Animal 2021, 15, 100198. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, T.; Liu, N.; Zhang, H.; Zhao, X.; Ma, Y. Characterization of GLOD4 in Leydig Cells of Tibetan Sheep During Different Stages of Maturity. Genes 2019, 10, 796. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Zhou, Z.; Tian, X.; Yang, P.; Fu, K. CYP19A1 May Influence Lambing Traits in Goats by Regulating the Biological Function of Granulosa Cells. Animals 2022, 12, 1911. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Zhong, L.; Zhu, Y.; Liu, G. Research on the isolation of mouse Leydig cells using differential digestion with a low concentration of collagenase. J. Reprod. Dev. 2011, 57, 433–436. [Google Scholar] [CrossRef]
- Duan, P.; Ha, M.; Huang, X.; Zhang, P.; Liu, C. Intronic miR-140-5p contributes to beta-cypermethrin-mediated testosterone decline. The Science of the Total Environment 2022, 806, 150517. [Google Scholar] [CrossRef]
- Lim, Y.; Gang, D.Y.; Lee, W.Y.; Yun, S.H.; Cho, Y.B.; Huh, J.W.; Park, Y.A.; Kim, H.C. Proteomic identification of arginine-methylated proteins in colon cancer cells and comparison of messenger RNA expression between colorectal cancer and adjacent normal tissues. Ann. Coloproctol. 2022, 38, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Muthaiyan, S.M.; Chaudhuri, J.; Sellegounder, D.; Sahu, A.K.; Guha, S.; Chamoli, M.; Hodge, B.; Bose, N.; Roberts, C.; Farrera, D.O.; et al. Methylglyoxal-derived hydroimidazolone, MG-H1, increases food intake by altering tyramine signaling via the GATA transcription factor ELT-3 in Caenorhabditis elegans. Elife 2023, 12, e82446. [Google Scholar] [CrossRef] [PubMed]
- Albee, A.J.; Kwan, A.L.; Lin, H.; Granas, D.; Stormo, G.D.; Dutcher, S.K. Identification of cilia genes that affect cell-cycle progression using whole-genome transcriptome analysis in Chlamydomonas reinhardtti. G3-Genes Genomes Genet. 2013, 3, 979–991. [Google Scholar] [CrossRef]
- Xie, M.C.; Ai, C.; Jin, X.M.; Liu, S.F.; Tao, S.X.; Li, Z.D.; Wang, Z. Cloning and characterization of chicken SPATA4 gene and analysis of its specific expression. Mol. Cell. Biochem. 2007, 306, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, B.; He, S.; Zhao, Y.; Wang, Z. Cloning and characterization of zebra fish SPATA4 gene and analysis of its gonad specific expression. Biochem.-Moscow 2005, 70, 638–644. [Google Scholar] [CrossRef]
- Liu, B.; Liu, S.; He, S.; Zhao, Y.; Hu, H.; Wang, Z. Cloning and expression analysis of gonadogenesis-associated gene SPATA4 from rainbow trout (Oncorhynchus mykiss). Journal of Biochemistry and Molecular Biology 2005, 38, 206–210. [Google Scholar] [CrossRef]
- Zhang, H.T.; Yan, Z.Q.; Hu, X.B.; Yang, S.L.; Gong, Y. Interaction of C17orf25 with ADP-ribose pyrophosphatase NUDT9 detected via yeast two-hybrid method. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 2003, 35, 747–751. [Google Scholar] [PubMed]
- Strzalka, W.; Ziemienowicz, A. Proliferating cell nuclear antigen (PCNA): a key factor in DNA replication and cell cycle regulation. Ann. Bot. 2011, 107, 1127–1140. [Google Scholar] [CrossRef]
- Swaffer, M.P.; Jones, A.W.; Flynn, H.R.; Snijders, A.P.; Nurse, P. CDK Substrate Phosphorylation and Ordering the Cell Cycle. Cell 2016, 167, 1750–1761. [Google Scholar] [CrossRef]
- Price, M.A. CKI, there’s more than one: casein kinase I family members in Wnt and Hedgehog signaling. Genes. Dev. 2006, 20, 399–410. [Google Scholar] [CrossRef] [PubMed]
- Lavi, O.; Ginsberg, D.; Louzoun, Y. Regulation of modular Cyclin and CDK feedback loops by an E2F transcription oscillator in the mammalian cell cycle. Mathematical Biosciences and Engineering : Mbe 2011, 8, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Boudreault, J.; Wang, N.; Poulet, S.; Daliah, G.; Yan, G.; Moamer, A.; Burgos, S.A.; Sabri, S.; Ali, S.; et al. Differential Regulation of Cancer Progression by CDK4/6 Plays a Central Role in DNA Replication and Repair Pathways. Cancer Research (Chicago, Ill.) 2021, 81, 1332–1346. [Google Scholar] [CrossRef] [PubMed]
- Moroy, T.; Geisen, C. Cyclin E. The International Journal of Biochemistry & Cell Biology 2004, 36, 1424–1439. [Google Scholar] [CrossRef]
- Omoboyowa, D.A.; Balogun, T.A.; Saibu, O.A.; Chukwudozie, O.S.; Alausa, A.; Olubode, S.O.; Aborode, A.T.; Batiha, G.E.; Bodun, D.S.; Musa, S.O. Structure-based discovery of selective CYP(17)A(1) inhibitors for Castration-resistant prostate cancer treatment. Biol. Methods Protoc. 2022, 7, bpab26. [Google Scholar] [CrossRef]
- Janer, G.; Leblanc, G.A.; Porte, C. A comparative study on androgen metabolism in three invertebrate species. Gen. Comp. Endocrinol. 2005, 143, 211–221. [Google Scholar] [CrossRef]
- Ohta, Y.; Kawate, N.; Inaba, T.; Morii, H.; Takahashi, K.; Tamada, H. Feeding hydroalcoholic extract powder of Lepidium meyenii (maca) enhances testicular gene expression of 3beta-hydroxysteroid dehydrogenase in rats. Andrologia 2017, 49, e12792. [Google Scholar] [CrossRef] [PubMed]
- Gunther, J.; Schuler, G.; Teppa, E.; Furbass, R. Charged Amino Acids in the Transmembrane Helix Strongly Affect the Enzyme Activity of Aromatase. International Journal of Molecular Sciences 2024, 25, 1440. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England Journal of Medicine 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, J.; Yang, L.; Zhang, H.; Zhang, Y.; Gao, D.; Jiang, H.; Li, Y.; Dong, H.; Ma, T.; et al. Glyphosate exposure attenuates testosterone synthesis via NR1D1 inhibition of StAR expression in mouse Leydig cells. The Science of the Total Environment 2021, 785, 147323. [Google Scholar] [CrossRef] [PubMed]

| Gene | Primer sequence (5′→3′) |
|---|---|
| Sh-GLOD4 | F:CACCGATATAAGTTCTATTTGCAGGATTCAAGAGATCCTGCAAATAGAACTTATATTTTTTTG R:GATCCAAAAAAATATAAGTTCTATTTGCAGGATCTCTTGAATCCTGCAAATAGAACTTATATC |
| Sh-NC | F:CACCGTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTG R:GATCCAAAAAATTCTCCGAACGTGTCACGTTCTCTTGAAACGTGACACGTTCGGAGAAC |
| Gene | Primer sequence (5′→3′) |
Gen Bank ID | Fragment Size(bP) | Tm/℃ |
|---|---|---|---|---|
| CYP17A1 | F:GCTCACCCTCGCCTETTTETT R:GTCTCCTGACACTGCTCACA |
NM_001314145.1 | 169 | 58 |
| CYP11A1 | F:CTCCAGAGGCAETAAAGAA R:TCAAAGGCAAAGTGAAACA |
NM_001287574.1 | 145 | 60 |
| 3BHSD | F:AGACCAGAAGTTCGGGAGGAA R:TCTCCCTGTAGGAGTTGGGC |
NM_001285716.1 | 292 | 60 |
| GLOD4 | F:AGCTCTGCACTTCGTGTTCA R: GCAATGCGTCCAAAACCTGT |
XM_013971906 |
554 | 60 |
| CDK6 | F:GTGGACCTCTGGAGCGTTGG R:TGCCTTGCTCATCAATGTCTGTTAC |
XM_018047426.1 | 223 | 58 |
| PCNA | F:GTAGCCGTGTCETTGCGACTCC R:GCTCTGTAGGTTCACGCCACTTG |
XM_005688167.3 | 145 | 60 |
| CylinE | F:GETGTCGGCTGCTTAGAET R:GTCTCCTGACACTGCTCACA |
XM_018062248.1 | 104 | 60 |
| β-actin | F:GETGTCGGCTGCTTAGAET R:CCAETCTCETCTCGTTTTCTG |
NM_00297986.1 | 139 | 60 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




