Submitted:
17 June 2024
Posted:
18 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
1.1. Vitronectin Structure and Physio/Patho-Logical Role in the Host
1.2. Vitronectin as Regulator of Complement System
1.3. Vitronectin as Mediator of Cell Migration and Adhesion
2. Bacterial Engagement of Vitronectin as a Weapon to Escape the Immune System
3. Bacterial Targeting of Vitronectin for Host Colonization
3.1. Gram-Positive
3.2. Gram-Negative
4. Vitronectin-Binding by Bacteria with a Yet to Be Defined Activity
5. Vitronectin Adsorption on Biomaterial Surfaces: A Double-Edged Weapon
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gibson AD, Peterson CB. Full-length and truncated forms of vitronectin provide insight into effects of proteolytic processing on function. Biochim Biophys Acta. 2001. 1545:289-304. [CrossRef]
- Hayman EG, Pierschbacher MD, Ohgren Y, Ruoslahti E. Serum spreading factor (vitronectin) is present at the cell surface and in tissues. Proc Natl Acad Sci U S A. 1983. 80:4003-4007. [CrossRef]
- Zhou, A. Functional structure of the somatomedin B domain of vitronectin. Protein Sci. 2007. 16:1502-1508. [CrossRef]
- Zhou A, Huntington JA, Pannu NS, Carrell RW, Read RJ. How vitronectin binds PAI-1 to modulate fibrinolysis and cell migration. Nat Struct Biol. 2003. 10:541-544. [CrossRef]
- Wei Y, Waltz DA, Rao N, Drummond RJ, Rosenberg S, Chapman HA. Identification of the urokinase receptor as an adhesion receptor for vitronectin. J Biol Chem. 1994. 269:32380-32388.
- Seiffert D, Smith JW. The cell adhesion domain in plasma vitronectin is cryptic. J Biol Chem. 1997. 272:13705-13710. [CrossRef]
- Gebb C, Hayman EG, Engvall E, Ruoslahti E. Interaction of vitronectin with collagen. J Biol Chem. 1986. 261:16698-16703.
- Chillakuri CR, Jones C, Mardon HJ. Heparin binding domain in vitronectin is required for oligomerization and thus enhances integrin mediated cell adhesion and spreading. FEBS Lett. 2010. 584:3287-3291. [CrossRef]
- Hunt LT, Barker WC, Chen HR. A domain structure common to hemopexin, vitronectin, interstitial collagenase, and a collagenase homolog. Protein Seq Data Anal. 1987. 1:21-26.
- Tschopp J, Masson D, Schäfer S, Peitsch M, Preissner KT. The heparin binding domain of S-protein/vitronectin binds to complement components C7, C8, and C9 and perforin from cytolytic T-cells and inhibits their lytic activities. Biochemistry. 1988. 27:4103-4109. [CrossRef]
- Preissner, KT. Specific binding of plasminogen to vitronectin. Evidence for a modulatory role of vitronectin on fibrin(ogen)-induced plasmin formation by tissue plasminogen activator. Biochem Biophys Res Commun. 1990. 168:966-971. [CrossRef]
- de Boer HC, Preissner KT, Bouma BN, de Groot PG. Binding of vitronectin-thrombin-antithrombin III complex to human endothelial cells is mediated by the heparin binding site of vitronectin. J Biol Chem. 1992. 267:2264-2268.
- Gechtman Z, Belleli A, Lechpammer S, Shaltiel S. The cluster of basic amino acids in vitronectin contributes to its binding of plasminogen activator inhibitor-1: evidence from thrombin-, elastase- and plasmin-cleaved vitronectins and anti-peptide antibodies. Biochem J. 1997. 325:339-349. [CrossRef]
- Stoop AA, Lupu F, Pannekoek H. Colocalization of thrombin, PAI-1, and vitronectin in the atherosclerotic vessel wall: A potential regulatory mechanism of thrombin activity by PAI-1/vitronectin complexes. Arterioscler Thromb Vasc Biol. 2000. 20:1143-1149. [CrossRef]
- Yoneda A, Ogawa H, Kojima K, Matsumoto I. Characterization of the ligand binding activities of vitronectin: interaction of vitronectin with lipids and identification of the binding domains for various ligands using recombinant domains. Biochemistry. 1998. 37:6351-6360. [CrossRef]
- Jenne D, Hille A, Stanley KK, Huttner WB. Sulfation of two tyrosine-residues in human complement S-protein (vitronectin). Eur J Biochem. 1989. 185:391-395. [CrossRef]
- Schvartz I, Seger D, Shaltiel S. Vitronectin. Int J Biochem Cell Biol. 1999. 31:539-544. [CrossRef]
- Wang C, Cui Y, Miao H, Sun T, Lu Y, Zhang Y. Circulating Vitronectin Predicts Liver Injury and Mortality in Children With Sepsis: A Prospective Observational Study. Clin Appl Thromb Hemost. 2020. 26:1076029620935201. [CrossRef]
- Semeraro N, Ammollo CT, Semeraro F, Colucci M. Sepsis, thrombosis and organ dysfunction. Thromb Res. 2012. 129:290-295. [CrossRef]
- Bera A, Subramanian M, Karaian J, et al. Functional role of vitronectin in breast cancer. PLoS One. 2020. 15:e0242141. Published 2020 Nov 19. [CrossRef]
- Burgos-Panadero R, Noguera I, Cañete A, Navarro S, Noguera R. Vitronectin as a molecular player of the tumor microenvironment in neuroblastoma. BMC Cancer. 2019. 19:479. Published 2019. 22 May. [CrossRef]
- Sarma JV, Ward PA. The complement system. Cell Tissue Res. 2011. 343:227-235. [CrossRef]
- Westra D, Volokhina EB, van der Molen RG, et al. Serological and genetic complement alterations in infection-induced and complement-mediated hemolytic uremic syndrome. Pediatr Nephrol. 2017. 32:297-309. [CrossRef]
- Laarman A, Milder F, van Strijp J, Rooijakkers S. Complement inhibition by gram-positive pathogens: molecular mechanisms and therapeutic implications. J Mol Med (Berl). 2010. 88:115-120. [CrossRef]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010. 11:785-797. [CrossRef]
- Podack ER, Preissner KT, Müller-Eberhard HJ. Inhibition of C9 polymerization within the SC5b-9 complex of complement by S-protein. Acta Pathol Microbiol Immunol Scand Suppl. 1984. 284:89-96.
- Mayasundari A, Whittemore NA, Serpersu EH, Peterson CB. The solution structure of the N-terminal domain of human vitronectin: proximal sites that regulate fibrinolysis and cell migration. J Biol Chem. 2004. 279:29359-29366. [CrossRef]
- Meredith JE Jr, Winitz S, Lewis JM, et al. The regulation of growth and intracellular signaling by integrins. Endocr Rev. 1996. 17:207-220. [CrossRef]
- Singh B, Su YC, Riesbeck K. Vitronectin in bacterial pathogenesis: a host protein used in complement escape and cellular invasion. Mol Microbiol. 2010. 78:545-560. [CrossRef]
- Hallström T, Singh B, Kraiczy P, et al. Conserved Patterns of Microbial Immune Escape: Pathogenic Microbes of Diverse Origin Target the Human Terminal Complement Inhibitor Vitronectin via a Single Common Motif. PLoS One. 2016. 11:e0147709. Published 2016 Jan 25. [CrossRef]
- Foxwell AR, Kyd JM, Cripps AW. Nontypeable Haemophilus influenzae: pathogenesis and prevention. Microbiol Mol Biol Rev. 1998. 62:294-308. [CrossRef]
- Hallström T, Blom AM, Zipfel PF, Riesbeck K. Nontypeable Haemophilus influenzae protein E binds vitronectin and is important for serum resistance. J Immunol. 2009. 183:2593-2601. [CrossRef]
- 33. Singh B, Jalalvand F, Mörgelin M, Zipfel P, Blom AM, Riesbeck K. Haemophilus influenzae, 8: the C-terminal domain of vitronectin and modulates the membrane attack complex. Mol Microbiol. 2011. 81, 2011. [CrossRef]
- 34. Su YC, Jalalvand F, Mörgelin M, Blom AM, Singh B, Riesbeck K. Haemophilus influenzae, 1: via the ubiquitous Protein F to subvert host innate immunity. Mol Microbiol. 2013. 87, 2013. [CrossRef]
- Singh B, Al-Jubair T, Mörgelin M, Thunnissen MM, Riesbeck K. The unique structure of Haemophilus influenzae protein E reveals multiple binding sites for host factors. Infect Immun. 2013. 81:801-814. [CrossRef]
- 36. Jalalvand F, Su YC, Mörgelin M, et al. Haemophilus influenzae, 8: binding to laminin and human pulmonary epithelial cells. J Infect Dis. 2013. 207, 2013. [CrossRef]
- 37. Al-Jubair T, Mukherjee O, Oosterhuis S, et al. Haemophilus influenzae, 5: Vitronectin Using Protein H To Resist Host Innate Immunity and Adhere to Pulmonary Epithelial Cells. J Immunol. 2015. 195, 2015. [CrossRef]
- Linke D, Riess T, Autenrieth IB, Lupas A, Kempf VA. Trimeric autotransporter adhesins: variable structure, common function. Trends Microbiol. 2006. 14:264-270. [CrossRef]
- Singh B, Su YC, Al-Jubair T, et al. A fine-tuned interaction between trimeric autotransporter haemophilus surface fibrils and vitronectin leads to serum resistance and adherence to respiratory epithelial cells. Infect Immun. 2014. 82:2378-2389. [CrossRef]
- Attia AS, Ram S, Rice PA, Hansen EJ. Binding of vitronectin by the Moraxella catarrhalis UspA2 protein interferes with late stages of the complement cascade. Infect Immun. 2006. 74:1597-1611. [CrossRef]
- Singh B, Blom AM, Unal C, Nilson B, Mörgelin M, Riesbeck K. Vitronectin binds to the head region of Moraxella catarrhalis ubiquitous surface protein A2 and confers complement-inhibitory activity. Mol Microbiol. 2010. 75:1426-1444. [CrossRef]
- Su YC, Hallström BM, Bernhard S, Singh B, Riesbeck K. Impact of sequence diversity in the Moraxella catarrhalis UspA2/UspA2H head domain on vitronectin binding and antigenic variation. Microbes Infect. 2013. 15:375-387. [CrossRef]
- Riley SP, Patterson JL, Nava S, Martinez JJ. Pathogenic Rickettsia species acquire vitronectin from human serum to promote resistance to complement-mediated killing. Cell Microbiol. 2014. 16:849-861. [CrossRef]
- Fish AI, Riley SP, Singh B, Riesbeck K, Martinez JJ. The Rickettsia conorii Adr1 Interacts with the C-Terminus of Human Vitronectin in a Salt-Sensitive Manner. Front Cell Infect Microbiol. 2017. 7:61. [CrossRef]
- 45. Sa E Cunha C, Griffiths NJ, Virji M. Neisseria meningitidis, e: to the sulphated tyrosines of activated vitronectin to attach to and invade human brain endothelial cells. PLoS Pathog. 2010. 6, 20 May 2010. [CrossRef]
- Andreae CA, Sessions RB, Virji M, Hill DJ. Bioinformatic analysis of meningococcal Msf and Opc to inform vaccine antigen design. PLoS One. 2018. 13:e0193940. Published 2018 Mar 16. [CrossRef]
- Hill DJ, Griffiths NJ, Borodina E, Andreae CA, Sessions RB, Virji M. Identification and therapeutic potential of a vitronectin binding region of meningococcal msf. PLoS One. 2015. 10:e0124133. [CrossRef]
- 48. Bartra SS, Ding Y, Miya Fujimoto L, et al. Yersinia pestis, 2: membrane protein to recruit vitronectin. Microbiology (Reading). 2015. 161, 2015. [CrossRef]
- Shin K, Lechtenberg BC, Fujimoto LM, et al. Structure of human Vitronectin C-terminal domain and interaction with Yersinia pestis outer membrane protein Ail. Sci Adv. 2019. 5:eaax5068. [CrossRef]
- Krukonis ES, Thomson JJ. Complement evasion mechanisms of the systemic pathogens Yersiniae and Salmonellae. FEBS Lett. 2020. 594:2598-2620. [CrossRef]
- Mühlenkamp MC, Hallström T, Autenrieth IB, et al. Vitronectin Binds to a Specific Stretch within the Head Region of Yersinia Adhesin A and Thereby Modulates Yersinia enterocolitica Host Interaction. J Innate Immun. 2017. 9:33-51. [CrossRef]
- Brooks MJ, Sedillo JL, Wagner N, et al. Modular arrangement of allelic variants explains the divergence in Moraxella catarrhalis UspA protein function. Infect Immun. 2008. 76:5330-5340. [CrossRef]
- Meuskens I, Leva-Bueno J, Millner P, Schütz M, Peyman SA, Linke D. The Trimeric Autotransporter Adhesin YadA of Yersinia enterocolitica Serotype O:9 Binds Glycan Moieties. Front Microbiol. 2022. 12:738818. [CrossRef]
- Richter C, Mukherjee O, Ermert D, et al. Moonlighting of Helicobacter pylori catalase protects against complement-mediated killing by utilising the host molecule vitronectin. Sci Rep. 2016. 6:24391. [CrossRef]
- 55. Hallström T, Uhde M, Singh B, Skerka C, Riesbeck K, Zipfel PF. Pseudomonas aeruginosa, e: Dehydrogenase (Lpd) to Bind to the Human Terminal Pathway Regulators Vitronectin and Clusterin to Inhibit Terminal Pathway Complement Attack. PLoS One. 2015. 10, 2015. [CrossRef]
- Paulsson M, Che KF, Ahl J, et al. Bacterial Outer Membrane Vesicles Induce Vitronectin Release Into the Bronchoalveolar Space Conferring Protection From Complement-Mediated Killing. Front Microbiol. 2018. 9:1559. [CrossRef]
- Li S, Wang Y, Yang R, et al. Outer membrane protein OMP76 of Riemerella anatipestifer contributes to complement evasion and virulence by binding to duck complement factor vitronectin. Virulence. 2023. 14:2223060. [CrossRef]
- da Silva LB, Miragaia Ldos S, Breda LC, et al. Pathogenic Leptospira species acquire factor H and vitronectin via the surface protein LcpA. Infect Immun. 2015. 83:888-897. [CrossRef]
- Sato K, Kumagai Y, Sekizuka T, et al. Vitronectin binding protein, BOM1093, confers serum resistance on Borrelia miyamotoi. Sci Rep. 2021. 11:5462. Published 2021 Mar 9. [CrossRef]
- Brown, EJ. Interaction of gram-positive microorganisms with complement. Curr Top Microbiol Immunol. 1985. 121:159-187. [CrossRef]
- Voss S, Hallström T, Saleh M, et al. The choline-binding protein PspC of Streptococcus pneumoniae interacts with the C-terminal heparin-binding domain of vitronectin. J Biol Chem. 2013. 288:15614-15627. [CrossRef]
- Janulczyk R, Iannelli F, Sjoholm AG, Pozzi G, Bjorck L. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J Biol Chem. 2000. 275:37257-37263. [CrossRef]
- Kohler S, Hallström T, Singh B, et al. Binding of vitronectin and Factor H to Hic contributes to immune evasion of Streptococcus pneumoniae serotype 3. Thromb Haemost. 2015. 113:125-142. [CrossRef]
- Voss S, Hallström T, Saleh M, et al. The choline-binding protein PspC of Streptococcus pneumoniae interacts with the C-terminal heparin-binding domain of vitronectin. J Biol Chem. 2013. 288:15614-15627. [CrossRef]
- Adegbola, RA. Bacterial adhesion and pathogenicity. Afr J Med Med Sci. 1988. 17:63-69.
- Speziale P, Pietrocola G, Rindi S, et al. Structural and functional role of Staphylococcus aureus surface components recognizing adhesive matrix molecules of the host. Future Microbiol. 2009. 4:1337-1352. [CrossRef]
- Scibelli A, Roperto S, Manna L, et al. Engagement of integrins as a cellular route of invasion by bacterial pathogens. Vet J. 2007. 173:482-491. [CrossRef]
- Valentin-Weigand P, Grulich-Henn J, Chhatwal GS, Müller-Berghaus G, Blobel H, Preissner KT. Mediation of adherence of streptococci to human endothelial cells by complement S protein (vitronectin). Infect Immun. 1988. 56:2851-2855. [CrossRef]
- Filippsen LF, Valentin-Weigand P, Blobel H, Preissner KT, Chhatwal GS. Role of complement S protein (vitronectin) in adherence of Streptococcus dysgalactiae to bovine epithelial cells. Am J Vet Res. 1990. 51:861-865.
- Bergmann S, Lang A, Rohde M, et al. Integrin-linked kinase is required for vitronectin-mediated internalization of Streptococcus pneumoniae by host cells. J Cell Sci. 2009. 122:256-267. [CrossRef]
- Buscetta M, Firon A, Pietrocola G, et al. PbsP, a cell wall-anchored protein that binds plasminogen to promote hematogenous dissemination of group B Streptococcus. Mol Microbiol. 2016. 101:27-41. [CrossRef]
- De Gaetano GV, Pietrocola G, Romeo L, et al. The Streptococcus agalactiae cell wall-anchored protein PbsP mediates adhesion to and invasion of epithelial cells by exploiting the host vitronectin/αv integrin axis. Mol Microbiol. 2018. 110:82-94. [CrossRef]
- Coates R, Moran J, Horsburgh MJ. Staphylococci: colonizers and pathogens of human skin. Future Microbiol. 2014. 9:75-91. [CrossRef]
- Chhatwal GS, Preissner KT, Müller-Berghaus G, Blobel H. Specific binding of the human S protein (vitronectin) to streptococci, Staphylococcus aureus, and Escherichia coli. Infect Immun. 1987. 55:1878-1883. [CrossRef]
- Paulsson M, Wadström T. Vitronectin and type-I collagen binding by Staphylococcus aureus and coagulase-negative staphylococci. FEMS Microbiol Immunol. 1990. 2:55-62. [CrossRef]
- Li DQ, Lundberg F, Ljungh A. Characterization of vitronectin-binding proteins of Staphylococcus epidermidis. Curr Microbiol. 2001. 42:361-367. [CrossRef]
- Heilmann C, Hussain M, Peters G, Götz F. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol. 1997. 24:1013-1024. [CrossRef]
- Zoll S, Schlag M, Shkumatov AV, et al. Ligand-binding properties and conformational dynamics of autolysin repeat domains in staphylococcal cell wall recognition. J Bacteriol. 2012. 194:3789-3802. [CrossRef]
- Kohler TP, Gisch N, Binsker U, et al. Repeating structures of the major staphylococcal autolysin are essential for the interaction with human thrombospondin 1 and vitronectin. J Biol Chem. 2014. 289:4070-4082. [CrossRef]
- Pietrocola G, Pellegrini A, Alfeo MJ, Marchese L, Foster TJ, Speziale P. The iron-regulated surface determinant B (IsdB) protein from Staphylococcus aureus acts as a receptor for the host protein vitronectin. J Biol Chem. 2020. 295:10008-10022. [CrossRef]
- Mathelié-Guinlet M, Viela F, Pietrocola G, Speziale P, Dufrêne YF. Nanonewton forces between Staphylococcus aureus surface protein IsdB and vitronectin. Nanoscale Adv. 2020. 2:5728-5736. [CrossRef]
- Mora-Uribe P, Miranda-Cárdenas C, Castro-Córdova P, et al. Characterization of the Adherence of Clostridium difficile Spores: The Integrity of the Outermost Layer Affects Adherence Properties of Spores of the Epidemic Strain R20291 to Components of the Intestinal Mucosa. Front Cell Infect Microbiol. 2016. 6:99. [CrossRef]
- Ikeda M, Enomoto N, Hashimoto D, et al. Nontypeable Haemophilus influenzae exploits the interaction between protein-E and vitronectin for the adherence and invasion to bronchial epithelial cells. BMC Microbiol. 2015. 15:263. [CrossRef]
- Zhang Y, Ying X, He Y, et al. Invasiveness of the Yersinia pestis ail protein contributes to host dissemination in pneumonic and oral plague. Microb Pathog. 2020. 141:103993. [CrossRef]
- Davies, JC. Pseudomonas aeruginosa in cystic fibrosis: pathogenesis and persistence. Paediatr Respir Rev. 2002. 3:128-134. [CrossRef]
- Leroy-Dudal J, Gagnière H, Cossard E, Carreiras F, Di Martino P. Role of alphavbeta5 integrins and vitronectin in Pseudomonas aeruginosa PAK interaction with A549 respiratory cells. Microbes Infect. 2004. 6:875-881. [CrossRef]
- Li Y, Wang J, Liu B, et al. DnaK Functions as a Moonlighting Protein on the Surface of Mycoplasma hyorhinis Cells. Front Microbiol. 2022. 13:842058. [CrossRef]
- Biswas R, Voggu L, Simon UK, Hentschel P, Thumm G, Götz F. Activity of the major staphylococcal autolysin Atl. FEMS Microbiol Lett. 2006. 259:260-268. [CrossRef]
- 89. Pathak H, Sokkalingam M, Prasanth L, Devi K, Joshi P. Staphylococcus aureus, 6: with caprine vitronectin without involving the heparin binding domain and the second arginine-glycine-aspartic acid motif of the host protein. Arch Microbiol. 2019. 201, 2019. [CrossRef]
- Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekötter A, Peters G. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology (Reading). 2003. 149:2769-2778. [CrossRef]
- ElTahir Y, Al-Araimi A, Nair RR, et al. Correction to: Binding of Brucella protein, Bp26, to select extracellular matrix molecules. BMC Mol Cell Biol. 2020. 21:16. [CrossRef]
- Leduc I, Olsen B, Elkins C. Localization of the domains of the Haemophilus ducreyi trimeric autotransporter DsrA involved in serum resistance and binding to the extracellular matrix proteins fibronectin and vitronectin. Infect Immun. 2009. 77:657-666. [CrossRef]
- Liang OD, Preissner KT, Chhatwal GS. The hemopexin-type repeats of human vitronectin are recognized by Streptococcus pyogenes. Biochem Biophys Res Commun. 1997. 234:445-449. [CrossRef]
- 94. Esgleas M, Lacouture S, Gottschalk M. Streptococcus suis, 3: to extracellular matrix proteins. FEMS Microbiol Lett. 2005. 244, 2005. [CrossRef]
- Abdallah MN, Tran SD, Abughanam G, et al. Biomaterial surface proteomic signature determines interaction with epithelial cells. Acta Biomater. 2017. 54:150-163. [CrossRef]
- Li T, Hao L, Li J, Du C, Wang Y. Insight into vitronectin structural evolution on material surface chemistries: The mediation for cell adhesion. Bioact Mater. 2020. 5:1044-1052. [CrossRef]
- Li P, Yin R, Cheng J, Lin J. Bacterial Biofilm Formation on Biomaterials and Approaches to Its Treatment and Prevention. Int J Mol Sci. 2023. 24:11680. [CrossRef]
- Kreve S, Reis ACD. Bacterial adhesion to biomaterials: What regulates this attachment? A review. Jpn Dent Sci Rev. 2021. 57:85-96. [CrossRef]
- Hessenauer MET, Lauber K, Zuchtriegel G, et al. Vitronectin promotes the vascularization of porous polyethylene biomaterials. Acta Biomater. 2018. 82:24-33. [CrossRef]
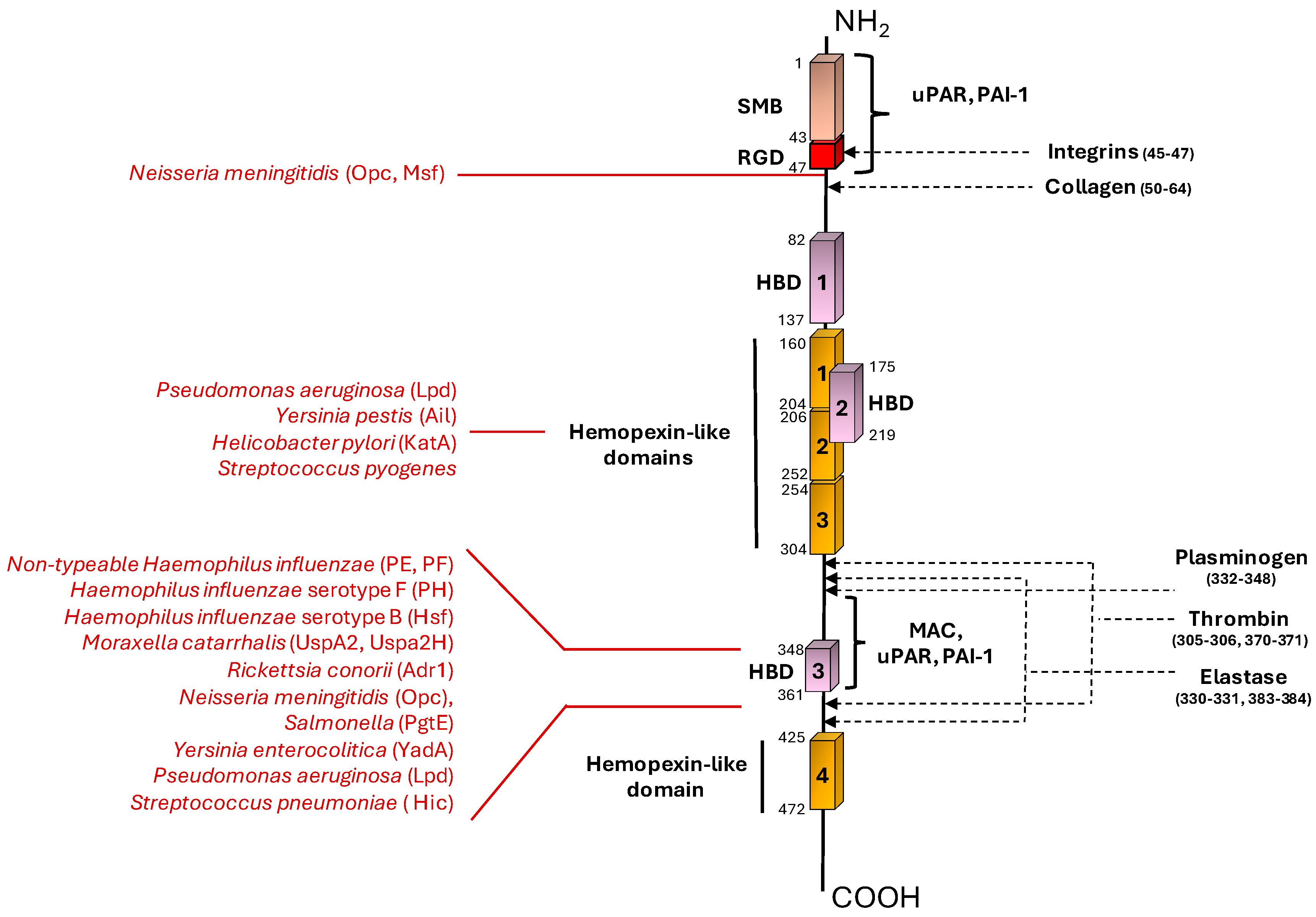
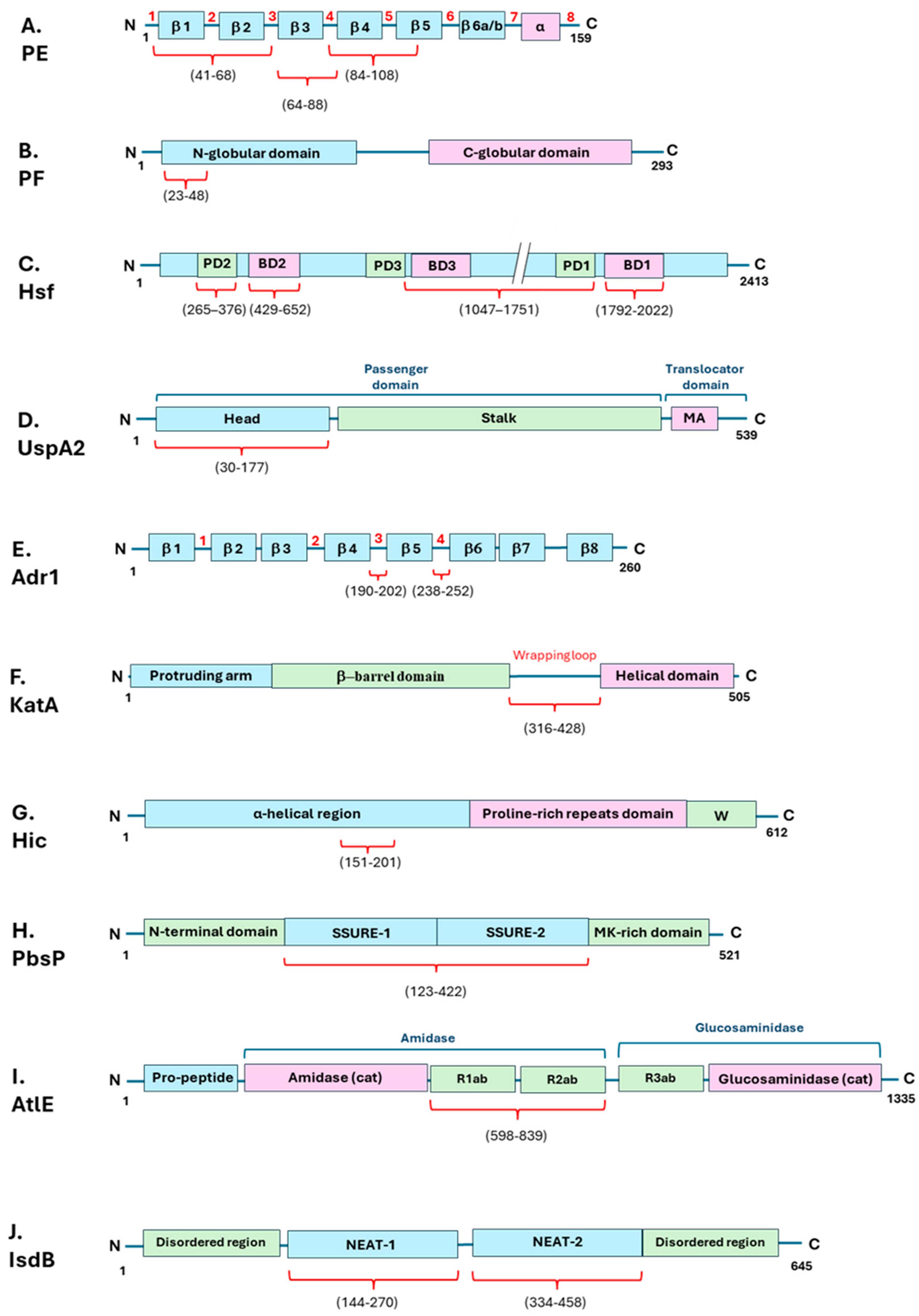
| Bacterial species | Bacterial Protein interacting with Vn | Bacterial protein region involved in Vn binding (amino acid residues) | Vn region bound (amino acid residues) | Role in Complement evasion | Role in Cell Adhesion / invasion | Ref. |
|---|---|---|---|---|---|---|
| Gram-negative | ||||||
| Nontypeable Haemophilus influenzae | PE | 84–108, 41–68, 64–88, |
HBD-3 (353-363) |
+ | U | [33,35] |
| “ | PF | 23-48 | HBD-3 (348-361), PAI-1 binding site (348-370) |
+ | U | [34,36] |
| Haemophilus influenzae serotype F | PH | U | C-terminal (352–362) |
+ | U | [37] |
| Haemophilus influenzae serotype B | Hsf | 265-376, 429-652, 1047-1751, 1792-2022 |
C-terminal (352–374) |
+ | + | [38,39] |
| Moraxella catarrhalis | UspA2 | 30-177 | HBD-3 (312-396) |
+ | U | [40,41,42] |
| “ | UspA2H | 30-177 | HBD-3 (312-396) |
+ | U | [42] |
| Rickettsia conorii | Adr1 | 190-202, 238-252 |
C-terminal (363-373) |
+ | U | [43,44] |
| Neisseria meningititis | Opc | U | N-terminal (43-68), HBD-3 |
+ | + | [45] |
| “ | Msf | 39-82 | N-terminal (43-68) |
+ | U | [46,47] |
| Salmonella | Pgte | U | C-terminal | + | U | [50] |
| Yersinia enterocolitica | YadA | Head domain | HBD-3 | + | U | [51,53] |
| Yersinia pestis | Ail | U | Hemopexin domain | + | U | [48,49] |
| Helicobacter pylori | KatA | 316-428 | C-terminal (229-339) |
+ | U | [54] |
| Pseudomonas aeruginosa | LpD | U | C-terminal (354-363) Hemopexin-like repeats (161-287) |
+ | U | [55] |
| Riemerella anatipestifer | OMP76 | U | U | + | U | [57] |
| Leptospira interrogans | Lcpa | U | HBD (s) | + | U | [58] |
| Brucella | Bp26 | 46–65, 96–115, 146–160, 176–190, 231–250) |
U | U | U | [91] |
| Borrelia miyamotoi | BOM1093 | 209-308 | U | + | U | [59] |
| Mycoplasma hyorhinis | DnaK | U | U | U | + | [87] |
| Haemophilus ducreyi | DsrA | C-terminal passenger domain | U | U | U | [92] |
| Gram-positive | ||||||
| Streptococcus pneumoniae | Hic | 151-201 | C-terminal (HBD-3) | + | U | [63,64] |
| Streptococcus dysagalactiae | U | U | U | U | + | [69] |
| Streptococcus pyogenes | U | U | Hemopexin-Type Repeats | U | U | [92] |
| Streptococcus suis | U | U | U | U | U | [93] |
| Streptococcus agalactiae | PbsP | 123-422 | U | U | + | [72] |
| Staphylococcus epidermidis | AtlE | 598-839 | U | U | + | [77,79] |
| “ | Aae | U | U | U | U | [90] |
| Staphylococcus aureus | AtlA | U | U | U | U | [88,89] |
| “ | IsdB | 144-270, 334-458 | HBD (s) | U | + | [80] |
| Clostridioides difficile | BclA3 | U | U | U | + | [82] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





