Submitted:
17 June 2024
Posted:
18 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
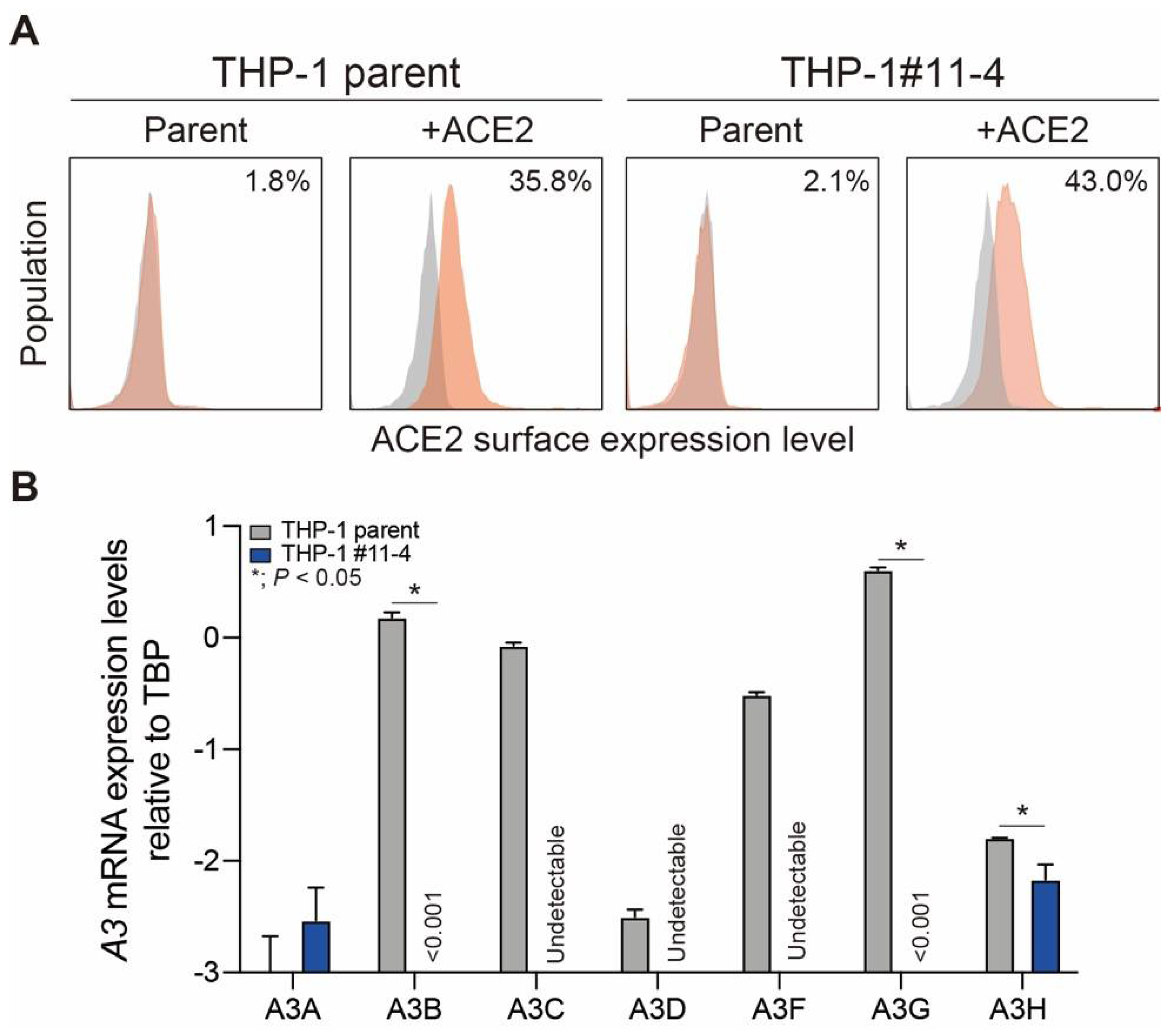
2.1. Creation of THP-1 and A3A-to-A3G-null THP-1 Cells Stably Expressing ACE2 Protein
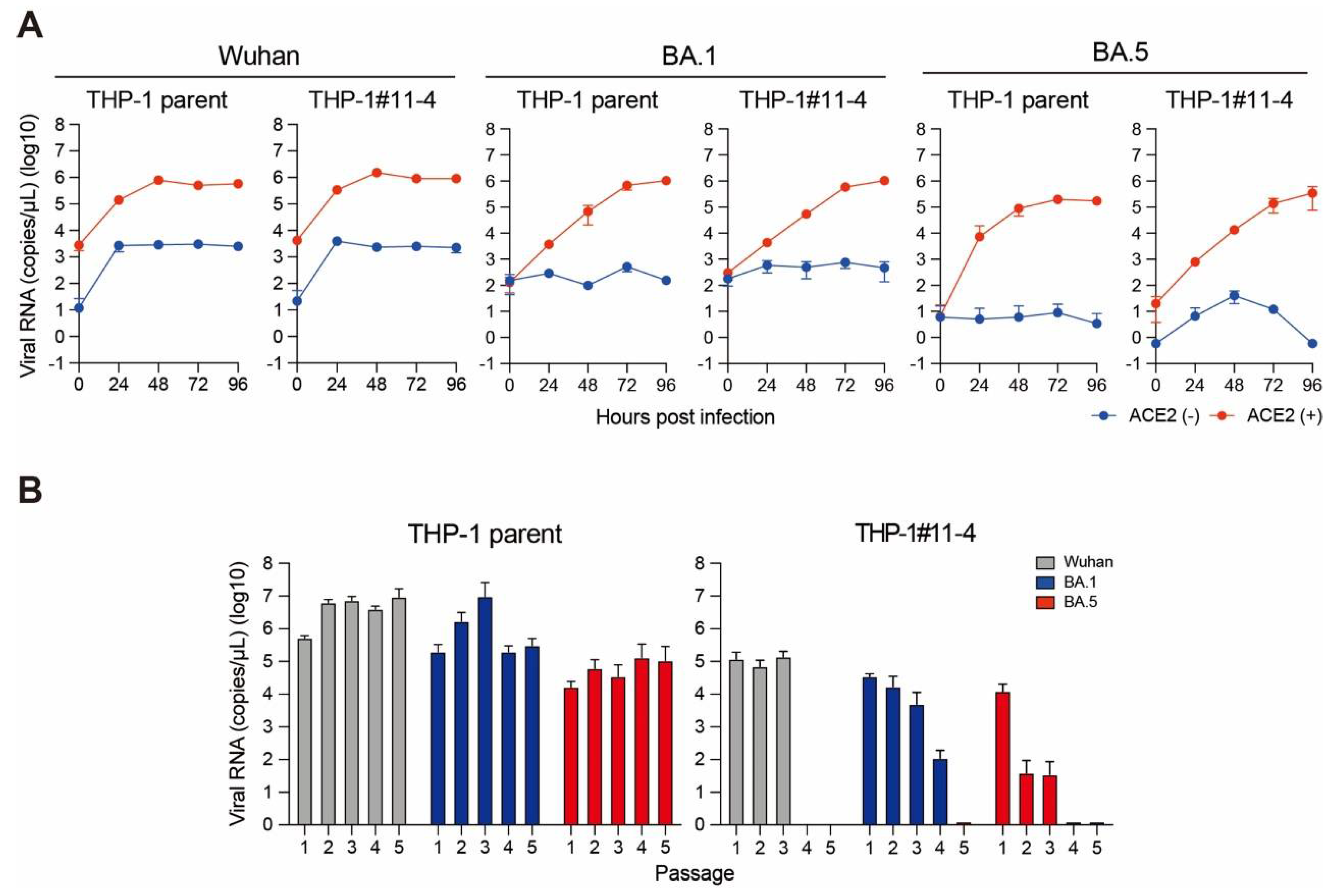
2.2. Effect of A3 Family Proteins on SARS-CoV-2 Replication
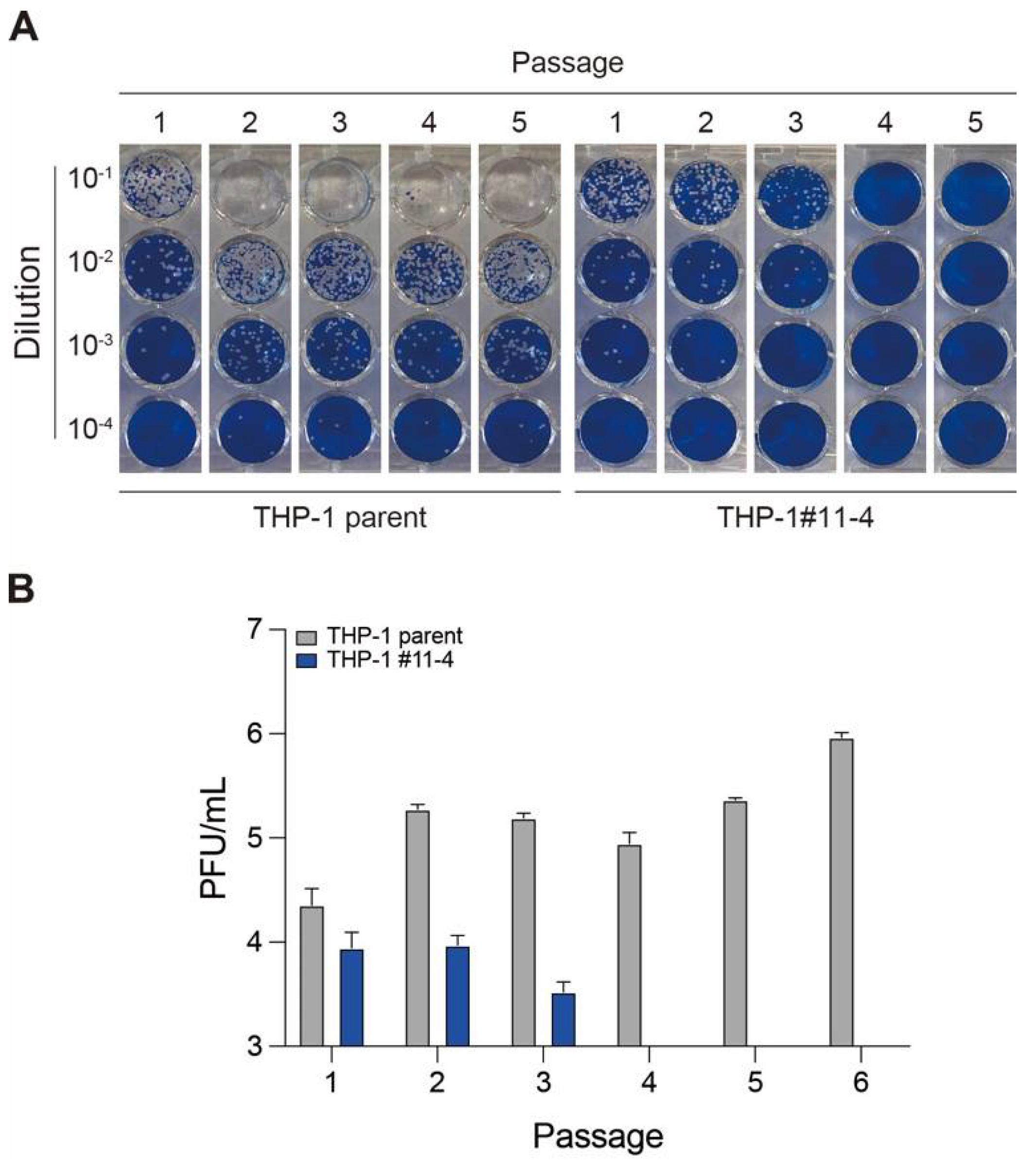
2.3. Effect of A3 Family Proteins on SARS-CoV-2 Infectivity
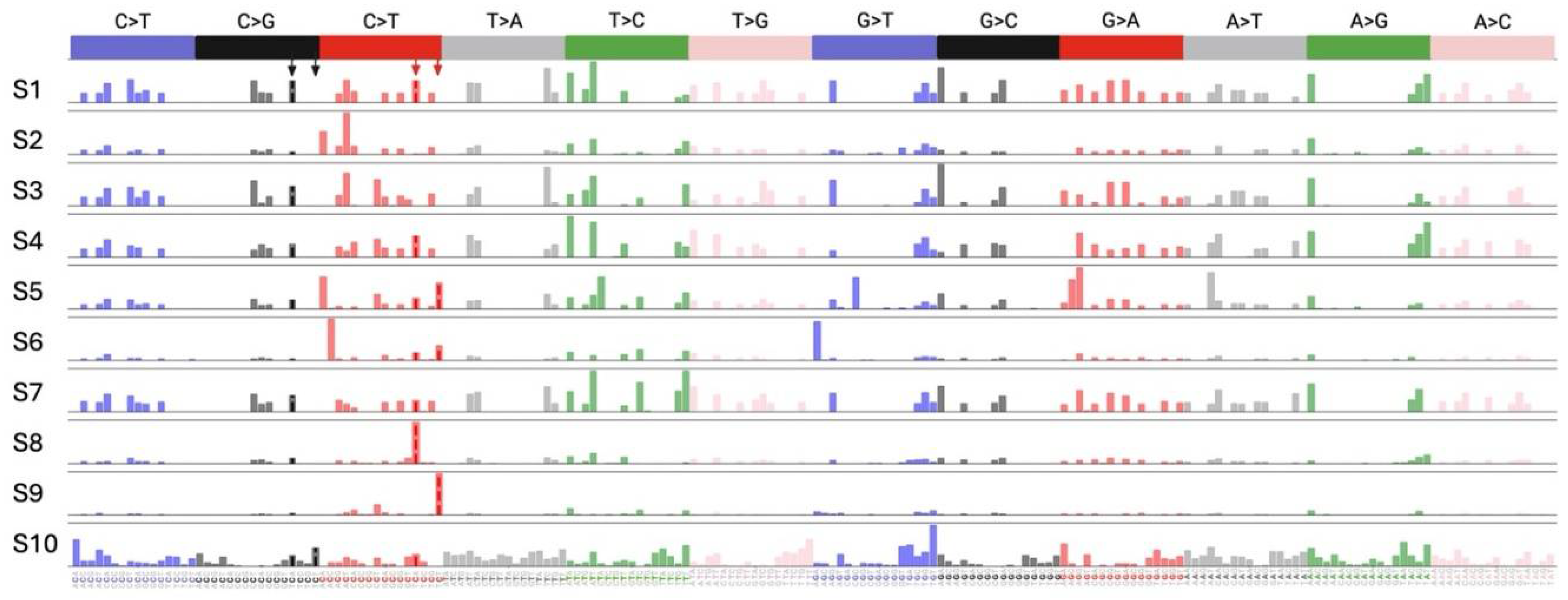
2.4. Effect of A3 Proteins on SARS-CoV-2 Mutagenesis
3. Discussion
4. Materials and Methods
4.1. Cell lines and Culture Conditions
4.2. Virus Preparation
4.3. A3 mRNA Quantification
4.4. ACE2 Transduction
4.5. SARS-CoV-2 Infection
4.6. RT-qPCR for SARS-CoV-2 RNA
4.7. Plaque Assay
4.8. SARS-CoV-2 Whole Genome Sequencing (WGS)
4.9. SARS-CoV-2 Mutational Signature Analysis
4.10. Statistical Analyses
5. Conclusion
Funding
Acknowledgements
Conflicts of Interest
References
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed]
- WHO. COVID-19 Public Health Emergency of International Concern (PHEIC) Global research and innovation forum (Feb 12, 2020), https://www.who.int/publications/m/item/covid-19-public-health-emergency-of-international-concern-(pheic)-global-research-and-innovation-forum.
- WHO. Statement on the fifteenth meeting of the IHR (2005) Emergency Committee on the COVID-19 pandemic (May 5, 2023), https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic.
- Desimmie, B.A.; Delviks-Frankenberrry, K.A.; Burdick, R.C.; Qi, D.; Izumi, T.; Pathak, V.K. Multiple APOBEC3 restriction factors for HIV-1 and one Vif to rule them all. J Mol Biol 2014, 426, 1220–1245. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.S.; Dudley, J.P. APOBECs and virus restriction. Virology 2015, 479-480C, 131–145. [Google Scholar] [CrossRef]
- Ikeda, T.; Yue, Y.; Shimizu, R.; Nasser, H. Potential utilization of APOBEC3-mediated mutagenesis for an HIV-1 functional cure. Front Microbiol 2021, 12, 686357. [Google Scholar] [CrossRef] [PubMed]
- Jonathan, M.; Ikeda, T. APOBEC3 family proteins as drivers of virus evolution. Frontiers in Virology 2023, 3. [Google Scholar] [CrossRef]
- Holmes, R.K.; Malim, M.H.; Bishop, K.N. APOBEC-mediated viral restriction: not simply editing? Trends Biochem Sci 2007, 32, 118–128. [Google Scholar] [CrossRef] [PubMed]
- Koito, A.; Ikeda, T. Apolipoprotein B mRNA-editing, catalytic polypeptide cytidine deaminases and retroviral restriction. Wiley interdisciplinary reviews. RNA 2012, 3, 529–541. [Google Scholar] [CrossRef]
- Cheng, A.Z.; Moraes, S.N.; Shaban, N.M.; Fanunza, E.; Bierle, C.J.; Southern, P.J.; Bresnahan, W.A.; Rice, S.A.; Harris, R.S. APOBECs and herpesviruses. Viruses 2021, 13. [Google Scholar] [CrossRef]
- Willems, L.; Gillet, N.A. APOBEC3 Interference during Replication of Viral Genomes. Viruses 2015, 7, 2999–3018. [Google Scholar] [CrossRef]
- Newman, E.N.; Holmes, R.K.; Craig, H.M.; Klein, K.C.; Lingappa, J.R.; Malim, M.H.; Sheehy, A.M. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Current biology : CB 2005, 15, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Holmes, R.K.; Koning, F.A.; Bishop, K.N.; Malim, M.H. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J Biol Chem 2007, 282, 2587–2595. [Google Scholar] [CrossRef] [PubMed]
- Hultquist, J.F.; Lengyel, J.A.; Refsland, E.W.; LaRue, R.S.; Lackey, L.; Brown, W.L.; Harris, R.S. Human and rhesus APOBEC3D, APOBEC3F, APOBEC3G, and APOBEC3H demonstrate a conserved capacity to restrict Vif-deficient HIV-1. J Virol 2011, 85, 11220–11234. [Google Scholar] [CrossRef]
- Wang, X.; Abudu, A.; Son, S.; Dang, Y.; Venta, P.J.; Zheng, Y.H. Analysis of human APOBEC3H haplotypes and anti-human immunodeficiency virus type 1 activity. J Virol 2011, 85, 3142–3152. [Google Scholar] [CrossRef]
- Refsland, E.W.; Hultquist, J.F.; Harris, R.S. Endogenous origins of HIV-1 G to A hypermutation and restriction in the nonpermissive T cell line CEM2n. PLoS Pathog 2012, 8, e1002800. [Google Scholar] [CrossRef] [PubMed]
- Ooms, M.; Brayton, B.; Letko, M.; Maio, S.M.; Pilcher, C.D.; Hecht, F.M.; Barbour, J.D.; Simon, V. HIV-1 Vif adaptation to human APOBEC3H haplotypes. Cell Host Microbe 2013, 14, 411–421. [Google Scholar] [CrossRef]
- Refsland, E.W.; Hultquist, J.F.; Luengas, E.M.; Ikeda, T.; Shaban, N.M.; Law, E.K.; Brown, W.L.; Reilly, C.; Emerman, M.; Harris, R.S. Natural polymorphisms in human APOBEC3H and HIV-1 Vif combine in primary T lymphocytes to affect viral G-to-A mutation levels and infectivity. PLoS genetics 2014, 10, e1004761. [Google Scholar] [CrossRef] [PubMed]
- Pollpeter, D.; Parsons, M.; Sobala, A.E.; Coxhead, S.; Lang, R.D.; Bruns, A.M.; Papaioannou, S.; McDonnell, J.M.; Apolonia, L.; Chowdhury, J.A.; et al. Deep sequencing of HIV-1 reverse transcripts reveals the multifaceted antiviral functions of APOBEC3G. Nat Microbiol 2018, 3, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Shimizu, R.; Nasser, H.; Carpenter, M.A.; Cheng, A.Z.; Brown, W.L.; Sauter, D.; Harris, R.S. APOBEC3 degradation is the primary function of HIV-1 Vif determining virion infectivity in the myeloid cell line THP-1. mBio 2023, e0078223. [Google Scholar] [CrossRef] [PubMed]
- Miyagi, E.; Opi, S.; Takeuchi, H.; Khan, M.; Goila-Gaur, R.; Kao, S.; Strebel, K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J Virol 2007, 81, 13346–13353. [Google Scholar] [CrossRef]
- Harris, R.S.; Bishop, K.N.; Sheehy, A.M.; Craig, H.M.; Petersen-Mahrt, S.K.; Watt, I.N.; Neuberger, M.S.; Malim, M.H. DNA deamination mediates innate immunity to retroviral infection. Cell 2003, 113, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Rathore, A.; Carpenter, M.A.; Demir, O.; Ikeda, T.; Li, M.; Shaban, N.M.; Law, E.K.; Anokhin, D.; Brown, W.L.; Amaro, R.E.; et al. The local dinucleotide preference of APOBEC3G can be altered from 5'-CC to 5'-TC by a single amino acid substitution. J Mol Biol 2013, 425, 4442–4454. [Google Scholar] [CrossRef] [PubMed]
- Poulain, F.; Lejeune, N.; Willemart, K.; Gillet, N.A. Footprint of the host restriction factors APOBEC3 on the genome of human viruses. PLoS Pathog 2020, 16, e1008718. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, J.P.; Guetard, D.; Henry, M.; Wain-Hobson, S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science 2008, 320, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Verhalen, B.; Starrett, G.J.; Harris, R.S.; Jiang, M. Functional Upregulation of the DNA Cytosine Deaminase APOBEC3B by Polyomaviruses. J Virol 2016, 90, 6379–6386. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Van Doorslaer, K.; Pandey, A.; Espinosa, J.M.; Pyeon, D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evol 2015, 1, vev015. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Kannisto, E.D.; Enriquez, S.M.; Gollnick, P.; Baysal, B.E. APOBEC3A cytidine deaminase induces RNA editing in monocytes and macrophages. Nat Commun 2015, 6, 6881. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Patnaik, S.K.; Taggart, R.T.; Baysal, B.E. The double-domain cytidine deaminase APOBEC3G is a cellular site-specific RNA editing enzyme. Sci Rep 2016, 6, 39100. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Baysal, B.E. Stem-loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. PeerJ 2017, 5, e4136. [Google Scholar] [CrossRef]
- Sharma, S.; Patnaik, S.K.; Kemer, Z.; Baysal, B.E. Transient overexpression of exogenous APOBEC3A causes C-to-U RNA editing of thousands of genes. RNA Biol 2017, 14, 603–610. [Google Scholar] [CrossRef]
- Sharma, S.; Wang, J.; Alqassim, E.; Portwood, S.; Cortes Gomez, E.; Maguire, O.; Basse, P.H.; Wang, E.S.; Segal, B.H.; Baysal, B.E. Mitochondrial hypoxic stress induces widespread RNA editing by APOBEC3G in natural killer cells. Genome Biol 2019, 20, 37. [Google Scholar] [CrossRef]
- Alqassim, E.Y.; Sharma, S.; Khan, A.; Emmons, T.R.; Cortes Gomez, E.; Alahmari, A.; Singel, K.L.; Mark, J.; Davidson, B.A.; Robert McGray, A.J.; et al. RNA editing enzyme APOBEC3A promotes pro-inflammatory M1 macrophage polarization. Commun Biol 2021, 4, 102. [Google Scholar] [CrossRef] [PubMed]
- Milewska, A.; Kindler, E.; Vkovski, P.; Zeglen, S.; Ochman, M.; Thiel, V.; Rajfur, Z.; Pyrc, K. APOBEC3-mediated restriction of RNA virus replication. Sci Rep 2018, 8, 5960. [Google Scholar] [CrossRef] [PubMed]
- Pauli, E.K.; Schmolke, M.; Hofmann, H.; Ehrhardt, C.; Flory, E.; Munk, C.; Ludwig, S. High level expression of the anti-retroviral protein APOBEC3G is induced by influenza A virus but does not confer antiviral activity. Retrovirology 2009, 6, 38. [Google Scholar] [CrossRef] [PubMed]
- Fehrholz, M.; Kendl, S.; Prifert, C.; Weissbrich, B.; Lemon, K.; Rennick, L.; Duprex, P.W.; Rima, B.K.; Koning, F.A.; Holmes, R.K.; et al. The innate antiviral factor APOBEC3G targets replication of measles, mumps and respiratory syncytial viruses. J Gen Virol 2012, 93, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hozumi, Y.; Yin, C.; Wei, G.W. Mutations on COVID-19 diagnostic targets. Genomics 2020. [Google Scholar] [CrossRef]
- Simmonds, P. Rampant C-->U Hypermutation in the Genomes of SARS-CoV-2 and Other Coronaviruses: Causes and Consequences for Their Short- and Long-Term Evolutionary Trajectories. mSphere 2020, 5. [Google Scholar] [CrossRef] [PubMed]
- Nakata, Y.; Ode, H.; Kubota, M.; Kasahara, T.; Matsuoka, K.; Sugimoto, A.; Imahashi, M.; Yokomaku, Y.; Iwatani, Y. Cellular APOBEC3A deaminase drives mutations in the SARS-CoV-2 genome. Nucleic Acids Res 2023, 51, 783–795. [Google Scholar] [CrossRef] [PubMed]
- Di Giorgio, S.; Martignano, F.; Torcia, M.G.; Mattiuz, G.; Conticello, S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci Adv 2020, 6, eabb5813. [Google Scholar] [CrossRef]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; de Cesare, M.; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-host diversity and transmission. Science 2021, 372. [Google Scholar] [CrossRef]
- Kosuge, M.; Furusawa-Nishii, E.; Ito, K.; Saito, Y.; Ogasawara, K. Point mutation bias in SARS-CoV-2 variants results in increased ability to stimulate inflammatory responses. Sci Rep 2020, 10, 17766. [Google Scholar] [CrossRef] [PubMed]
- Siqueira, J.D.; Goes, L.R.; Alves, B.M.; de Carvalho, P.S.; Cicala, C.; Arthos, J.; Viola, J.P.B.; de Melo, A.C.; Soares, M.A. SARS-CoV-2 genomic analyses in cancer patients reveal elevated intrahost genetic diversity. Virus Evol 2021, 7, veab013. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Calabrese, P.; Wang, S.; Qin, C.; Rao, Y.; Feng, P.; Chen, X.S. The roles of APOBEC-mediated RNA editing in SARS-CoV-2 mutations, replication and fitness. Sci Rep 2022, 12, 14972. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Molan, A.M.; Jarvis, M.C.; Carpenter, M.A.; Salamango, D.J.; Brown, W.L.; Harris, R.S. HIV-1 restriction by endogenous APOBEC3G in the myeloid cell line THP-1. J Gen Virol 2019, 100, 1140–1152. [Google Scholar] [CrossRef]
- Browne, E.P.; Allers, C.; Landau, N.R. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology 2009, 387, 313–321. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.D.; Ikeda, T.; Moghadasi, S.A.; Martin, A.S.; Brown, W.L.; Harris, R.S. Natural APOBEC3C variants can elicit differential HIV-1 restriction activity. Retrovirology 2018, 15, 78. [Google Scholar] [CrossRef] [PubMed]
- Gillick, K.; Pollpeter, D.; Phalora, P.; Kim, E.Y.; Wolinsky, S.M.; Malim, M.H. Suppression of HIV-1 infection by APOBEC3 proteins in primary human CD4(+) T cells is associated with inhibition of processive reverse transcription as well as excessive cytidine deamination. J Virol 2013, 87, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, T.; Symeonides, M.; Albin, J.S.; Li, M.; Thali, M.; Harris, R.S. HIV-1 adaptation studies reveal a novel Env-mediated homeostasis mechanism for evading lethal hypermutation by APOBEC3G. PLoS Pathog 2018, 14, e1007010. [Google Scholar] [CrossRef]
- Bishop, K.N.; Verma, M.; Kim, E.Y.; Wolinsky, S.M.; Malim, M.H. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog 2008, 4, e1000231. [Google Scholar] [CrossRef]
- Shaban, N.M.; Shi, K.; Lauer, K.V.; Carpenter, M.A.; Richards, C.M.; Salamango, D.; Wang, J.; Lopresti, M.W.; Banerjee, S.; Levin-Klein, R.; et al. The Antiviral and Cancer Genomic DNA Deaminase APOBEC3H Is Regulated by an RNA-Mediated Dimerization Mechanism. Mol Cell 2018, 69, 75–86. [Google Scholar] [CrossRef]
- Iwatani, Y.; Chan, D.S.; Wang, F.; Stewart-Maynard, K.; Sugiura, W.; Gronenborn, A.M.; Rouzina, I.; Williams, M.C.; Musier-Forsyth, K.; Levin, J.G. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res 2007, 35, 7096–7108. [Google Scholar] [CrossRef]
- Li, X.Y.; Guo, F.; Zhang, L.; Kleiman, L.; Cen, S. APOBEC3G inhibits DNA strand transfer during HIV-1 reverse transcription. J Biol Chem 2007, 282, 32065–32074. [Google Scholar] [CrossRef]
- Belanger, K.; Savoie, M.; Rosales Gerpe, M.C.; Couture, J.F.; Langlois, M.A. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res 2013, 41, 7438–7452. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, D.H.; Gummuluru, S.; Hu, J. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J Virol 2007, 81, 4465–4472. [Google Scholar] [CrossRef]
- Narvaiza, I.; Linfesty, D.C.; Greener, B.N.; Hakata, Y.; Pintel, D.J.; Logue, E.; Landau, N.R.; Weitzman, M.D. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog 2009, 5, e1000439. [Google Scholar] [CrossRef]
- Stavrou, S.; Zhao, W.; Blouch, K.; Ross, S.R. Deaminase-Dead Mouse APOBEC3 Is an In Vivo Retroviral Restriction Factor. J Virol 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Boi, S.; Kolokithas, A.; Shepard, J.; Linwood, R.; Rosenke, K.; Van Dis, E.; Malik, F.; Evans, L.H. Incorporation of mouse APOBEC3 into murine leukemia virus virions decreases the activity and fidelity of reverse transcriptase. J Virol 2014, 88, 7659–7662. [Google Scholar] [CrossRef]
- Wang, X.; Ao, Z.; Chen, L.; Kobinger, G.; Peng, J.; Yao, X. The cellular antiviral protein APOBEC3G interacts with HIV-1 reverse transcriptase and inhibits its function during viral replication. J Virol 2012, 86, 3777–3786. [Google Scholar] [CrossRef]
- Manjunath, L.; Oh, S.; Ortega, P.; Bouin, A.; Bournique, E.; Sanchez, A.; Martensen, P.M.; Auerbach, A.A.; Becker, J.T.; Seldin, M.; et al. APOBEC3B drives PKR-mediated translation shutdown and protects stress granules in response to viral infection. Nat Commun 2023, 14, 820. [Google Scholar] [CrossRef]
- Chiu, Y.L.; Witkowska, H.E.; Hall, S.C.; Santiago, M.; Soros, V.B.; Esnault, C.; Heidmann, T.; Greene, W.C. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc Natl Acad Sci U S A 2006, 103, 15588–15593. [Google Scholar] [CrossRef]
- Kozak, S.L.; Marin, M.; Rose, K.M.; Bystrom, C.; Kabat, D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J Biol Chem 2006, 281, 29105–29119. [Google Scholar] [CrossRef]
- Gallois-Montbrun, S.; Kramer, B.; Swanson, C.M.; Byers, H.; Lynham, S.; Ward, M.; Malim, M.H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J Virol 2007, 81, 2165–2178. [Google Scholar] [CrossRef]
- Niewiadomska, A.M.; Tian, C.; Tan, L.; Wang, T.; Sarkis, P.T.; Yu, X.F. Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J Virol 2007, 81, 9577–9583. [Google Scholar] [CrossRef]
- Soros, V.B.; Yonemoto, W.; Greene, W.C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog 2007, 3, e15. [Google Scholar] [CrossRef]
- Gallois-Montbrun, S.; Holmes, R.K.; Swanson, C.M.; Fernandez-Ocana, M.; Byers, H.L.; Ward, M.A.; Malim, M.H. Comparison of cellular ribonucleoprotein complexes associated with the APOBEC3F and APOBEC3G antiviral proteins. J Virol 2008, 82, 5636–5642. [Google Scholar] [CrossRef]
- Xiao, X.; Yang, H.; Arutiunian, V.; Fang, Y.; Besse, G.; Morimoto, C.; Zirkle, B.; Chen, X.S. Structural determinants of APOBEC3B non-catalytic domain for molecular assembly and catalytic regulation. Nucleic Acids Res 2017, 45, 7494–7506. [Google Scholar] [CrossRef]
- Shirakawa, K.; Takaori-Kondo, A.; Yokoyama, M.; Izumi, T.; Matsui, M.; Io, K.; Sato, T.; Sato, H.; Uchiyama, T. Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nat Struct Mol Biol 2008, 15, 1184–1191. [Google Scholar] [CrossRef]
- Maeda, K.; Almofty, S.A.; Singh, S.K.; Eid, M.M.; Shimoda, M.; Ikeda, T.; Koito, A.; Pham, P.; Goodman, M.F.; Sakaguchi, N. GANP Interacts with APOBEC3G and Facilitates Its Encapsidation into the Virions To Reduce HIV-1 Infectivity. J Immunol 2013, 191, 6030–6039. [Google Scholar] [CrossRef]
- Berger, G.; Durand, S.; Fargier, G.; Nguyen, X.N.; Cordeil, S.; Bouaziz, S.; Muriaux, D.; Darlix, J.L.; Cimarelli, A. APOBEC3A is a specific inhibitor of the early phases of HIV-1 infection in myeloid cells. PLoS Pathog 2011, 7, e1002221. [Google Scholar] [CrossRef]
- Matsuyama, S.; Nao, N.; Shirato, K.; Kawase, M.; Saito, S.; Takayama, I.; Nagata, N.; Sekizuka, T.; Katoh, H.; Kato, F.; et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 2020, 117, 7001–7003. [Google Scholar] [CrossRef]
- Fukuda, M.; Islam, M.S.; Shimizu, R.; Nasser, H.; Rabin, N.N.; Takahashi, Y.; Sekine, Y.; Lindoy, L.F.; Fukuda, T.; Ikeda, T.; et al. Lethal Interactions of SARS-CoV-2 with Graphene Oxide: Implications for COVID-19 Treatment. Acs Appl Nano Mater 2021, 4, 11881–11887. [Google Scholar] [CrossRef]
- Begum, M.M.; Ichihara, K.; Takahashi, O.; Nasser, H.; Jonathan, M.; Tokunaga, K.; Yoshida, I.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; et al. Virological characteristics correlating with SARS-CoV-2 spike protein fusogenicity. Frontiers in Virology 2024, 4. [Google Scholar] [CrossRef]
- Suzuki, R.; Yamasoba, D.; Kimura, I.; Wang, L.; Kishimoto, M.; Ito, J.; Morioka, Y.; Nao, N.; Nasser, H.; Uriu, K.; et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature 2022, 603, 700–705. [Google Scholar] [CrossRef]
- Kimura, I.; Yamasoba, D.; Tamura, T.; Nao, N.; Suzuki, T.; Oda, Y.; Mitoma, S.; Ito, J.; Nasser, H.; Zahradnik, J.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 subvariants, including BA.4 and BA.5. Cell 2022, 185, 3992–4007. [Google Scholar] [CrossRef]
- Saito, A.; Tamura, T.; Zahradnik, J.; Deguchi, S.; Tabata, K.; Anraku, Y.; Kimura, I.; Ito, J.; Yamasoba, D.; Nasser, H.; et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2.75 variant. Cell Host Microbe 2022. [Google Scholar] [CrossRef]
- Islam, M.S.; Rabin, N.N.; Begum, M.M.; Goto, N.; Tagawa, R.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; Sekine, Y.; Ikeda, T.; et al. SARS-CoV-2 inactivation: Assessing the efficacy of GO-anchored filters versus various commercial masks. RSC Appl Interfaces 2024. [Google Scholar] [CrossRef]
- Islam, M.S.; Fukuda, M.; Hossain, M.J.; Rabin, N.N.; Tagawa, R.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; Sekine, Y.; Ikeda, T.; et al. SARS-CoV-2 suppression depending on the pH of graphene oxide nanosheets. Nanoscale Adv 2023, 5, 2413–2417. [Google Scholar] [CrossRef]
- Tamura, T.; Ito, J.; Uriu, K.; Zahradnik, J.; Kida, I.; Anraku, Y.; Nasser, H.; Shofa, M.; Oda, Y.; Lytras, S.; et al. Virological characteristics of the SARS-CoV-2 XBB variant derived from recombination of two Omicron subvariants. Nat Commun 2023, 14, 2800. [Google Scholar] [CrossRef]
- Ito, J.; Suzuki, R.; Uriu, K.; Itakura, Y.; Zahradnik, J.; Kimura, K.T.; Deguchi, S.; Wang, L.; Lytras, S.; Tamura, T.; et al. Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant. Nat Commun 2023, 14, 2671. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endopoints. Am J Epidemiol 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Refsland, E.W.; Stenglein, M.D.; Shindo, K.; Albin, J.S.; Brown, W.L.; Harris, R.S. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res 2010, 38, 4274–4284. [Google Scholar] [CrossRef]
- 10.1093/nar/gkq174.
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136. [Google Scholar] [CrossRef]
- Meng, B.; Abdullahi, A.; Ferreira, I.; Goonawardane, N.; Saito, A.; Kimura, I.; Yamasoba, D.; Gerber, P.P.; Fatihi, S.; Rathore, S.; et al. Altered TMPRSS2 usage by SARS-CoV-2 Omicron impacts infectivity and fusogenicity. Nature 2022, 603, 706–714. [Google Scholar] [CrossRef]
- Kimura, I.; Yamasoba, D.; Nasser, H.; Ito, H.; Zahradnik, J.; Wu, J.; Fujita, S.; Uriu, K.; Sasaki, J.; Tamura, T.; et al. Multiple mutations of SARS-CoV-2 Omicron BA.2 variant orchestrate its virological characteristics. J Virol 2023. [Google Scholar] [CrossRef]
- Tamura, T.; Irie, T.; Deguchi, S.; Yajima, H.; Tsuda, M.; Nasser, H.; Mizuma, K.; Plianchaisuk, A.; Suzuki, S.; Uriu, K.; et al. Virological characteristics of the SARS-CoV-2 Omicron XBB.1.5 variant. Nat Commun 2024, 15, 1176. [Google Scholar] [CrossRef]
- Tamura, T.; Mizuma, K.; Nasser, H.; Deguchi, S.; Padilla-Blanco, M.; Oda, Y.; Uriu, K.; Tolentino, J.E.M.; Tsujino, S.; Suzuki, R.; et al. Virological characteristics of the SARS-CoV-2 BA.2.86 variant. Cell Host Microbe 2024, 32, 170–180. [Google Scholar] [CrossRef]
- Kimura, I.; Yamasoba, D.; Nasser, H.; Zahradnik, J.; Kosugi, Y.; Wu, J.; Nagata, K.; Uriu, K.; Tanaka, Y.L.; Ito, J.; et al. The SARS-CoV-2 spike S375F mutation characterizes the Omicron BA.1 variant. iScience 2022, 25, 105720. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I.; et al. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef] [PubMed]
- Rajib, S.A.; Ogi, Y.; Hossain, M.B.; Ikeda, T.; Tanaka, E.; Kawaguchi, T.; Satou, Y. A SARS-CoV-2 Delta variant containing mutation in the probe binding region used for RT-qPCR test in Japan exhibited atypical PCR amplification and might induce false negative result. J Infect Chemother 2022, 28, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Tao, Q.; Weaver, S.; Sanderford, M.; Caraballo-Ortiz, M.A.; Sharma, S.; Pond, S.L.K.; Miura, S. An Evolutionary Portrait of the Progenitor SARS-CoV-2 and Its Dominant Offshoots in COVID-19 Pandemic. Mol Biol Evol 2021, 38, 3046–3059. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep 2013, 3, 246–259. [Google Scholar] [CrossRef]
- Lamb, K.D.; Luka, M.M.; Saathoff, M.; Orton, R.J.; Phan, M.V.T.; Cotten, M.; Yuan, K.; Robertson, D.L. Mutational signature dynamics indicate SARS-CoV-2's evolutionary capacity is driven by host antiviral molecules. PLoS Comput Biol 2024, 20, e1011795. [Google Scholar] [CrossRef]
- Zhu, B.; Xiao, Y.; Yeager, M.; Clifford, G.; Wentzensen, N.; Cullen, M.; Boland, J.F.; Bass, S.; Steinberg, M.K.; Raine-Bennett, T.; et al. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat Commun 2020, 11, 886. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




