Submitted:
10 June 2024
Posted:
12 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. RNA-Seq Datasets of Wildtype and miRNA-Biogenesis-Deficient Cells
2.2. Comparative Polysome Profiling Datasets of Wildtype and miRNA-Biogenesis- Deficient Cells
2.3. Evolutionarily Conserved miRNA Binding Sites Count
2.4. Statistical Analysis
3. Results
3.1. Description of RNA-Seq and Polysome Profiling Datasets Used in the Previous Studies
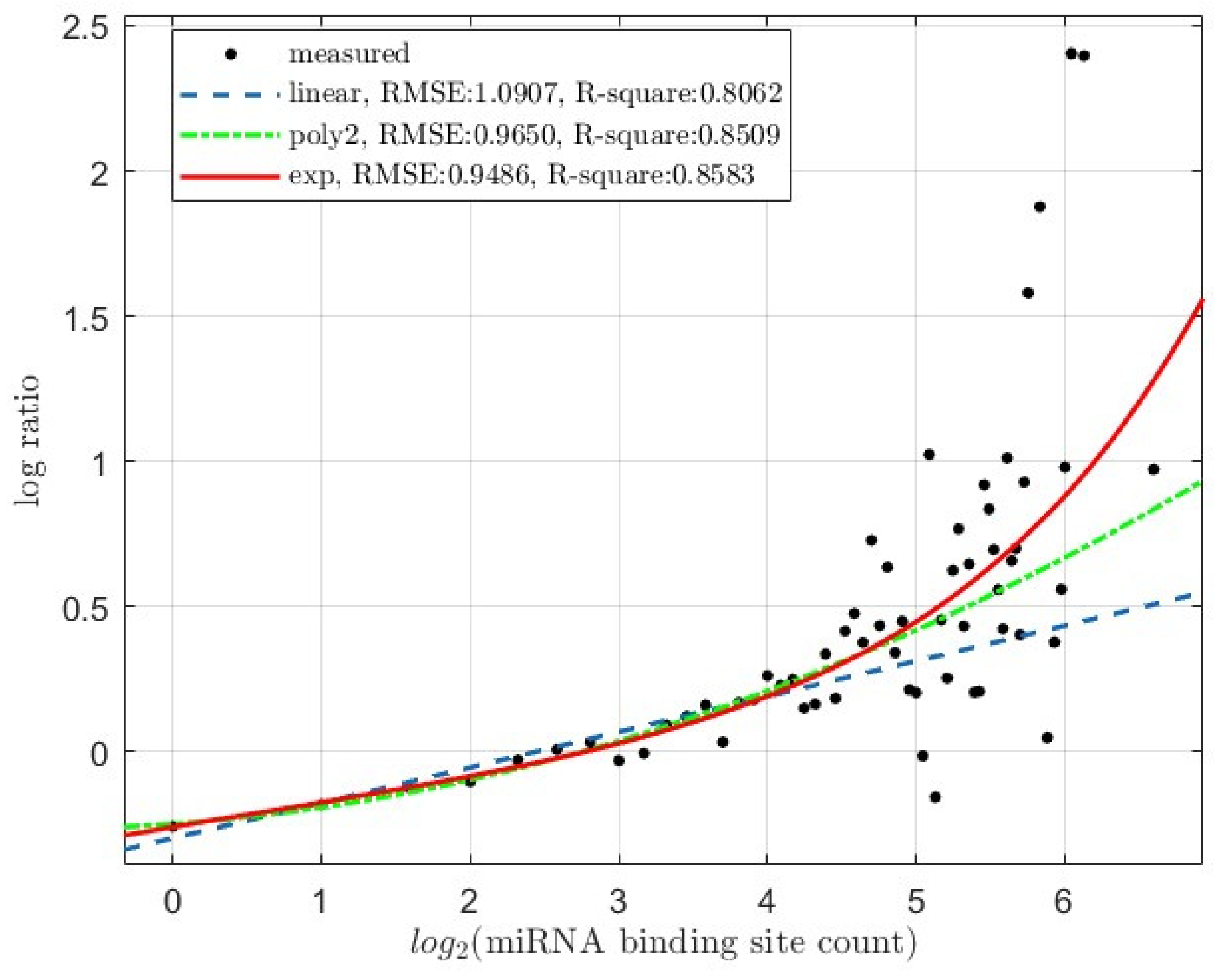
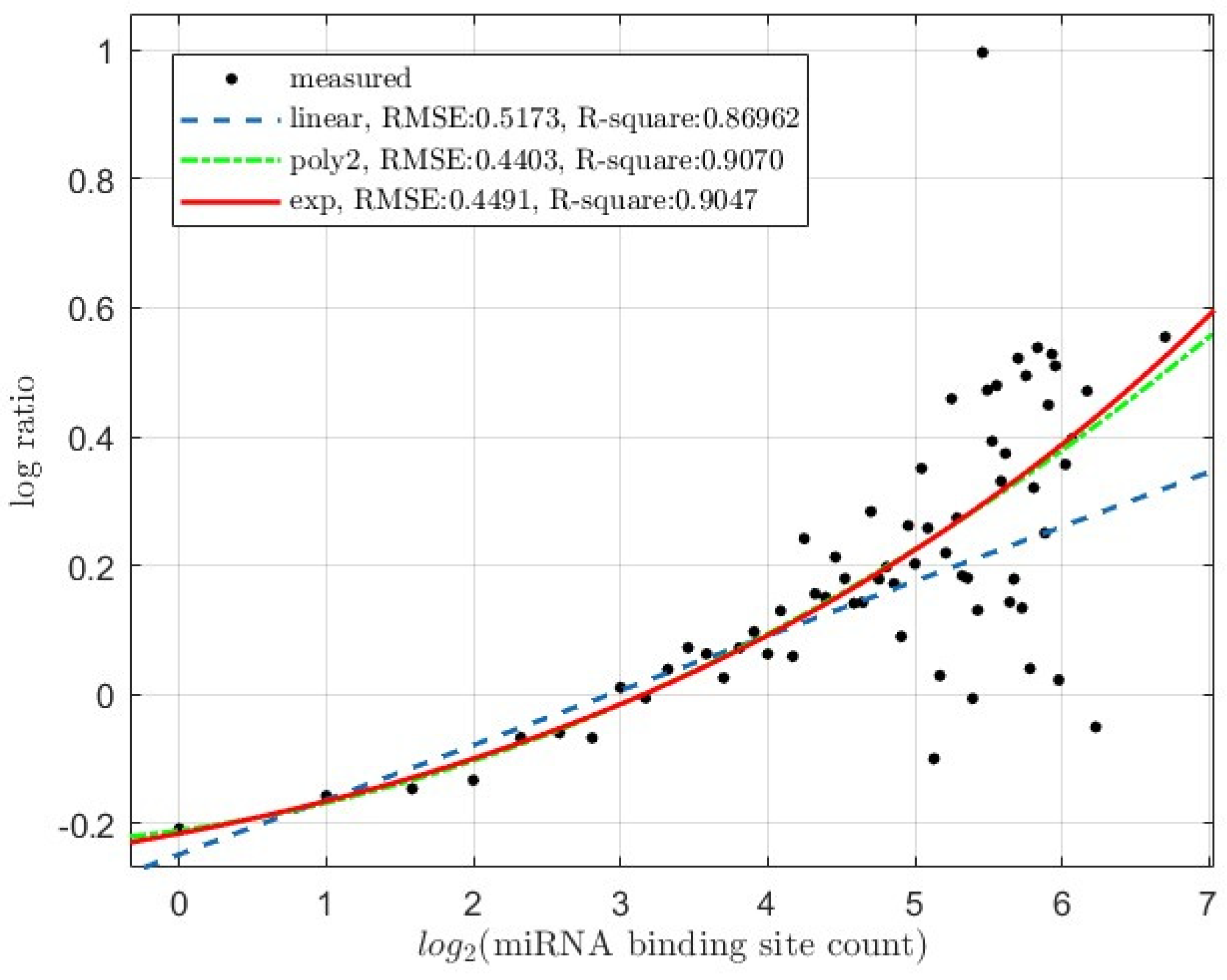
3.2. The levels of miRNA Regulatory Activity Correlates with the miRNA Binding Site Count
3.2.1. Target mRNA Degradation
3.2.2. Target mRNA Translation Inhibition
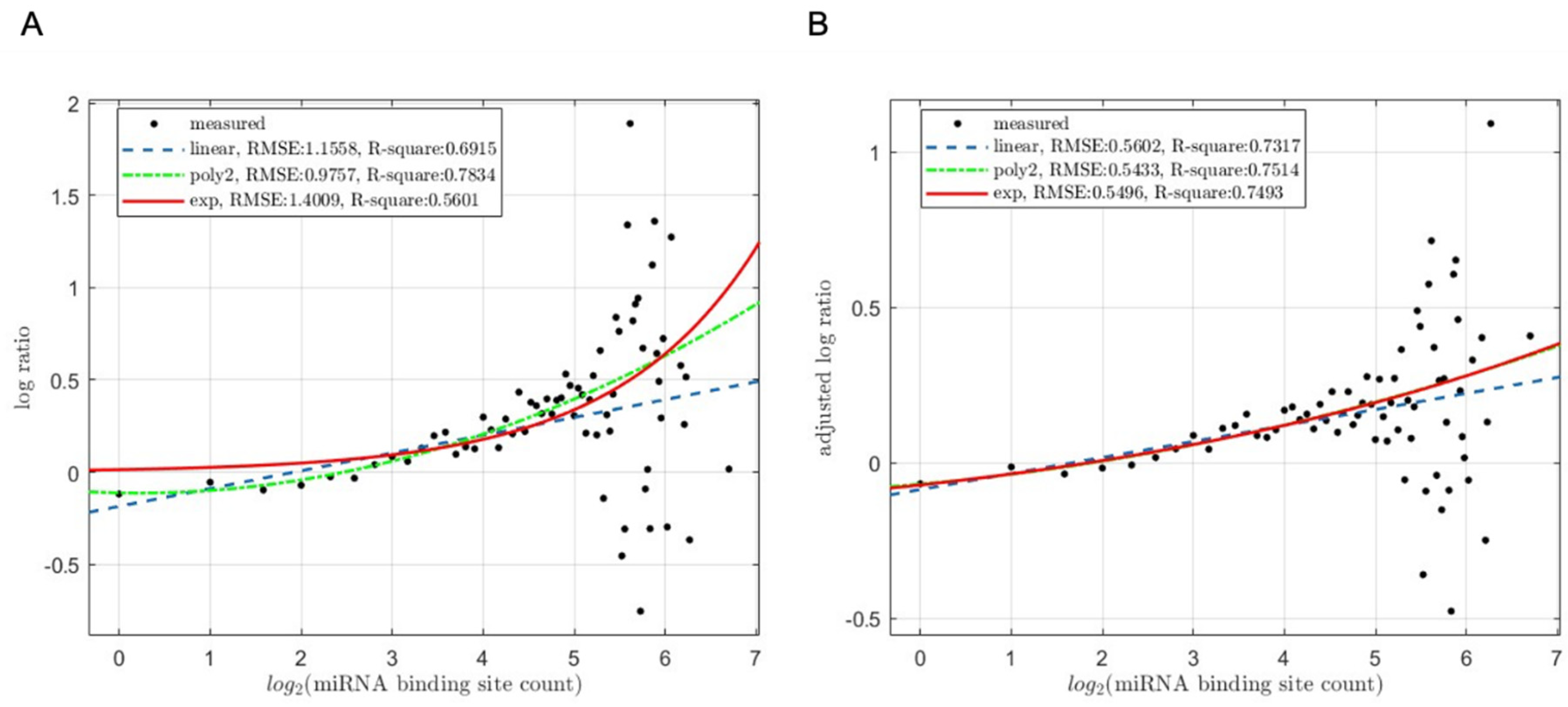
3.3. Reflection of the Synergistic, Instead of Additive, Interactions among miRNA Binding Sites
3.3.1. miRNA-Mediated Target mRNA Degradation
3.3.2. miRNA-Mediated Target mRNA Translation Inhibition
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, L.; Hannon, G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nature reviews genetics 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef]
- MacRae, I.J.; Ma, E.; Zhou, M.; Robinson, C.V.; Doudna, J.A. In vitro reconstitution of the human RISC-loading complex. Proceedings of the National Academy of Sciences 2008, 105, 512–517. [Google Scholar] [CrossRef]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Berezikov, E.; Chung, W.-J.; Willis, J.; Cuppen, E.; Lai, E.C. Mammalian mirtron genes. Molecular cell 2007, 28, 328–336. [Google Scholar] [CrossRef]
- Okamura, K.; Hagen, J.W.; Duan, H.; Tyler, D.M.; Lai, E.C. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 2007, 130, 89–100. [Google Scholar] [CrossRef]
- Cifuentes, D.; Xue, H.; Taylor, D.W.; Patnode, H.; Mishima, Y.; Cheloufi, S.; Ma, E.; Mane, S.; Hannon, G.J.; Lawson, N.D. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science 2010, 328, 1694–1698. [Google Scholar] [CrossRef]
- Cheloufi, S.; Dos Santos, C.O.; Chong, M.M.; Hannon, G.J. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature 2010, 465, 584–589. [Google Scholar] [CrossRef]
- Yang, J.-S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proceedings of the National Academy of Sciences 2010, 107, 15163–15168. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Jin, D.-Y.; McManus, M.T.; Mourelatos, Z. Precursor microRNA-programmed silencing complex assembly pathways in mammals. Molecular cell 2012, 46, 507–517. [Google Scholar] [CrossRef] [PubMed]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 2011, 12, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Qi, Y. RNAi in plants: an argonaute-centered view. The Plant Cell 2016, 28, 272–285. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Schmitter, D.; Filkowski, J.; Sewer, A.; Pillai, R.S.; Oakeley, E.J.; Zavolan, M.; Svoboda, P.; Filipowicz, W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic acids research 2006, 34, 4801–4815. [Google Scholar] [CrossRef]
- Iwakawa, H.O.; Tomari, Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol 2015, 25, 651–665. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front Endocrinol (Lausanne) 2018, 9, 402. [Google Scholar] [CrossRef]
- Jackson, R.J.; Hellen, C.U.; Pestova, T.V. The mechanism of eukaryotic translation initiation and principles of its regulation. Nature reviews Molecular cell biology 2010, 11, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef] [PubMed]
- DeVeale, B.; Swindlehurst-Chan, J.; Blelloch, R. The roles of microRNAs in mouse development. Nature Reviews Genetics 2021, 22, 307–323. [Google Scholar] [CrossRef] [PubMed]
- Nigi, L.; Grieco, G.E.; Ventriglia, G.; Brusco, N.; Mancarella, F.; Formichi, C.; Dotta, F.; Sebastiani, G. MicroRNAs as regulators of insulin signaling: research updates and potential therapeutic perspectives in type 2 diabetes. International journal of molecular sciences 2018, 19, 3705. [Google Scholar] [CrossRef] [PubMed]
- Im, H.-I.; Kenny, P.J. MicroRNAs in neuronal function and dysfunction. Trends in neurosciences 2012, 35, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Rottiers, V.; Näär, A.M. MicroRNAs in metabolism and metabolic disorders. Nature reviews Molecular cell biology 2012, 13, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Svoronos, A.A.; Engelman, D.M.; Slack, F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res 2016, 76, 3666–3670. [Google Scholar] [CrossRef] [PubMed]
- Briskin, D.; Wang, P.Y.; Bartel, D.P. The biochemical basis for the cooperative action of microRNAs. Proc Natl Acad Sci U S A 2020, 117, 17764–17774. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, D. The Pattern of microRNA Binding Site Distribution. Genes (Basel) 2017, 8. [Google Scholar] [CrossRef]
- Wang, D.; Wang, T.; Gill, A.; Hilliard, T.; Chen, F.; Karamyshev, A.L.; Zhang, F. Uncovering the cellular capacity for intensive and specific feedback self-control of the argonautes and MicroRNA targeting activity. Nucleic Acids Res 2020, 48, 4681–4697. [Google Scholar] [CrossRef]
- Tian, S.; Wang, J.; Zhang, F.; Wang, D. Comparative Analysis of microRNA Binding Site Distribution and microRNA-Mediated Gene Expression Repression of Oncogenes and Tumor Suppressor Genes. Genes 2022, 13, 481. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Tian, S.; Tikhonova, E.B.; Karamyshev, A.L.; Wang, J.J.; Zhang, F.; Wang, D. The Enrichment of miRNA-Targeted mRNAs in Translationally Less Active over More Active Polysomes. Biology 2023, 12, 1536. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proceedings of the National Academy of Sciences 2016, 113, E1881–E1889. [Google Scholar] [CrossRef] [PubMed]
- Jeong, G.; Lim, Y.-H.; Kim, Y.-K. Precise mapping of the transcription start sites of human microRNAs using DROSHA knockout cells. BMC genomics 2016, 17, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.X.; Do, B.T.; Webster, D.E.; Khavari, P.A.; Chang, H.Y. Dicer-microRNA-Myc circuit promotes transcription of hundreds of long noncoding RNAs. Nat Struct Mol Biol 2014, 21, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature protocols 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Gandin, V.; Sikström, K.; Alain, T.; Morita, M.; McLaughlan, S.; Larsson, O.; Topisirovic, I. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. Journal of visualized experiments: JoVE 2014. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Padawer, T.; Leighty, R.E.; Wang, D. Duplicate gene enrichment and expression pattern diversification in multicellularity. Nucleic Acids Res 2012, 40, 7597–7605. [Google Scholar] [CrossRef]
- Welte, T.; Goulois, A.; Stadler, M.B.; Hess, D.; Soneson, C.; Neagu, A.; Azzi, C.; Wisser, M.J.; Seebacher, J.; Schmidt, I.; et al. Convergence of multiple RNA-silencing pathways on GW182/TNRC6. Mol Cell 2023, 83, 2478–2492 e2478. [Google Scholar] [CrossRef]
- Wang, D. Discrepancy between mRNA and protein abundance: insight from information retrieval process in computers. Computational biology and chemistry 2008, 32, 462–468. [Google Scholar] [CrossRef]
- Jiang, W.; Guo, Z.; Lages, N.; Zheng, W.J.; Feliers, D.; Zhang, F.; Wang, D. A multi-parameter analysis of cellular coordination of major transcriptome regulation mechanisms. Scientific reports 2018, 8, 5742. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, K.; Kim, K.; Sciulli, C.; Dowd, S.R.; Minden, J.S.; Carthew, R.W. Targets of microRNA regulation in the Drosophila oocyte proteome. Proceedings of the National Academy of Sciences 2005, 102, 12023–12028. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ingolia, N.T.; Weissman, J.S.; Bartel, D.P. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 2010, 466, 835–840. [Google Scholar] [CrossRef] [PubMed]
- Eichhorn, S.W.; Guo, H.; McGeary, S.E.; Rodriguez-Mias, R.A.; Shin, C.; Baek, D.; Hsu, S.-h.; Ghoshal, K.; Villén, J.; Bartel, D.P. mRNA destabilization is the dominant effect of mammalian microRNAs by the time substantial repression ensues. Molecular cell 2014, 56, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Béthune, J.; Artus-Revel, C.G.; Filipowicz, W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO reports 2012, 13, 716–723. [Google Scholar] [CrossRef]
- Djuranovic, S.; Nahvi, A.; Green, R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 2012, 336, 237–240. [Google Scholar] [CrossRef]
- Bazzini, A.A.; Lee, M.T.; Giraldez, A.J. Ribosome profiling shows that miR-430 reduces translation before causing mRNA decay in zebrafish. Science 2012, 336, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Freimer, J.W.; Hu, T.J.; Blelloch, R. Decoupling the impact of microRNAs on translational repression versus RNA degradation in embryonic stem cells. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Hon, L.S.; Zhang, Z. The roles of binding site arrangement and combinatorial targeting in microRNA repression of gene expression. Genome Biol 2007, 8, R166. [Google Scholar] [CrossRef] [PubMed]
- Pasquinelli, A.E. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012, 13, 271–282. [Google Scholar] [CrossRef] [PubMed]
| Samples | Total Reads | NCBI SRA ID | NCBI GEO ID | ||
|---|---|---|---|---|---|
| mESCs GSE55338 |
Dcr KO1 | 185,933,955 | SRR1176702 | GSM1334360 | |
| Dcr KO3 | 207,509,665 | SRR1176703 | GSM1334361 | ||
| Dcr KO4 | 186,427,767 | SRR1176704 | GSM1334362 | ||
| Dcr WT rep1 | 89,965,743 | SRR1176705 | GSM1334363 | ||
| Dcr WT rep1 | 170,831,915 | SRR1176706 | GSM1334364 | ||
| HCT116 GSE80258 |
WT | 198,937,194 | SRR3380994 | GSM2122814 | |
| DroKO | 196,007,170 | SRR3380995 | GSM2122815 | ||
| HCT116 GSE134818 |
WT-Heavy | 35,352,261 | SRR9829887 | GSM3972384 | |
| WT-Light | 35,351,828 | SRR9829888 | GSM3972385 | ||
| KO-Heavy | 35,272,114 | SRR9829889 | GSM3972386 | ||
| KO-Light | 37,403,191 | SRR9829890 | GSM3972387 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





