Submitted:
12 June 2024
Posted:
12 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Animal Breeding
2.2. Daily Health Monitoring
2.3. Grouping
2.4. High-throughput 16S Ribosomal RNA Gene Sequencing
2.5. Bioinformatic Analysis
2.6. Statistical Analysis
2.7. Nucleotide Sequence Accession Numbers
3. Results
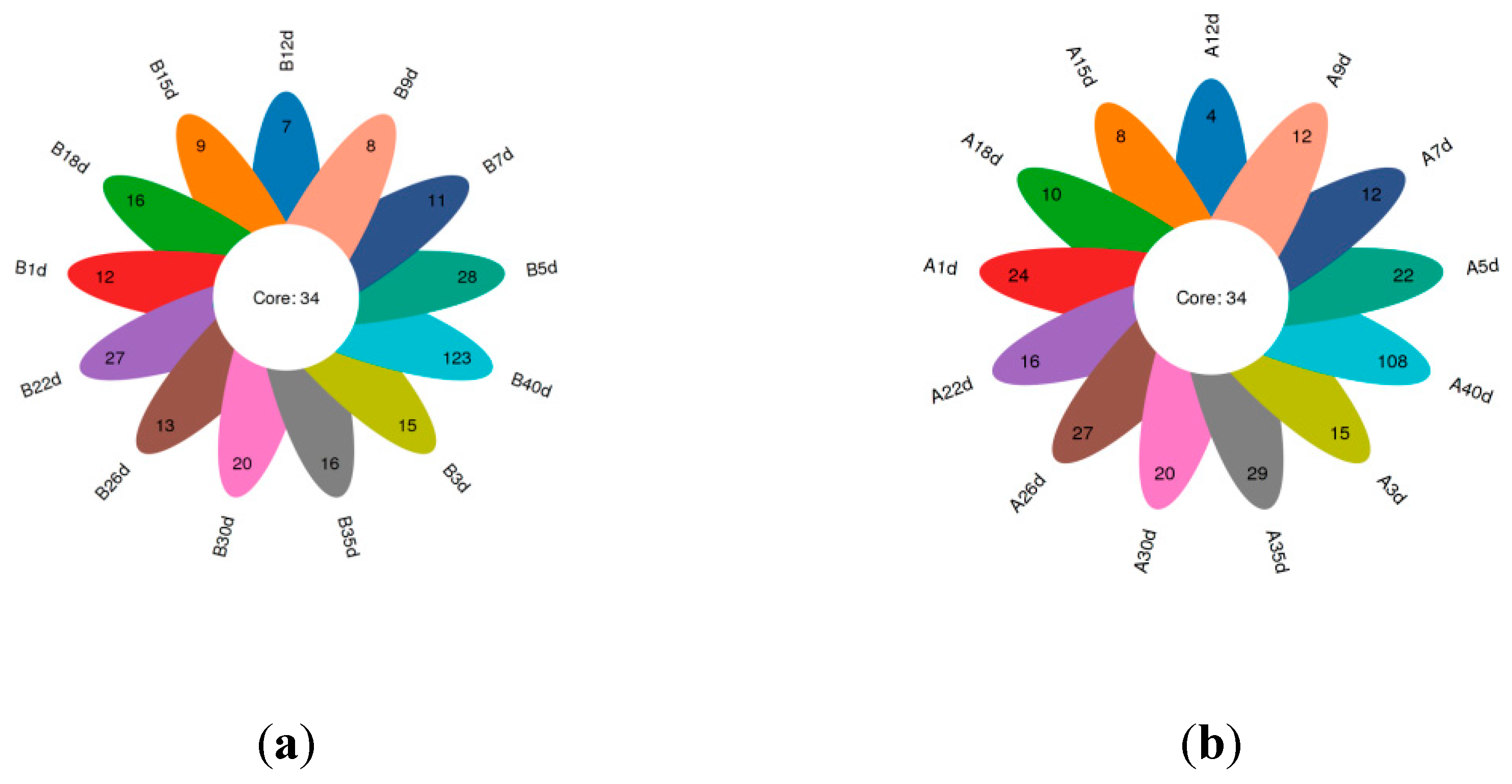
3.1. Data Collection and Diversity of Gut Microbiota
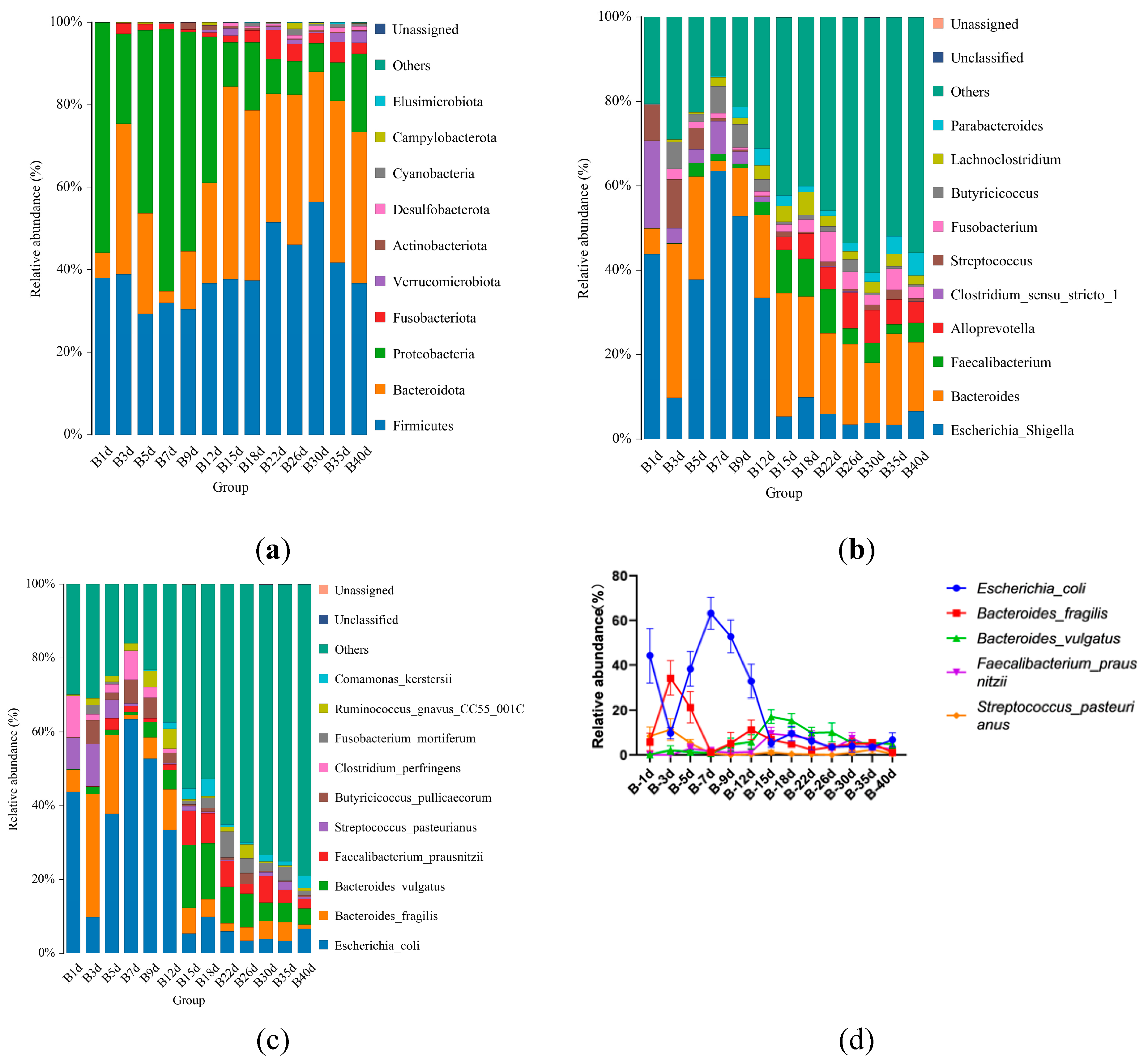
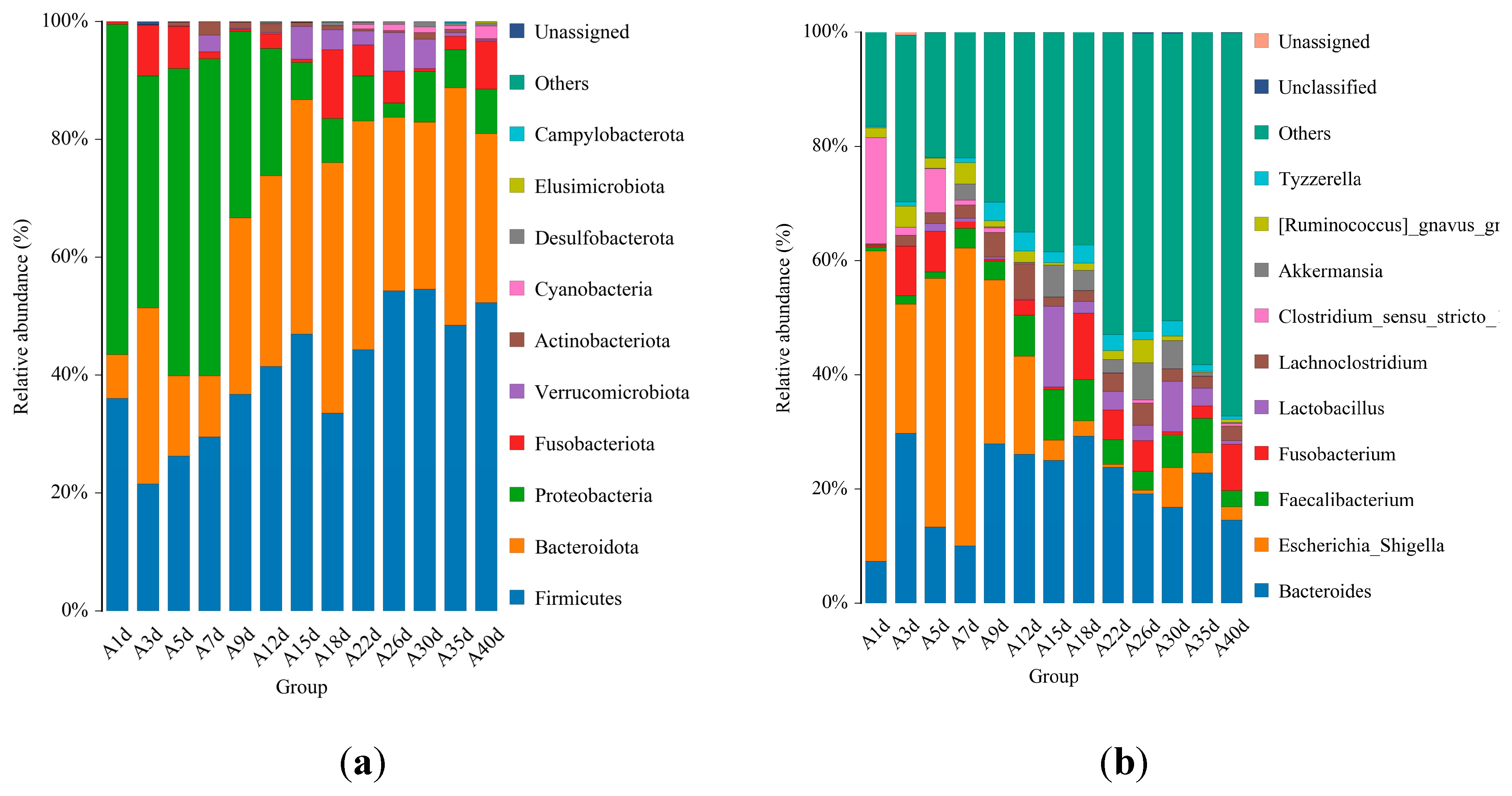
3.2. Taxonomic Composition of Healthy Simmental Calves during Pre-Weaning Period
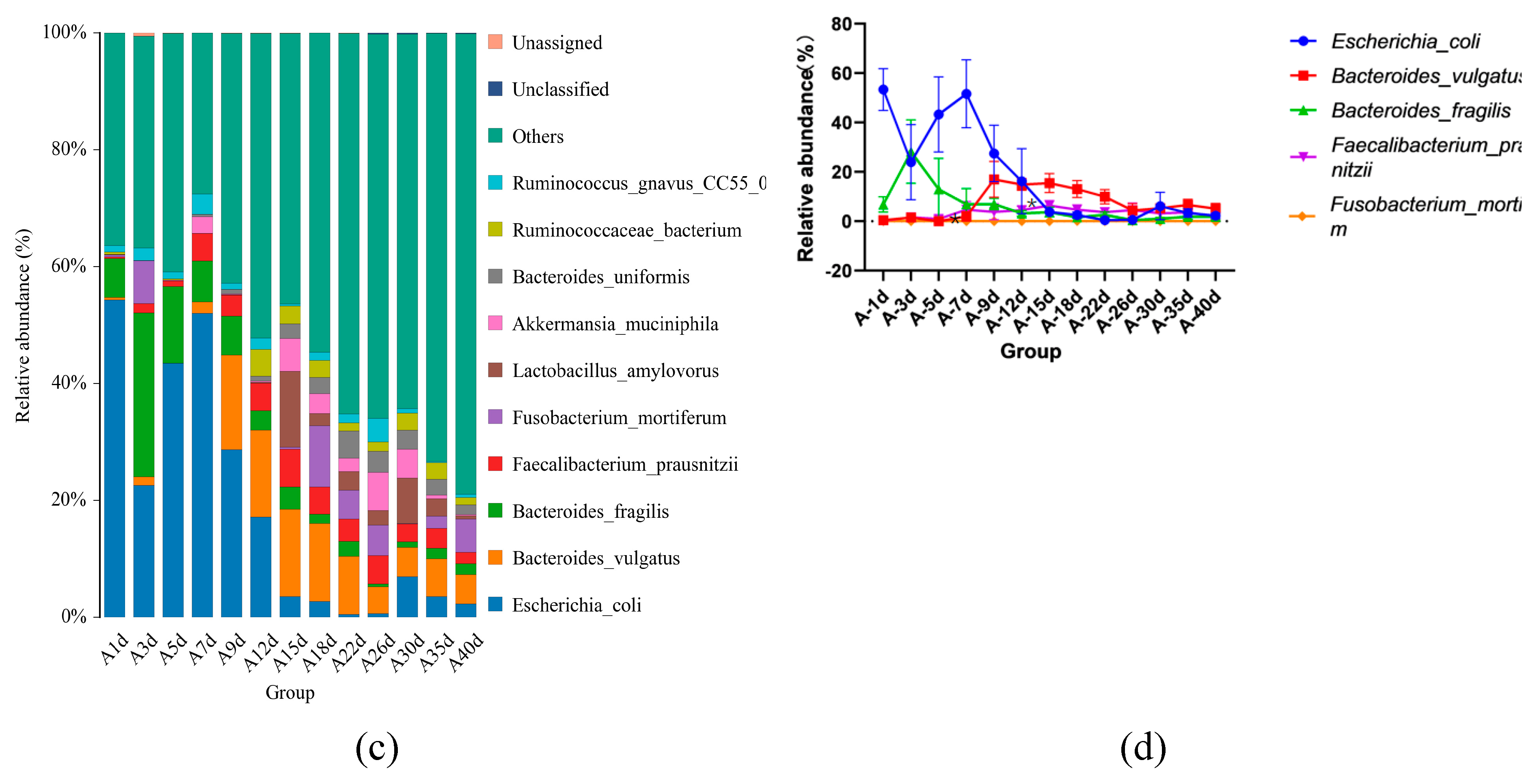
3.3. Taxonomic Composition of Simmental Calves Infected with Diarrhea during Pre-Weaning Period
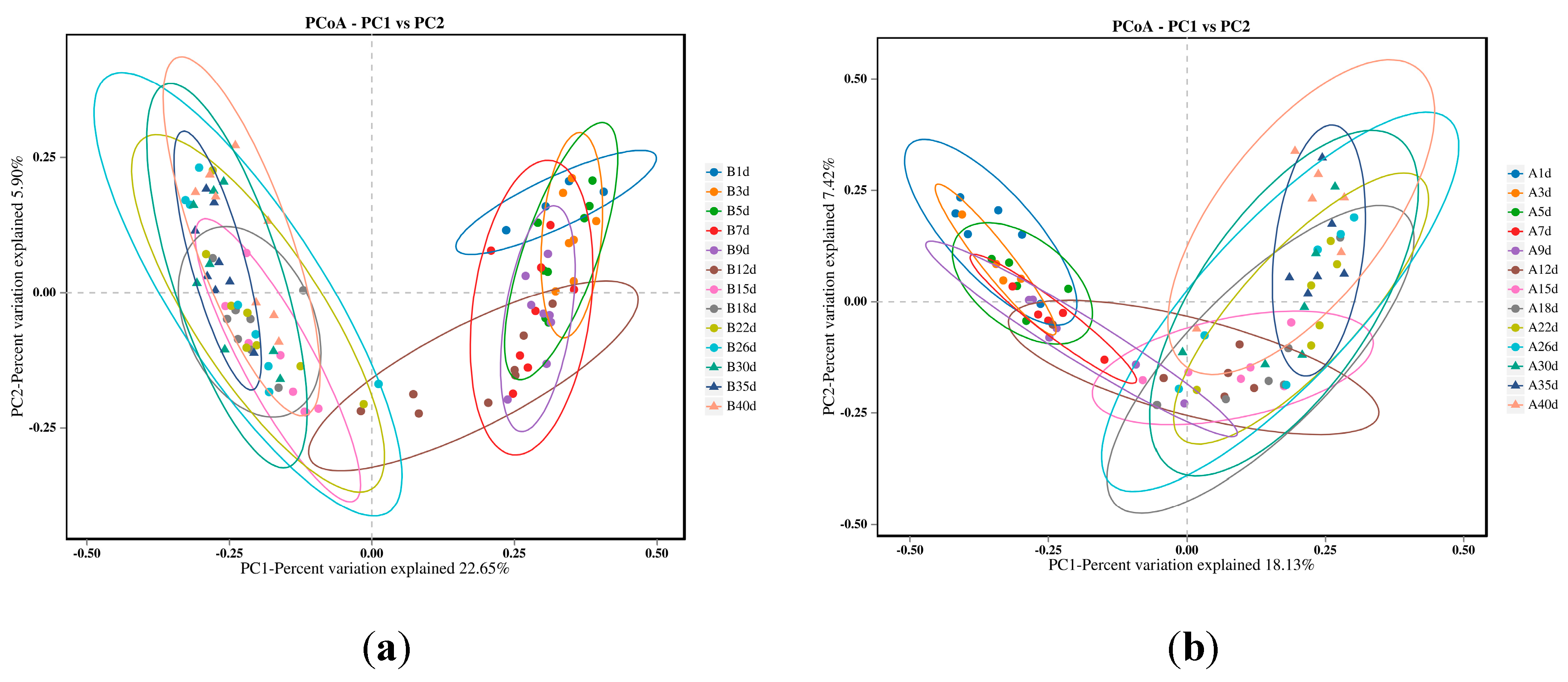
3.4. Similarity Analysis of Gut Microbiota of Simmental Calves during Pre-Weaning Period
3.5. Differential Analysis of Intestinal Microbiota of Simmental Calves between Healthy and Diarrheal Groups
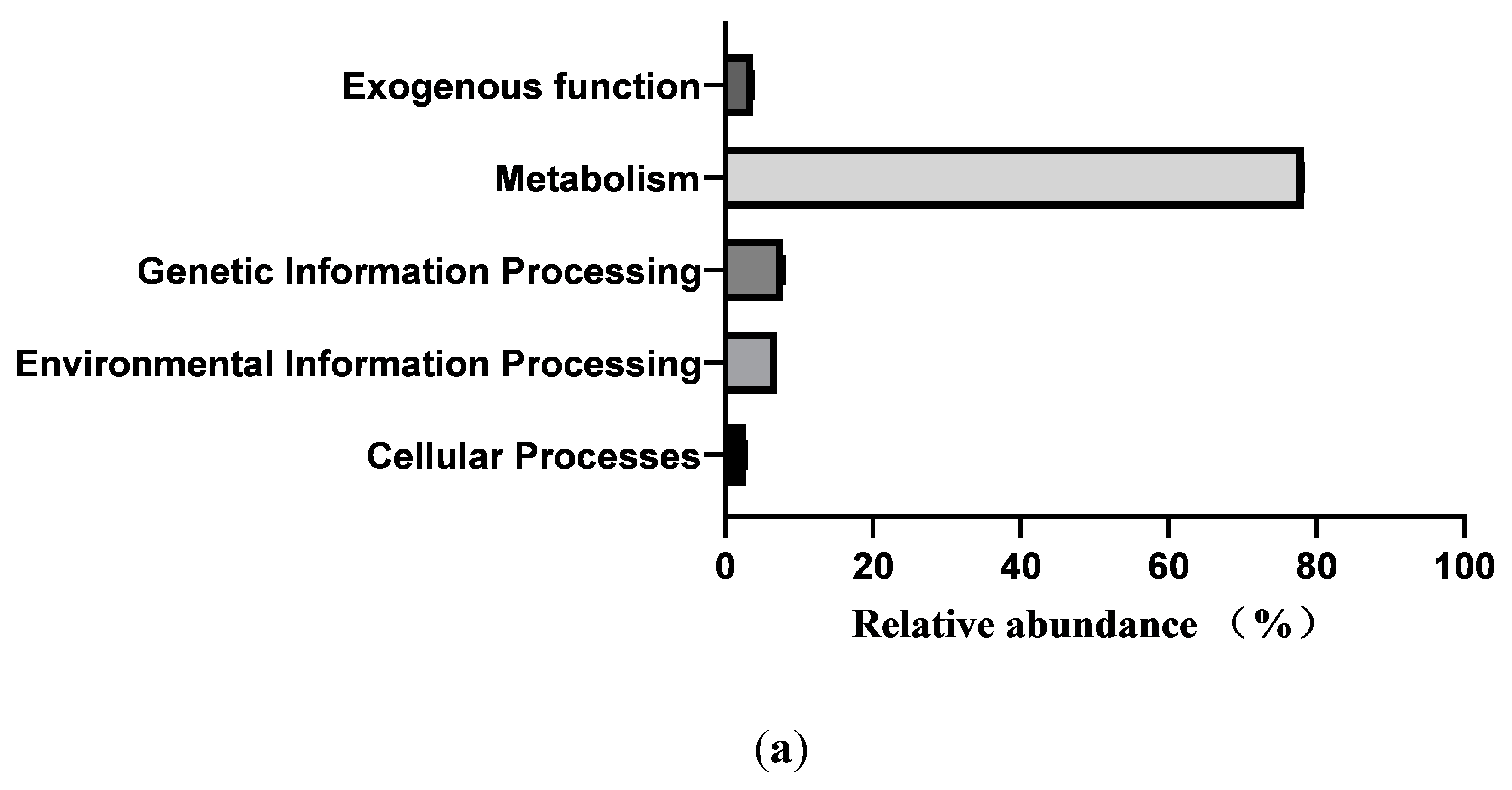
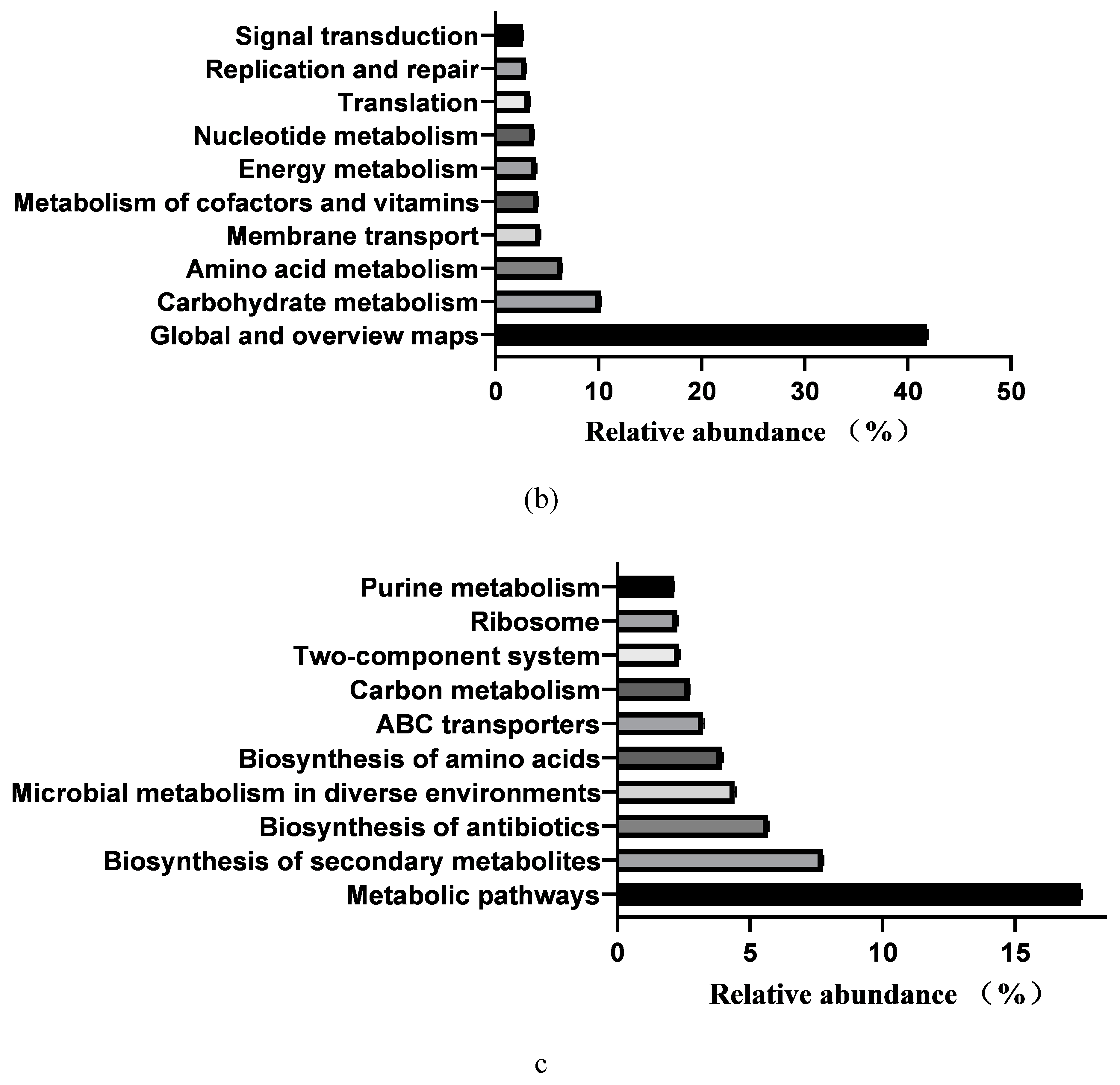
3.6. Prediction of Intestinal Microbial Functions of Simmental Calves
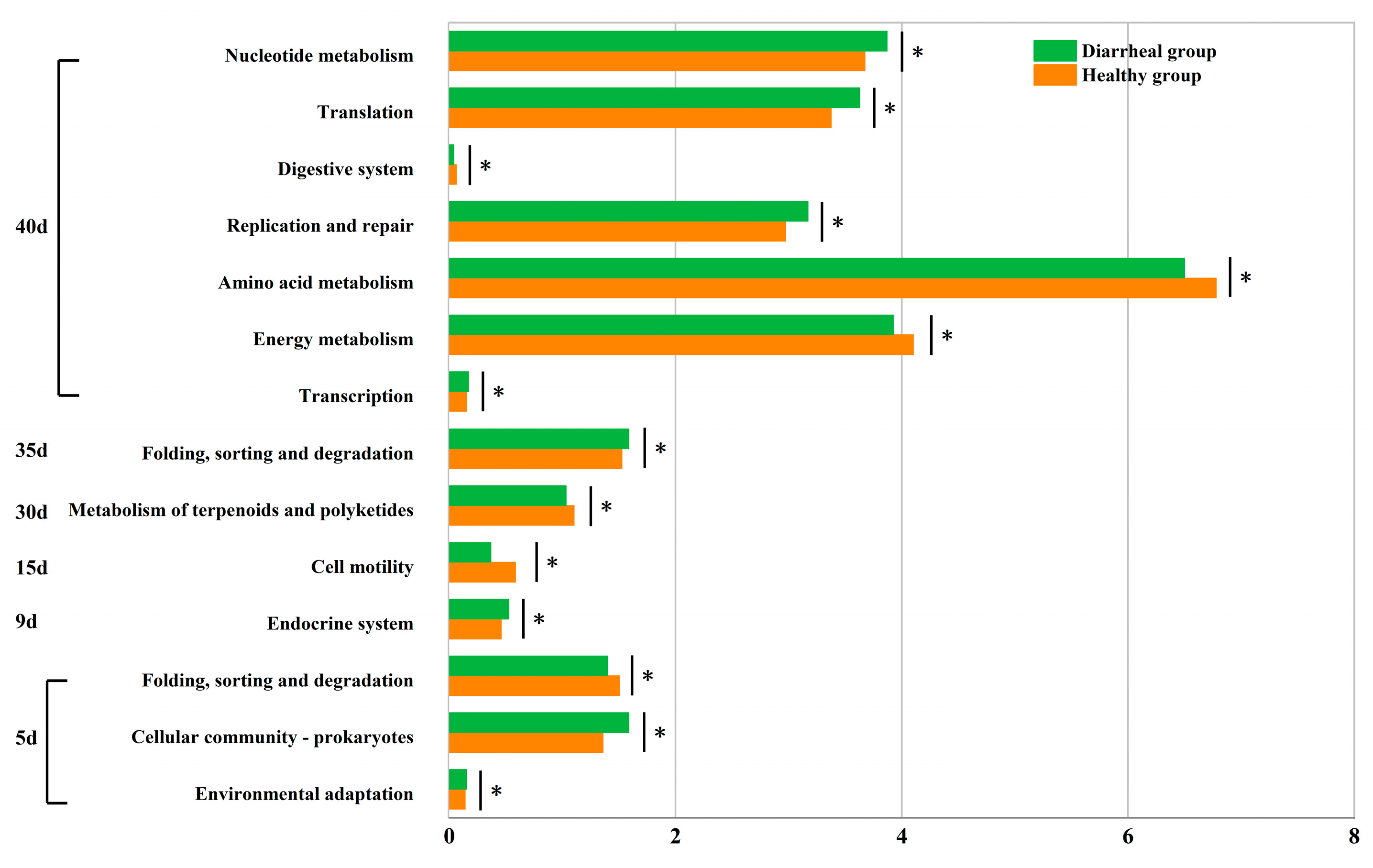
3.7. Differences in Microbial Functions of Calves between Healthy and Diarrheal Groups
4. Discussion
5. Conclusions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.; Wang, Z.; Hu, R.; Peng, Q.; Xue, B.; Wang, L. Comparison of carcass characteristics and meat quality between Simmental crossbred cattle, cattle-yaks and Xuanhan yellow cattle. J Sci Food Agric. 2021, 101, 3927–3932. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Shen, Y.; You, L; Zhang, Y; Su, Z; Peng, G; Deng, J; Zuo, Z; Zhong, Z; Ren, Z; Yu, S; Zong, X; Zhu, Y; Cao, S. Pueraria lobata polysaccharides alleviate neonatal calf diarrhea by modulating gut microbiota and metabolites. Front Vet Sci. 2023, 9, 1024392. [Google Scholar] [CrossRef] [PubMed]
- Tamrat, H.; Mekonnen, N.; Ferede, Y.; Cassini, R.; Belayneh, N. Epidemiological study on calf diarrhea and coccidiosis in dairy farms in Bahir Dar, North West Ethiopia. Ir Vet J. 2020, 73, 14. [Google Scholar] [CrossRef] [PubMed]
- Kim, HS.; Whon, TW.; Sung, H.; Jeong, YS.; Jung, ES.; Shin, N.R.; Hyun, D.W.; Kim, P.S.; Lee, J. Y, Lee, C.H, Bae, J.W. Longitudinal evaluation of fecal microbiota transplantation for ameliorating calf diarrhea and improving growth performance. Nat Commun. 2021, 12, 161. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Gao, Y.; Hu, M.; Hou, J.; Yang, L.; Wang, X.; Du, W.; Liu, J.; Xu, Q. Colonization and development of the gut microbiome in calves. J Anim Sci Biotechnol. 2023, 14, 46. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Tu, Y.; Zhang, N.; Wang, S.; Zhang, F.; Suen, G.; Shao, D.; Li, S.; Diao, Q. Multiomics analysis reveals the presence of a microbiome in the gut of fetal lambs. Gut. 2021, 70, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Guzman, CE.; Wood, J.L.; Egidi, E.; White-Monsant, A.C.; Semenec, L.; Grommen, S.V.H.; Hill-Yardin, E.L.; De Groef, B.; Franks, A.E. A pioneer calf foetus microbiome. Sci Rep. 2020, 10, 17712. [Google Scholar] [CrossRef]
- Schaedler,R. W.; Dubos,R.; Costello, R. The development of the bacterial flora in the gastrointestinal tract of mice. J Exp Med. 1965, 122, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Uyeno, Y.; Sekiguchi, Y.; Kamagata, Y. rRNA-based analysis to monitor succession of faecal bacterial communities in Holstein calves. Lett Appl Microbiol. 2010, 51, 570–577. [Google Scholar] [CrossRef] [PubMed]
- Malmuthuge, N.; Griebel, P.J.; Guan, le.L. Taxonomic identification of commensal bacteria associated with the mucosa and digesta throughout the gastrointestinal tracts of preweaned calves. Appl Environ Microbiol. 2014, 80, 2021–2028. [Google Scholar] [CrossRef]
- Vlková, E.; Trojanová, I.; Rada, V. Distribution of bifidobacteria in the gastrointestinal tract of calves. Folia Microbiol (Praha). 2006, 51, 325–328. [Google Scholar] [CrossRef] [PubMed]
- Guo,S.H.H.Study on intestinal flora diversity and fecal metabolome of calves with diarrhea.2021. Ningxia university, MA thesis.
- Penati, M.; Sala, G.; Biscarini, F.; Boccardo, A.; Bronzo, V.; Castiglioni, B.; Cremonesi, P.; Moroni, P.; Pravettoni, D.; Addis, M.F. Feeding Pre-weaned Calves With Waste Milk Containing Antibiotic Residues Is Related to a Higher Incidence of Diarrhea and Alterations in the Fecal Microbiota. Front Vet Sci. 2021, 8, 650150. [Google Scholar] [CrossRef]
- Lépine, A.F.P.; de Wit, N.; Oosterink, E.; Wichers, H.; Mes, J.; de Vos, P. Lactobacillus acidophilus Attenuates Salmonella-Induced Stress of Epithelial Cells by Modulating Tight-Junction Genes and Cytokine Responses. Front Microbiol. 2018, 9, 1439. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Mustapha, A.; Lin, M.; Zheng, G. Biocontrol of the internalization of Salmonella enterica and Enterohaemorrhagic Escherichia coli in mung bean sprouts with an endophytic Bacillus subtilis. Int J Food Microbiol. 2017, 250, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Memon, F.U.; Yang, Y.; Leghari, I.H.; Lv, F.; Soliman, A.M.; Zhang, W.; Si, H. Transcriptome Analysis Revealed Ameliorative Effects of Bacillus Based Probiotic on Immunity, Gut Barrier System, and Metabolism of Chicken under an Experimentally Induced Eimeria tenella Infection. Genes. 2021, 12, 536. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Nie, C.; Luo, R.; Qi, F.; Bai, X.; Chen, H.; Niu, J.; Chen, C.; Zhang, W. Effects of Multispecies Probiotic on Intestinal Microbiota and Mucosal Barrier Function of Neonatal Calves Infected With E. coli K99. Front Microbiol. 2022, 12, 813245. [Google Scholar] [CrossRef] [PubMed]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef] [PubMed]
- Stöber,M.“Kennzeichen, Anamnese, Grundregeln der Untersuchungstechnik, Allgemeine Untersuchung,” in Die klinische Untersuchung des Rindes, eds G. Dirksen, H. -D. Grunder, M. Stöber, Stuttgart (Erlangen: Enke Verlag), 2012, 75–141.
- Lesmeister,KE. ; Heinrichs,A.J.; Effects of corn processing on growth characteristics, rumen development, and rumen parameters in neonatal dairy calves. J Dairy Sci. 2004, 87, 3439–3450. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, MR.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. Erratum in: Nat Biotechnol. 2019 Sep;37(9):1091. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–6. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, P.; Dingkao, R.; Du, M.; Ahmad, A.A.; Liang, Z.; Zheng, J.; Shen, J.; Yan, P.; Ding, X. Fecal Microbiota Dynamics Reveal the Feasibility of Early Weaning of Yak Calves under Conventional Grazing System. Biology. 2021, 11, 31. [Google Scholar] [CrossRef]
- Song, Y.; Malmuthuge, N.; Steele, M.A.; Guan, L.L. Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol Ecol. 2018, 94. [Google Scholar] [CrossRef] [PubMed]
- Klein-Jöbstl, D.; Schornsteiner, E.; Mann, E.; Wagner, M.; Drillich, M.; Schmitz-Esser, S. Pyrosequencing reveals diverse fecal microbiota in Simmental calves during early development. Front Microbiol. 2014, 5, 622. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, Y.; Huang, K.; Yang, B.; Zhang, Y.; Yu, Z.; Wang, J. Fecal microbiota dynamics and its relationship to diarrhea and health in dairy calves. J Anim Sci Biotechnol. 2022, 13, 132. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Abenthum, A.; Matthes, JM.; Kleeberger, D.; Ege, M.J.; Hölzel, C.; Bauer, J.; Schwaiger, K. Development and genetic influence of the rectal bacterial flora of newborn calves. Vet Microbiol. 2012, 161, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Willems, A.; Collins, MD. 16S rRNA gene similarities indicate that Hallella seregens (Moore and Moore) and Mitsuokella dentalis (Haapsalo et al.) are genealogically highly related and are members of the genus Prevotella: emended description of the genus Prevotella (Shah and Collins) and description of Prevotella dentalis comb. nov. Int J Syst Bacteriol. 1995, 45, 832–836. [Google Scholar] [CrossRef] [PubMed]
- Trachsel, J.; Humphrey, S.; Allen, H.K. Butyricicoccus porcorum sp. nov., a butyrate-producing bacterium from swine intestinal tract. Int J Syst Evol Microbiol. 2018, 68, 1737–1742. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Tickle, T.L.; Sokol, H.; Gevers, D.; Devaney, K.L.; Ward, D.V.; Reyes, J.A.; Shah, S.A.; LeLeiko, N.; Snapper, S.B.; Bousvaros, A.; Korzenik, J.; Sands, B.E.; Xavier, R. J, Huttenhower, C. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012, 13, R79. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Nishida, A.; Fujimoto, T.; Fujii, M.; Shioya, M.; Imaeda, H.; Inatomi, O.; Bamba, S.; Sugimoto, M.; Andoh, A. Reduced Abundance of Butyrate-Producing Bacteria Species in the Fecal Microbial Community in Crohn's Disease. Digestion. 2016, 93, 59–65. [Google Scholar] [CrossRef]
- Foditsch,C. ;Pereira,R.V.;Ganda,E.K.;Gomez,M.S.;Marques,E.C.;Santin,T.;Bicalho,R.C.Oral Administration of Faecalibacterium prausnitzii Decreased the Incidence of Severe Diarrhea and Related Mortality Rate and Increased Weight Gain in Preweaned Dairy Heifers. PLoS One. 2015, 10, e0145485. [Google Scholar]
- Li, Y.; Li, X.; Nie, C.; Wu, Y.; Luo, R.; Chen, C.; Niu, J.; Zhang, W. Effects of two strains of Lactobacillus isolated from the feces of calves after fecal microbiota transplantation on growth performance, immune capacity, and intestinal barrier function of weaned calves. Front Microbiol. 2023, 14, 1249628. [Google Scholar] [CrossRef] [PubMed]
- Foditsch, C.; Santos, T.M.; Teixeira, A.G.; Pereira, R.V.; Dias, J.M.; Gaeta, N.; Bicalho, R.C. Isolation and characterization of Faecalibacterium prausnitzii from calves and piglets. PLoS One. 2014, 9, e116465. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Feng, S.; Wang, Z.; He, J.; Zhang, Z.; Zou, H.; Wu, Z.; Liu, X.; Wei, H.; Tao, S. Limosilactobacillus mucosae-derived extracellular vesicles modulates macrophage phenotype and orchestrates gut homeostasis in a diarrheal piglet model. NPJ Biofilms Microbiomes. 2023, 9, 33. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, R.; Burdukiewicz, M.; Schierack, P. A systematic review and meta-analysis of the epidemiology of pathogenic Escherichia coli of calves and the role of calves as reservoirs for human pathogenic E. coli. Front Cell Infect Microbiol. 2015, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Blanton, L.V.; Frese, S.A.; Charbonneau, M.; Mills, D.A.; Gordon, J.I. Cultivating healthy growth and nutrition through the gut microbiota. Cell. 2015, 161, 36–48. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- McNeil, NI. The contribution of the large intestine to energy supplies in man. Am J Clin Nutr. 1984, 39, 338–342. [Google Scholar] [CrossRef] [PubMed]
| Healthy group | 1d | 3d | 5d | 7d | 9d | 12d | 15d | 18d | 22d | 26d | 30d | 35d | 40d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1d | 0 | ||||||||||||
| 3d | r=0.398* | 0 | |||||||||||
| 5d | r=0.402* | r=-0.001 | 0 | ||||||||||
| 7d | r=0.340* | r=0.363* | r=0.328* | 0 | |||||||||
| 9d | r=0.718* | r=0.544* | r=0.491* | r=-0.021 | 0 | ||||||||
| 12d | r=0.861* | r=0.789* | r=0.667* | r=0.199* | r=0.137* | 0 | |||||||
| 15d | r=0.889* | r=1* | r=0.988* | r=0.871* | r=0.954* | r=0.646* | 0 | ||||||
| 18d | r=1* | r=1* | r=0.996* | r=0.947* | r=0.992* | r=0.845* | r=0.027 | 0 | |||||
| 22d | r=1* | r=0.985* | r=0.978* | r=0.925* | r=0.951* | r=0.801* | r=0.388* | r=0.122 | 0 | ||||
| 26d | r=0.985* | r=0.961* | r=0.962* | r=0.900* | r=0.934* | r=0.735* | r=0.314* | r=0.068 | r=-0.109 | 0 | |||
| 30d | r=1* | r=1* | r=0.996* | r=0.965* | r=0.996* | r=0.907* | r=0.525* | r=0.213* | r=-0.034 | r=-0.634 | 0 | ||
| 35d | r=1* | r=1* | r=0.999* | r=0.996* | r=1* | r=0.969* | r=0.824* | r=0.422* | r=0.181* | r=0.007 | r=-0.047 | 0 | |
| 40d | r=1* | r=0.998* | r=0.991* | r=0.980* | r=0.996* | r=0.906* | r=0.611* | r=0.413* | r=0.252* | r=0.014 | r=0.091 | r=0.064 | 0 |
| Diarrhea group | 1d | 3d | 5d | 7d | 9d | 12d | 15d | 18d | 22d | 26d | 30d | 35d | 40d |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1d | 0 | ||||||||||||
| 3d | r=0.017 | 0 | |||||||||||
| 5d | r=0.362* | r=0.125 | 0 | ||||||||||
| 7d | r=0.533* | r=0.409* | r=0.022 | 0 | |||||||||
| 9d | r=0.635* | r=0.619* | r=0.024 | r=-0.192 | 0 | ||||||||
| 12d | r=0.861* | r=0.833* | r=0.709* | r=0.347* | r=0.128 | 0 | |||||||
| 15d | r=0.938* | r=1* | r=0.924* | r=0.728* | r=0.425* | r=-0.099 | 0 | ||||||
| 18d | r=0.942* | r=0.976* | r=0.887* | r=0.728* | r=0.474* | r=0.039 | r=-0.110 | 0 | |||||
| 22d | r=0.983* | r=0.996* | r=0.979* | r=0.867* | r=0.648* | r=0.109 | r=0.051 | r=-0.011 | 0 | ||||
| 26d | r=0.935* | r=0.976* | r=0.864* | r=0.715* | r=0.611* | r=0.094 | r=0.132 | r=-0.057 | r=-0.185 | 0 | |||
| 30d | r=0.972* | r=0.968* | r=0.949* | r=0.888* | r=0.709* | r=0.240* | r=0.174 | r=-0.002 | r=-0.067 | r=-0.204 | 0 | ||
| 35d | r=0.998* | r=1* | r=0.997* | r=0.992* | r=0.898* | r=0.583* | r=0.462* | r=0.302* | r=-0.019 | r=-0.093 | r=-0.004 | 0 | |
| 40d | r=0.959* | r=0.968* | r=0.947* | r=0.912* | r=0.863* | r=0.561* | r=0.581* | r=0.435* | r=0.146 | r=0.056 | r=0.105 | r=-0.098 | 0 |
| Day | Bacteria | Group | P-value | |
| A | B | |||
| 1d | L.johnsonii | 0.012±0.010% | 0.030±0.019% | 0.088 |
| Limosilactobacillus | 0 | 0.035±0.021% | 0.088 | |
| P.mirabilis | 0.073±0.071% | 0.043±0.017% | 0.088 | |
| 3d | P.russellii | 0.023±0.018% | 1.330±0.720% | 0.008 |
| F.prausnitzii | 0.029±0.023% | 1.619±1.461% | 0.014 | |
| E.ramosum | 0.778±0.416% | 2.283±0.611% | 0.023 | |
| L.johnsonii | 0.002±0.002% | 1.208±1.113% | 0.023 | |
| 5d | F.umbilicata | 0 | 0.269±0.159% | 0.010 |
| G.genomosp._3 | 0.057±0.037% | 2.764±1.556% | 0.019 | |
| C.pharyngocola | 0.309±0.309% | 1.376±0.609% | 0.040 | |
| 7d | S.mitis | 0.380±0.380% | 0.005±0.008% | 0.013 |
| E.ramosum | 0.638±0.252% | 0.087±0.053% | 0.019 | |
| P.mirabilis | 0.180±0.099% | 0.026±0.017% | 0.019 | |
| A.muciniphila | 2.967±2.760% | 0.004±0.003% | 0.040 | |
| L.amylovorus | 0.011±0.004% | 0.002±0.001% | 0.040 | |
| 9d | P.russellii | 0.032±0.026% | 6.153±5.149% | 0.004 |
| L.murinus | 8.123±7.134% | 0.128±0.127% | 0.010 | |
| F.necrophorum | 0.071±0.042% | 0.001±0.001% | 0.012 | |
| S.mitis | 0.012±0.018% | 8.893±0.003% | 0.012 | |
| A.muciniphila | 0.159±0.096% | 0.008±0.006% | 0.020 | |
| B.vulgatus | 16.951±7.283% | 4.473±2.922% | 0.020 | |
| L.johnsonii | 0.262±0.253% | 2.409±1.656% | 0.020 | |
| P.dorei | 0.003±0.003% | 0.005±0.005% | 0.020 | |
| K.pneumoniae | 1.384±1.285% | 0.003±0.002% | 0.028 | |
| E.coli | 27.465±11.439% | 52.753±7.410% | 0.039 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





