Submitted:
31 May 2024
Posted:
03 June 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Isolation and Purification of Bacteriophage
2.3. Host Adsorption Assay
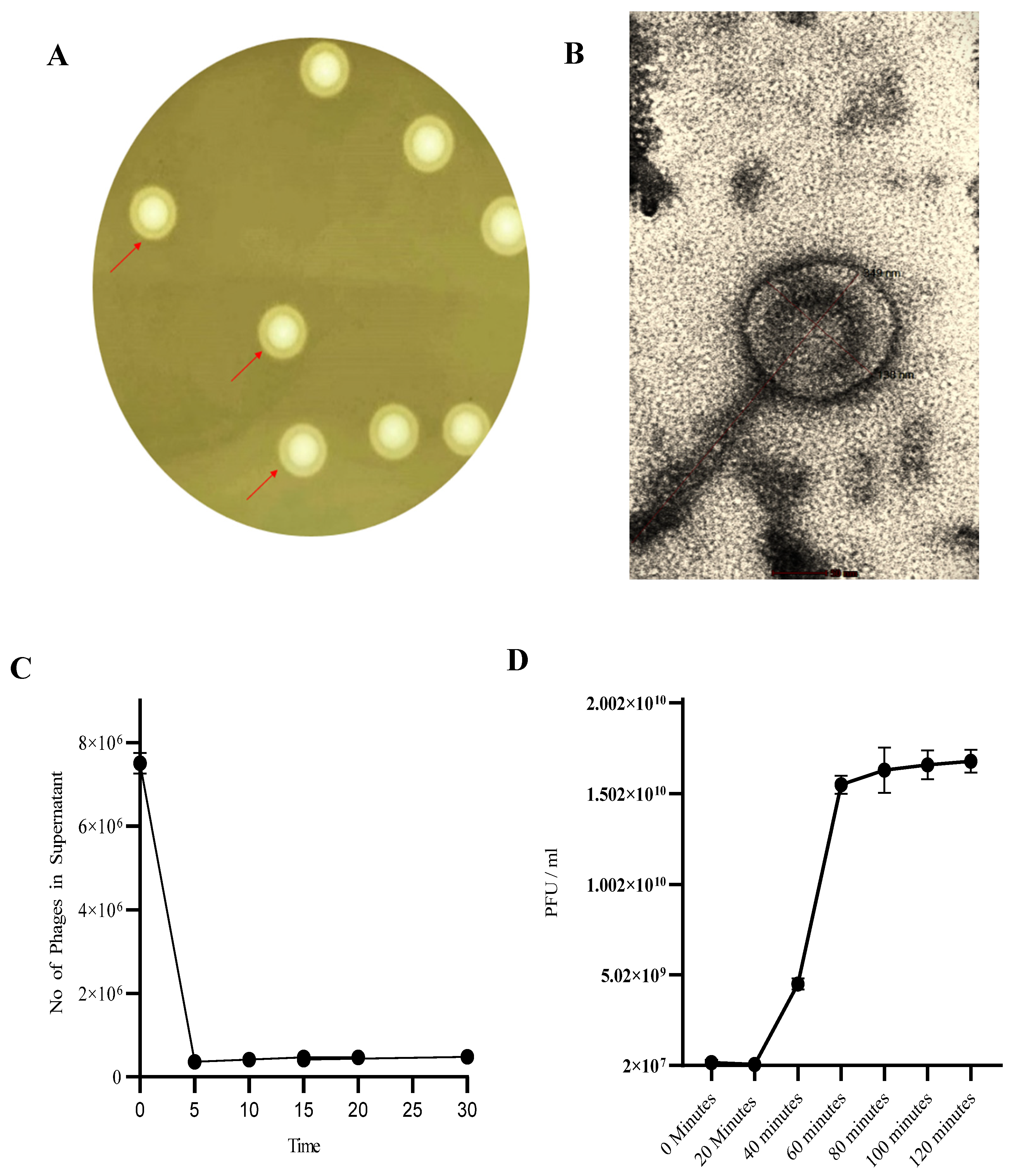
2.4. Determination of Phage Life Cycle by One Step Growth Curve
2.5. Host Range of the Bacteriophage
2.6. Assessment of Bacteriolytic Activity
2.7. Phage Stability
2.8. Transmission Electron Microscopy
2.9. DNA Extraction
2.10. Bacteriophage Whole Genome Sequencing, Annotation and Bioinformatics
2.11. Assessment of Anti-Biofilm Activity of SAKp02
2.12. Scanning Electron Microscopy
2.13. Statistical Analysis
3. Results
3.1. Phage SAKp02 Forms Depolymerase-Induced Bull’s Eye Plaques with a Short Adsorption Time and a 20-Minute Latency
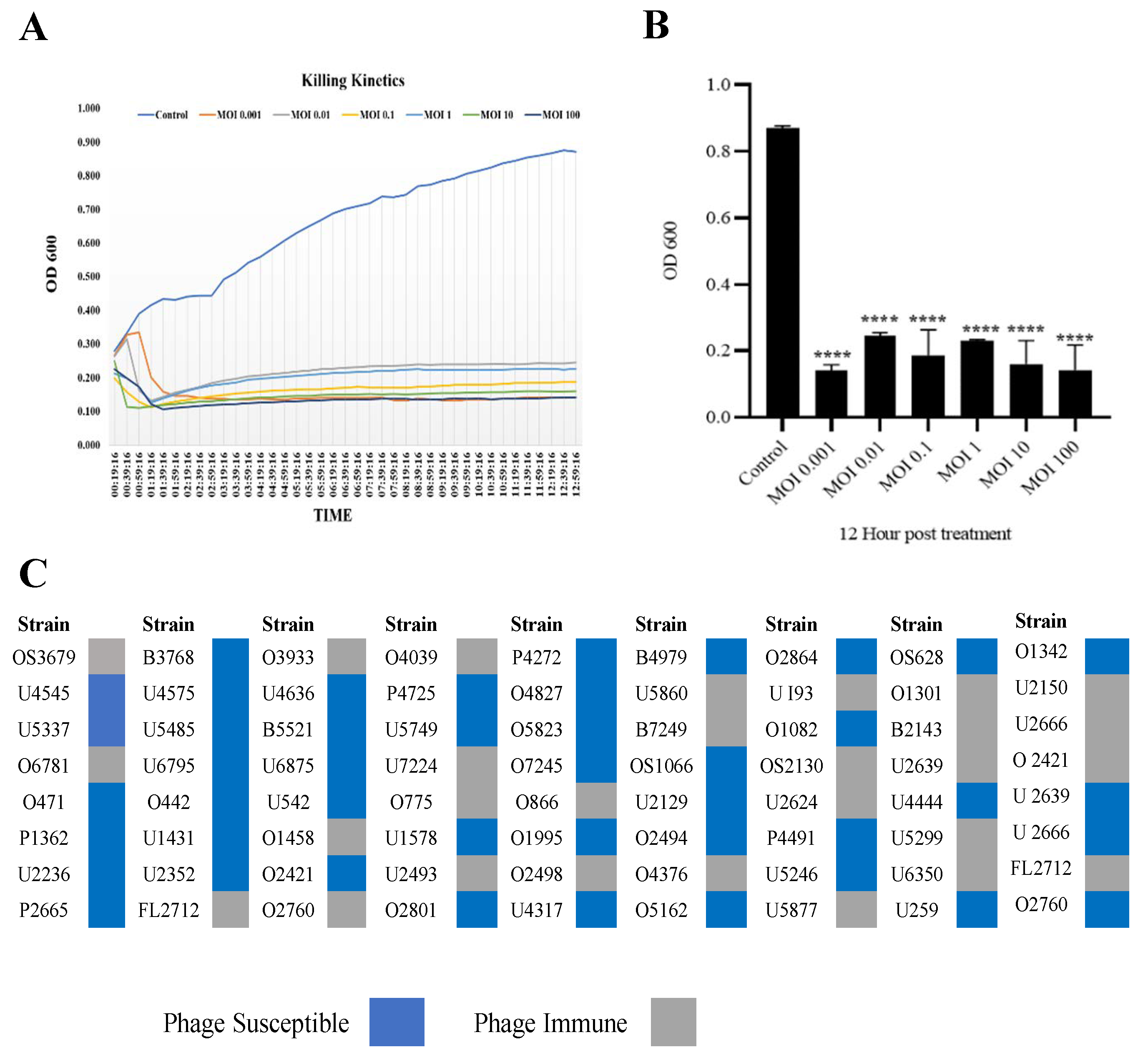
3.2. Phage SAKp02 Is Strongly Bacteriolytic with a Broad Host Range
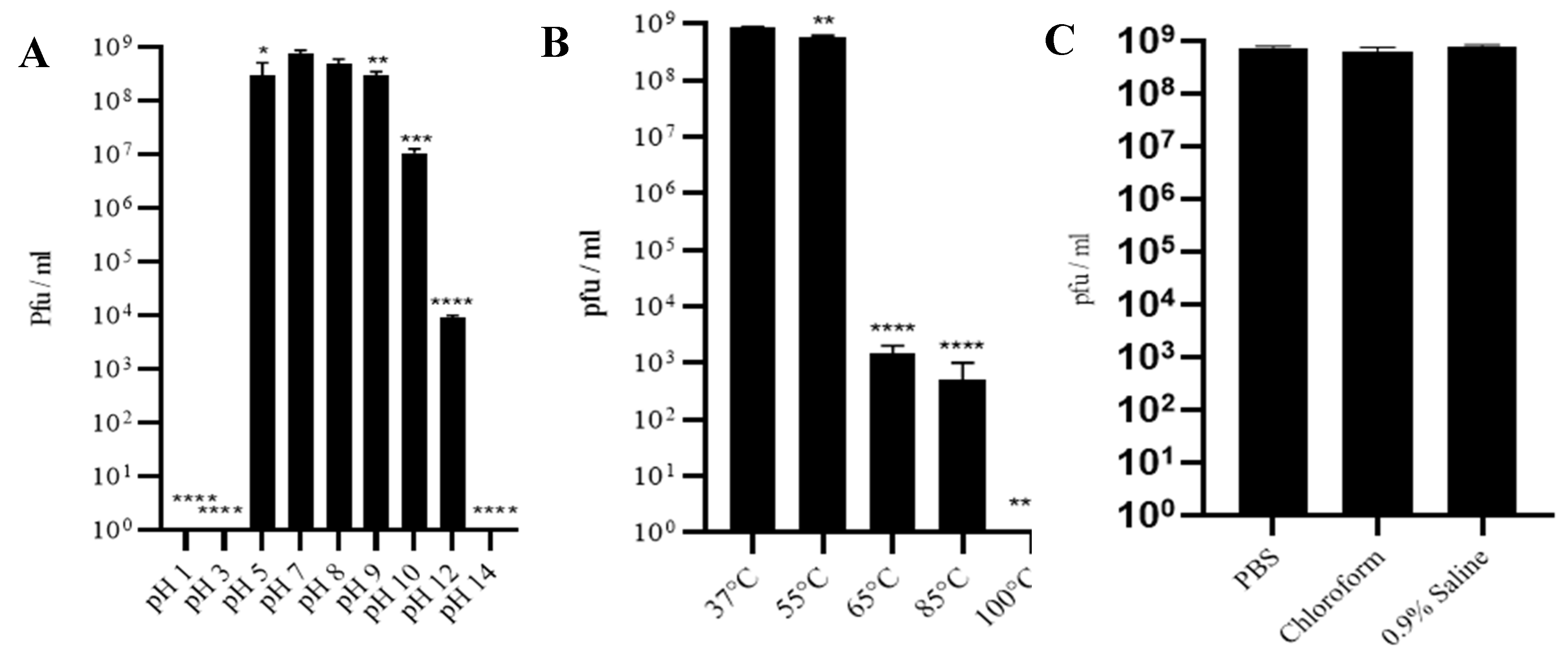
3.3. Phage SAKp02 Is Stable at Different pH, Temperature and Solvent
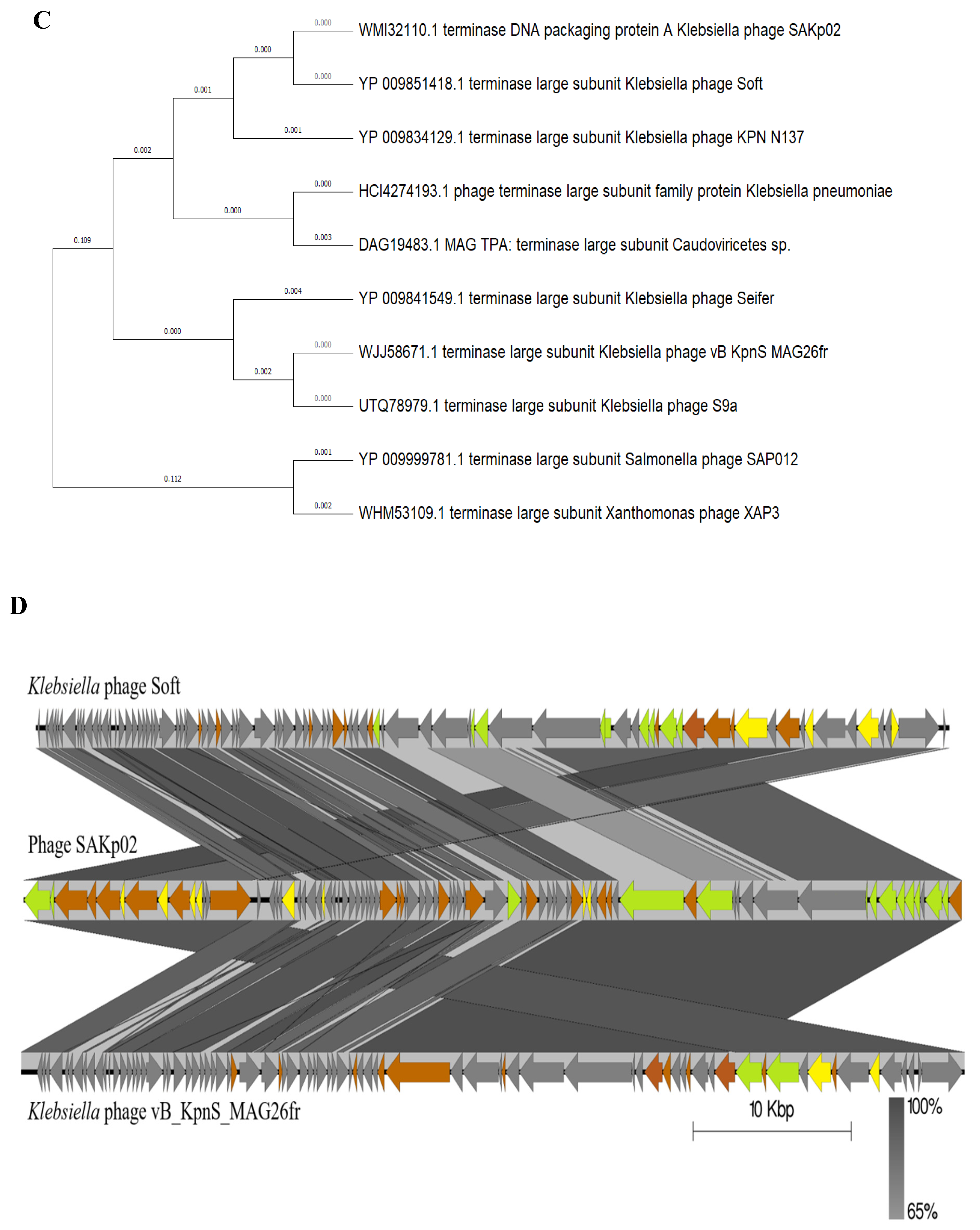
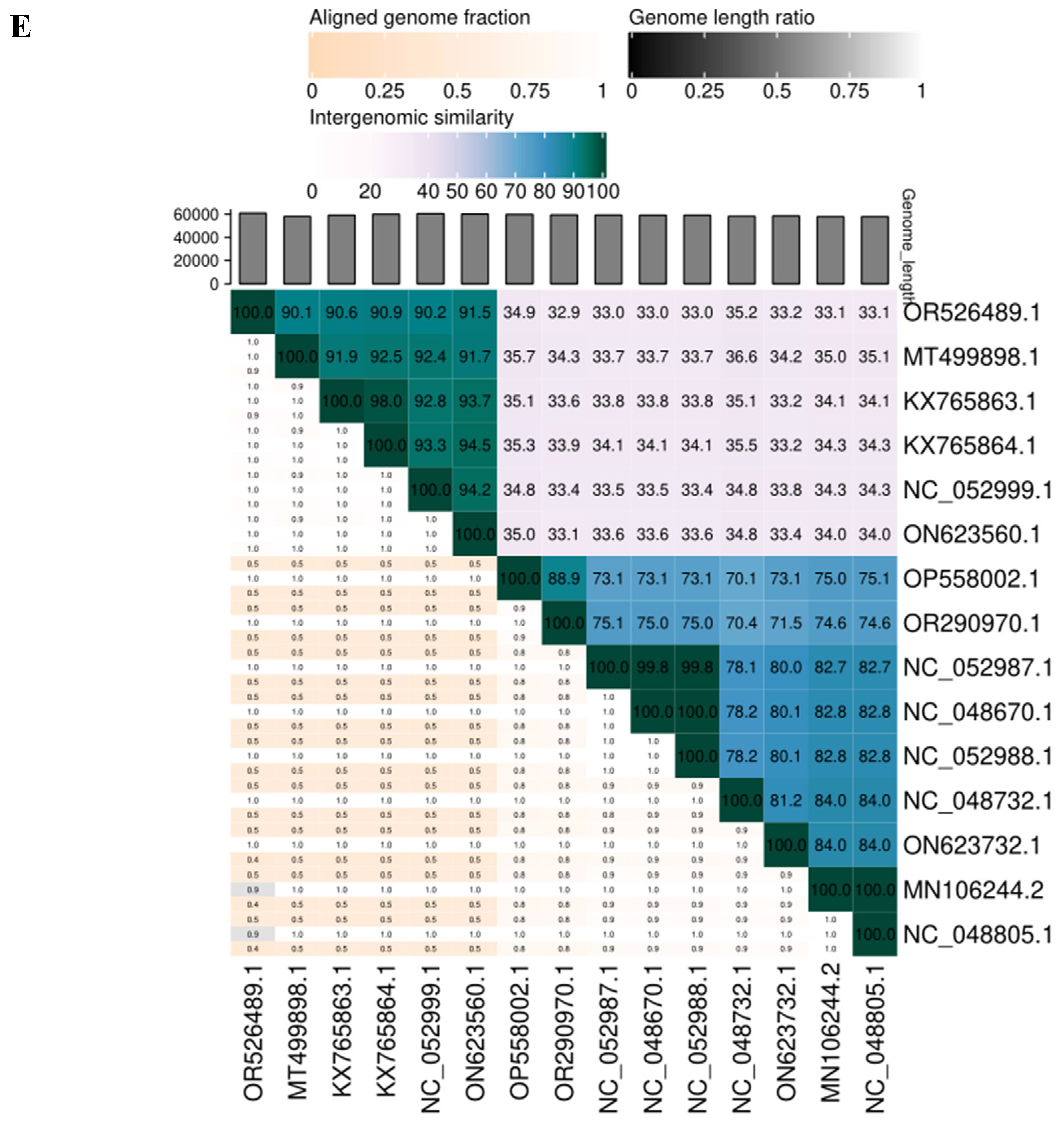
3.4. SAKp02 Belongs to the Casjensviridae Family of Viruses
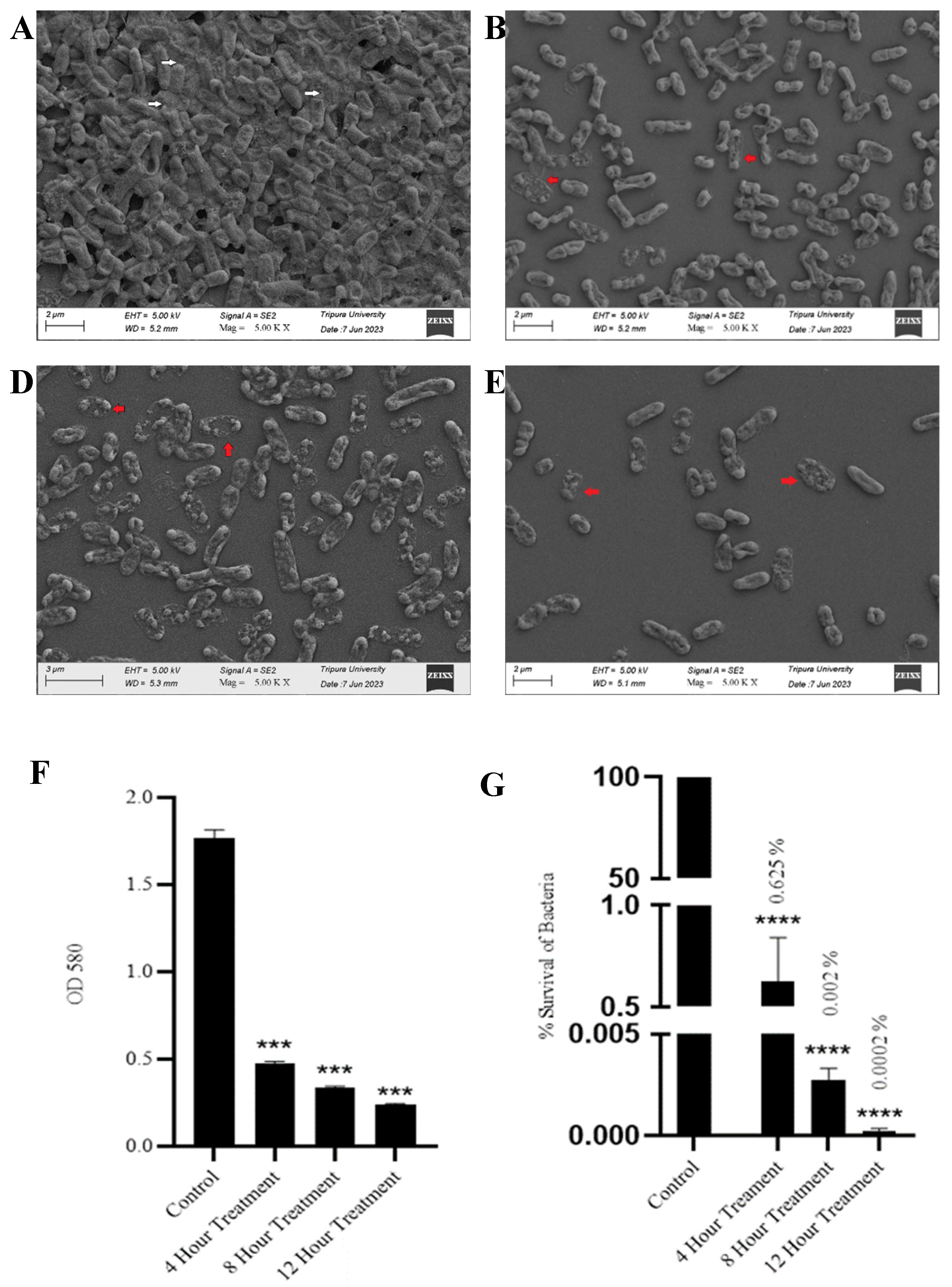
3.5. SAKp02 Effectively Dismantle K. pneumoniae Biofilm and Reduce Viable Cell Count
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ESKAPE | Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp |
| WHO | World Health Organization |
| MMMH | Madras Medical Mission Hospital |
| ORF | Open Reading Frame |
| CDS | Coding Sequence |
| cKp | Classic K. pneumoniae |
| hyKp | Hypervirulent K. pneumoniae |
| EPS | Extracellular Polymeric Substance |
| BP | Base Pair |
| ESBL | Extended-Spectrum Beta-lactamase |
| MDR | Multi Drug Resistance |
| XDR | Extremely Drug Resistance |
| TEM | Transmission Electron Microscopy |
| SEM | Scanning Electron Microscopy |
| OD | Optical Density |
References
- Podschun, R.; Ullmann, U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical Microbiology Reviews 1998, 11, 589–603. [Google Scholar] [CrossRef]
- Luo, K.; Tang, J.; Qu, Y.; Yang, X.; Zhang, L.; Chen, Z.; Kuang, L.; Su, M.; Mu, D. Nosocomial infection by Klebsiella pneumoniae among neonates: a molecular epidemiological study. The Journal of Hospital Infection 2021, 108, 174–180. [Google Scholar] [CrossRef]
- Mohd Asri, N.A.; Ahmad, S.; Mohamud, R.; Mohd Hanafi, N.; Mohd Zaidi, N.F.; Irekeola, A.A.; Shueb, R.H.; Yee, L.C.; Mohd Noor, N.; Mustafa, F.H.; Yean, C.Y.; Yusof, N.Y. Global Prevalence of Nosocomial Multidrug-Resistant Klebsiella pneumoniae: A Systematic Review and Meta-Analysis. Antibiotics 2021, 10, 1508. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, G.; Midiri, A.; Gerace, E.; Biondo, C. Bacterial Antibiotic Resistance: The Most Critical Pathogens. Pathogens 2021, 10, 1310. [Google Scholar] [CrossRef]
- Chang, D.; Sharma, L.; Dela Cruz, C.S.; Zhang, D. Clinical Epidemiology, Risk Factors, and Control Strategies of Klebsiella pneumoniae Infection. Frontiers in Microbiology 2021, 12, 750662. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Ferrer, S.; Peñaloza, H.F.; Budnick, J.A.; Bain, W.G.; Nordstrom, H.R.; Lee, J.S.; Van Tyne, D. Finding Order in the Chaos: Outstanding Questions in Klebsiella pneumoniae Pathogenesis. Infection and Immunity 2021, 89, e00693-20. [Google Scholar] [CrossRef]
- Wyres, K.L.; Wick, R.R.; Judd, L.M.; Froumine, R.; Tokolyi, A.; Gorrie, C.L.; Lam MM, C.; Duchêne, S.; Jenney, A.; Holt, K.E. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genetics 2019, 15, e1008114. [Google Scholar] [CrossRef]
- Zhu, J.; Wang, T.; Chen, L.; Du, H. Virulence Factors in Hypervirulent Klebsiella pneumoniae. Frontiers in Microbiology 2021, 12, 642484. [Google Scholar] [CrossRef] [PubMed]
- Desai, S.; Sanghrajka, K.; Gajjar, D. High Adhesion and Increased Cell Death Contribute to Strong Biofilm Formation in Klebsiellapneumoniae. Pathogens 2019, 8, 277. [Google Scholar] [CrossRef]
- Guerra, M.E.S.; Destro, G.; Vieira, B.; Lima, A.S.; Ferraz, L.F.C.; Hakansson, A.P.; Darrieux, M.; Converso, T.R. Klebsiella pneumoniae Biofilms and Their Role in Disease Pathogenesis. Frontiers in Cellular and Infection Microbiology 2022, 12, 877995. [Google Scholar] [CrossRef]
- Ochońska, D.; Ścibik, Ł.; Brzychczy-Włoch, M. Biofilm Formation of Clinical Klebsiella pneumoniae Strains Isolated from Tracheostomy Tubes and Their Association with Antimicrobial Resistance, Virulence and Genetic Diversity. Pathogens 2021, 10, 1345. [Google Scholar] [CrossRef] [PubMed]
- Stahlhut, S.G.; Struve, C.; Krogfelt, K.A.; Reisner, A. Biofilm formation of Klebsiella pneumoniae on urethral catheters requires either type 1 or type 3 fimbriae. FEMS Immunology and Medical Microbiology 2012, 65, 350–359. [Google Scholar] [CrossRef]
- Yang, D.; Zhang, Z. Biofilm-forming Klebsiella pneumoniae strains have greater likelihood of producing extended-spectrum beta-lactamases. The Journal of Hospital Infection 2008, 68, 369–371. [Google Scholar] [CrossRef] [PubMed]
- Anderl, J.N.; Franklin, M.J.; Stewart, P.S. Role of antibiotic penetration limitation in Klebsiella pneumoniae biofilm resistance to ampicillin and ciprofloxacin. Antimicrobial Agents and Chemotherapy 2000, 44, 1818–1824. [Google Scholar] [CrossRef] [PubMed]
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiology Reviews 2014, 38, 916–931. [Google Scholar] [CrossRef]
- Mangalea, M.R.; Duerkop, B.A. Fitness Trade-Offs Resulting from Bacteriophage Resistance Potentiate Synergistic Antibacterial Strategies. Infection and Immunity 2020, 88, e00926-19. [Google Scholar] [CrossRef]
- Chan, B.; Sistrom, M.; Wertz, J.; et al. Phage selection restores antibiotic sensitivity in MDR Pseudomonas aeruginosa. Sci Rep 2016, 6, 26717. [Google Scholar] [CrossRef]
- Chang, C.; Yu, X.; Guo, W.; Guo, C.; Guo, X.; Li, Q.; Zhu, Y. Bacteriophage-Mediated Control of Biofilm: A Promising New Dawn for the Future. Frontiers in Microbiology 2022, 13, 825828. [Google Scholar] [CrossRef]
- Oliveira, A.; Sillankorva, S.; Quinta, R.; Henriques, A.; Sereno, R.; Azeredo, J. Isolation and characterization of bacteriophages for avian pathogenic E. coli strains. Journal of Applied Microbiology 2009, 106, 1919–1927. [Google Scholar] [CrossRef]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods in Molecular Biology 2009, 501, 69–76. [Google Scholar] [CrossRef]
- Sinha, A.; Eniyan, K.; Manohar, P.; Ramesh, N.; Bajpai, U. Characterization and genome analysis of B1 sub-cluster mycobacteriophage PDRPxv. Virus Research 2020, 279, 197884. [Google Scholar] [CrossRef]
- Amarillas, L.; Rubí-Rangel, L.; Chaidez, C.; González-Robles, A.; Lightbourn-Rojas, L.; León-Félix, J. Isolation and Characterization of phiLLS, a Novel Phage with Potential Biocontrol Agent against Multidrug-Resistant Escherichia coli. Frontiers in Microbiology 2017, 8, 1355. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Das, M.; Bhowmick, T.S.; Koley, H.; Atterbury, R.; Chakrabarti, A.K.; Sarkar, B.L. Isolation and Characterization of Novel Broad Host Range Bacteriophages of Vibrio cholerae O1 from Bengal. Journal of Global Infectious Diseases 2018, 10, 84–88. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Lebeaux, D.; Ghigo, J.M.; Beloin, C. Full and broad-spectrum in vivo eradication of catheter-associated biofilms using gentamicin-EDTA antibiotic lock therapy. Antimicrobial Agents and Chemotherapy 2012, 56, 6310–6318. [Google Scholar] [CrossRef] [PubMed]
- Chauhan, A.; Ghigo, J.M.; Beloin, C. Study of in vivo catheter biofilm infections using pediatric central venous catheter implanted in rat. Nature Protocols 2016, 11, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Mitiku, A.; Aklilu, A.; Tsalla, T.; Woldemariam, M.; Manilal, A.; Biru, M. Magnitude and antimicrobial susceptibility profiles of Gram-Negative bacterial isolates among patients suspected of urinary tract infections in Arba Minch General Hospital, southern Ethiopia. PloS one 2022, 17, e0279887. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lu, H.; Zhang, S.; Shi, Y.; Chen, Q. Phages against Pathogenic Bacterial Biofilms and Biofilm-Based Infections: A Review. Pharmaceutics 2022, 14, 427. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. Bacteriophage Adsorption: Likelihood of Virion Encounter with Bacteria and Other Factors Affecting Rates. Antibiotics 2023, 12, 723. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zheng, P.; Ji, W.; Fu, Q.; Wang, H.; Yan, Y.; Sun, J. SLPW: A Virulent Bacteriophage Targeting Methicillin-Resistant Staphylococcus aureus In vitro and In vivo. Frontiers in Microbiology 2016, 7, 934. [Google Scholar] [CrossRef]
- Li, N.; Zeng, Y.; Wang, M.; Bao, R.; Chen, Y.; Li, X.; Pan, J.; Zhu, T.; Hu, B.; Tan, D. Characterization of Phage Resistance and Their Impacts on Bacterial Fitness in Pseudomonas aeruginosa. Microbiology Spectrum 2022, 10, e0207222. [Google Scholar] [CrossRef]
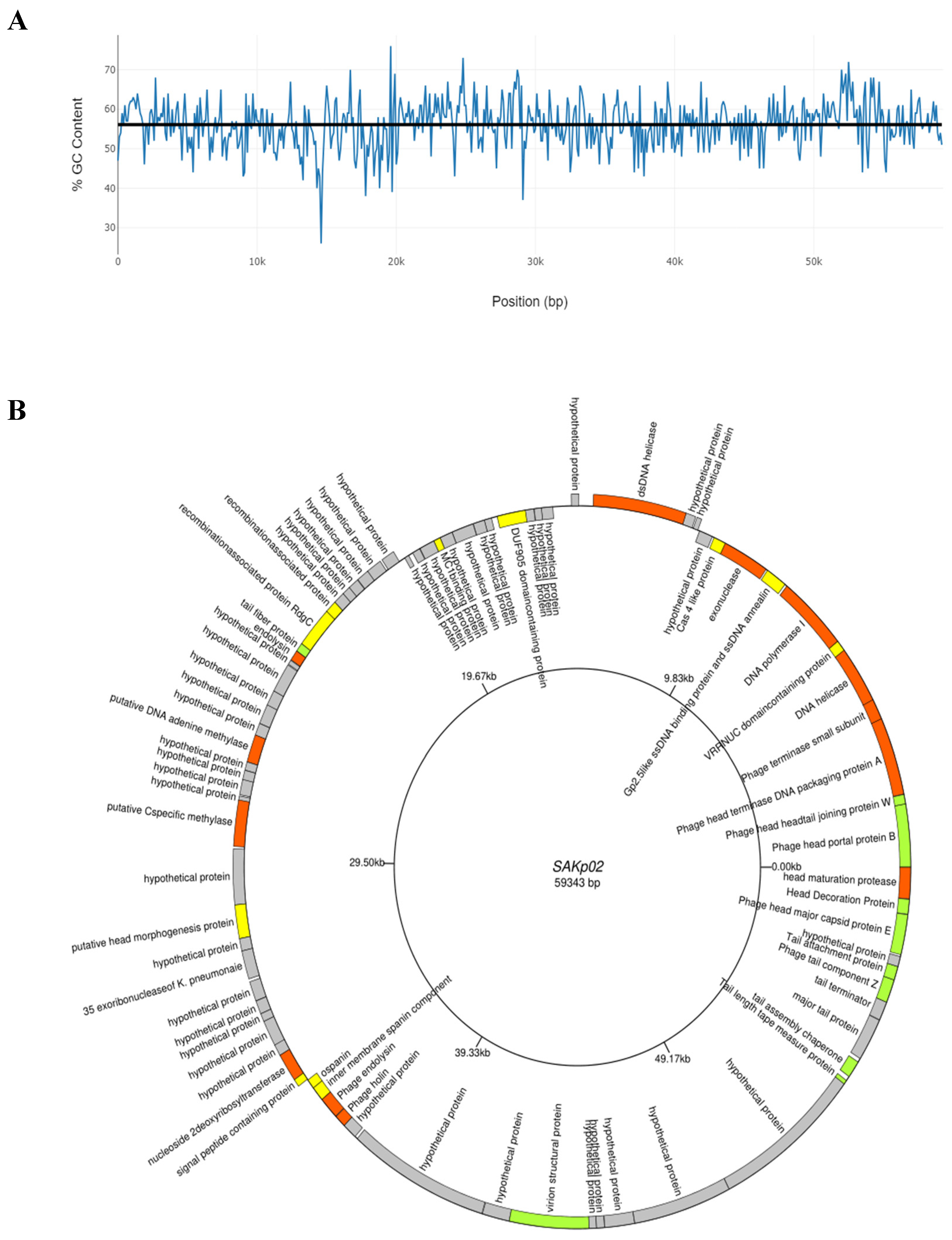
| Sr. | Start | Stop | Size(nt) | Strand | Function |
| 1 | 1665 | 1 | 1664 | - | Phage head, portal protein B |
| 2 | 1916 | 1665 | 251 | - | Phage head, head-tail joining protein W |
| 3 | 4003 | 1928 | 2075 | - | Phage head, terminase DNA packaging protein A |
| 4 | 4562 | 3993 | 569 | - | Phage terminase, small subunit |
| 5 | 6087 | 4546 | 1541 | - | DNA helicase |
| 6 | 6376 | 6080 | 296 | - | VRR-NUC domain-containing protein |
| 7 | 8418 | 6376 | 2042 | - | DNA polymerase I |
| 8 | 9105 | 8485 | 620 | - | Gp2.5-like ssDNA binding protein and ssDNA annealing protein |
| 9 | 10501 | 9152 | 1349 | - | Exonuclease |
| 10 | 10844 | 10494 | 350 | - | Cas 4 like protein |
| 11 | 11291 | 10899 | 392 | - | Hypothetical protein |
| 12 | 11344 | 11481 | 137 | + | Hypothetical protein |
| 13 | 11521 | 11802 | 281 | + | Hypothetical protein |
| 14 | 11799 | 14390 | 2591 | + | dsDNA helicase |
| 15 | 14793 | 15005 | 212 | + | Hypothetical protein |
| 16 | 15860 | 15531 | 329 | - | Hypothetical protein |
| 17 | 16063 | 15857 | 206 | - | Hypothetical protein |
| 18 | 16326 | 16075 | 251 | - | Hypothetical protein |
| 19 | 17131 | 16316 | 815 | - | DUF905 domain-containing protein |
| 20 | 17510 | 17298 | 212 | - | Hypothetical protein |
| 21 | 17833 | 17507 | 326 | - | Hypothetical protein |
| 22 | 18472 | 17843 | 629 | - | Hypothetical protein |
| 23 | 18840 | 18475 | 365 | - | Hypothetical protein |
| 24 | 19040 | 18843 | 197 | - | MC1-binding protein |
| 25 | 19477 | 19040 | 437 | - | Hypothetical protein |
| 26 | 19714 | 19490 | 224 | - | Hypothetical protein |
| 27 | 19976 | 19848 | 128 | - | Hypothetical protein |
| 28 | 20158 | 20535 | 377 | + | Hypothetical protein |
| 29 | 20595 | 21044 | 449 | + | Hypothetical protein |
| 30 | 21022 | 21366 | 344 | + | Hypothetical protein |
| 31 | 21379 | 21681 | 302 | + | Hypothetical protein |
| 32 | 21674 | 21907 | 233 | + | Hypothetical protein |
| 33 | 21904 | 22218 | 314 | + | Hypothetical protein |
| 34 | 22215 | 22547 | 332 | + | Hypothetical protein |
| 35 | 22537 | 23607 | 1070 | + | Exonuclease |
| 36 | 23609 | 23836 | 227 | + | Tail fiber protein |
| 37 | 23838 | 24110 | 272 | + | Endolysin |
| 38 | 24126 | 24242 | 116 | + | Hypothetical protein |
| 39 | 24266 | 25033 | 767 | + | Hypothetical protein |
| 40 | 25030 | 25437 | 407 | + | Hypothetical protein |
| 41 | 25427 | 25936 | 509 | + | Hypothetical protein |
| 42 | 25942 | 26262 | 320 | + | Hypothetical protein |
| 43 | 26255 | 26965 | 710 | + | Putative DNA adenine methylase |
| 44 | 26968 | 27183 | 215 | + | Hypothetical protein |
| 45 | 27187 | 27450 | 263 | + | Hypothetical protein |
| 46 | 27428 | 27820 | 392 | + | Hypothetical protein |
| 47 | 27855 | 27980 | 125 | + | Hypothetical protein |
| 48 | 27958 | 29142 | 1184 | + | Putative C-specific methylase |
| 49 | 29203 | 30630 | 1427 | + | Hypothetical protein |
| 50 | 30630 | 31493 | 863 | + | Putative head morphogenesis protein |
| 51 | 31495 | 31815 | 320 | + | Hypothetical protein |
| 52 | 31812 | 32546 | 734 | + | 3'-5' exoribonuclease of K. pneumonaie |
| 53 | 32621 | 33133 | 512 | + | Hypothetical protein |
| 54 | 33137 | 33484 | 347 | + | Hypothetical protein |
| 55 | 33487 | 33696 | 209 | + | Hypothetical protein |
| 56 | 33698 | 34381 | 683 | + | Hypothetical protein |
| 57 | 34365 | 34664 | 299 | + | Hypothetical protein |
| 58 | 34661 | 35404 | 743 | + | Nucleoside 2-deoxyribosyltransferase |
| 59 | 35394 | 35594 | 200 | + | Signal peptide containing protein |
| 60 | 35881 | 35618 | 263 | - | O-Spanin |
| 61 | 36267 | 35878 | 389 | - | Hypothetical protein |
| 62 | 36851 | 36267 | 584 | - | Phage endolysin |
| 63 | 37196 | 36864 | 332 | - | Phage holin |
| 64 | 37615 | 37196 | 419 | - | Hypothetical protein |
| 65 | 41762 | 37695 | 4067 | - | Hypothetical protein |
| 66 | 42560 | 41781 | 779 | - | Hypothetical protein |
| 67 | 44839 | 42563 | 2276 | - | Vrion structural protein |
| 68 | 45051 | 44839 | 212 | - | Hypothetical protein |
| 69 | 45278 | 45051 | 227 | - | Hypothetical protein |
| 70 | 46139 | 45288 | 851 | - | Hypothetical protein |
| 71 | 48969 | 46144 | 2825 | - | Hypothetical protein |
| 72 | 53274 | 48973 | 4301 | - | Hypothetical protein |
| 73 | 53380 | 53267 | 113 | - | Tail length tape measure protein |
| 74 | 53964 | 53506 | 458 | - | Tail assembly chaperone |
| 75 | 55210 | 54071 | 1139 | - | Major tail protein |
| 76 | 55727 | 55224 | 503 | - | Tail terminator |
| 77 | 56344 | 55724 | 620 | - | Phage tail, component Z |
| 78 | 56709 | 56344 | 365 | - | Tail attachment protein |
| 79 | 56980 | 56711 | 269 | - | Hypothetical protein |
| 80 | 58095 | 57031 | 1064 | - | Phage head, major capsid protein E |
| 81 | 58497 | 58108 | 389 | - | Phage head, head-DNA stabilization protein D |
| 82 | 59343 | 58510 | 833 | - | Phage head, head-tail preconnector protease C |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





