Submitted:
31 May 2024
Posted:
31 May 2024
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Isolation, Identification, and Characterization of Strains
2.2. Tested Prebiotics and Cultivation Media Composition
2.3 Testing of Bacterial Growth
2.4 Statistical Analyses
3. Results
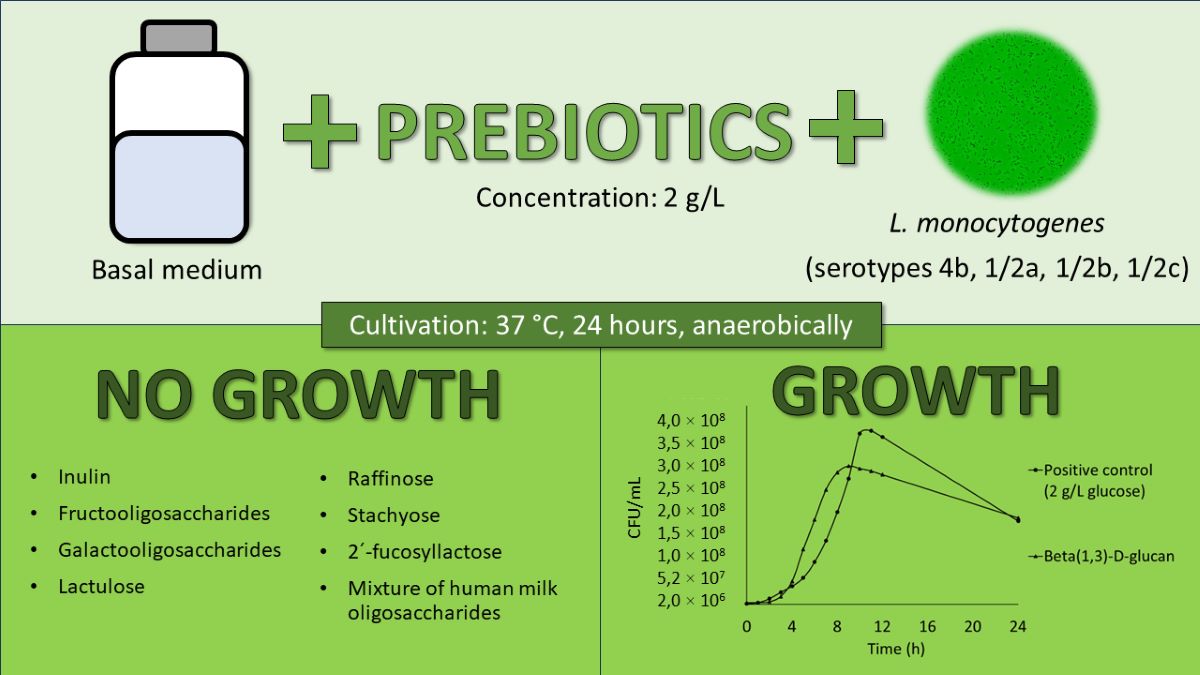
3.1. Utilization of Water-Soluble Prebiotics
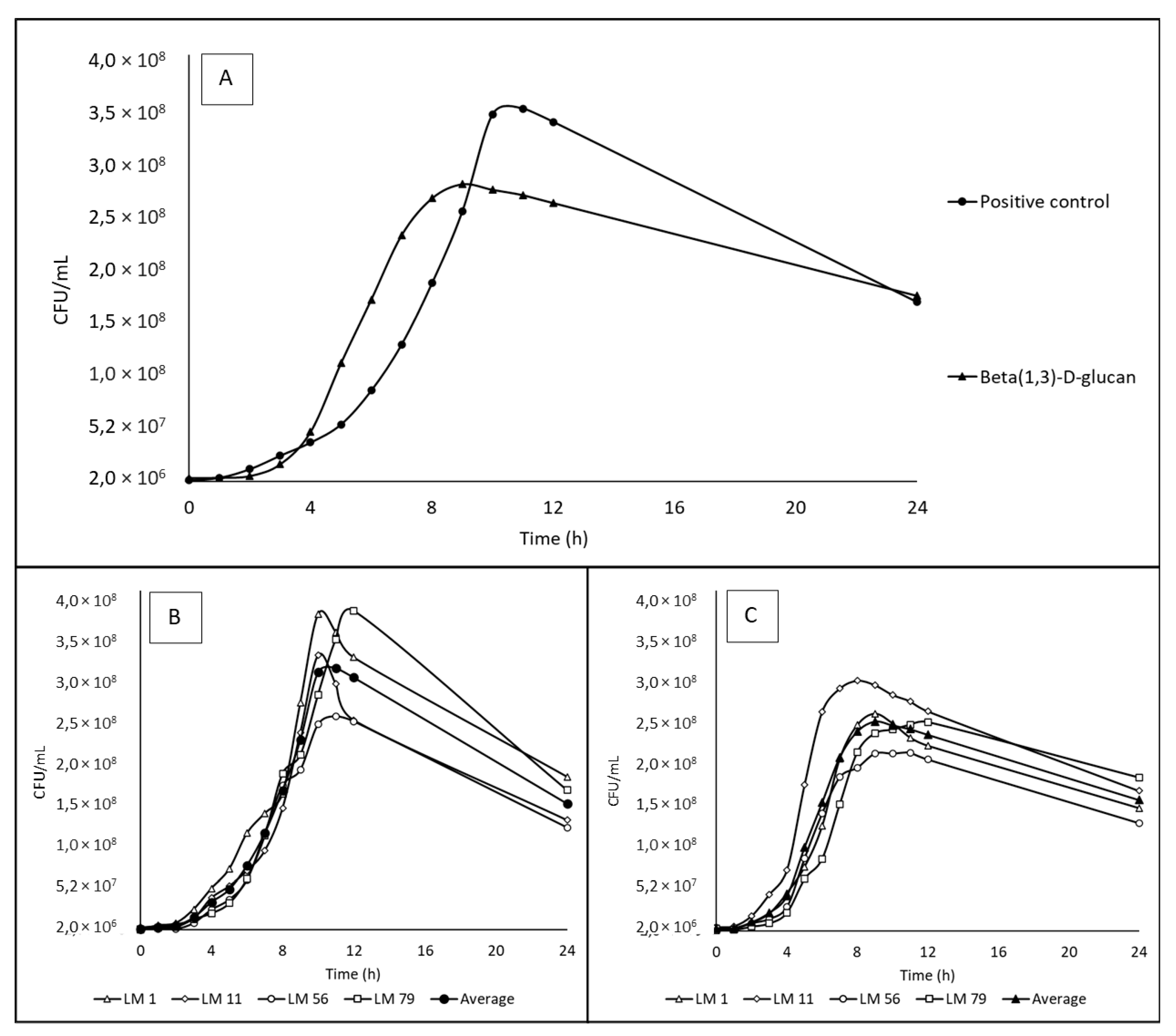
3.2. Utilization of beta(1,3)-D-glucan
3.3. Growth CURVES and rates
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Farber, J.M.; Peterkin, P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol Rev 1991, 55, 476–511. [Google Scholar] [CrossRef]
- Shamloo, E.; Hosseini, H.; Moghadam, A.Z.; Larsen, H.M.; Haslberger, A.; Alebouyeh, M. Importance of Listeria monocytogenes in food safety: a review of its prevalence, detection, and antibiotic resistance. Iran J Vet Res 2019, 20, 241–254. [Google Scholar]
- Bhunia, A.K. Foodborne microbial pathogens, mechanisms and pathogenesis, 2nd ed.; Springer: Switzerland, 2018; p. 365. ISBN 978-1-4939-7347-7. [Google Scholar]
- Azari, S.; Johnson, L.J.; Webb, A.; Kozlowski, S.M.; Zhang, X.; Rood, K.; Amer, A.; Seveau, S. Hofbauer cells spread Listeria monocytogenes among placental cells and undergo pro-inflammatory reprogramming while retaining production of tolerogenic factors. mBio 2021, 12. [Google Scholar] [CrossRef]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union one health 2020 zoonoses report. EFSA Journal 2021, 19, 6971. [Google Scholar] [CrossRef]
- Das, S.; Surendran, P.K.; Thampuran, N. Detection and differentiation of Listeria monocytogenes and Listeria innocua by multiplex PCR. Fishery Technology 2010, 47, 91–94. [Google Scholar]
- Nadon, C.A.; Woodward, D.L.; Young, C.; Rodgers, F.G.; Wiedmann, M. Correlations between molecular subtyping and serotyping of Listeria monocytogenes. J Clin Microbiol 2001, 39, 2704–2707. [Google Scholar] [CrossRef]
- Bortolussi, R. Public Health - Listeriosis: a primer. Can Med Assoc J 2008, 179, 795–797. [Google Scholar] [CrossRef]
- Miceli, A.; Settanni, L. Influence of agronomic practices and pre-harvest conditions on the attachment and development of Listeria monocytogenes in vegetables. Annals of Microbiology 2019, 69, 185–199. [Google Scholar] [CrossRef]
- Simonetti, T.; Peter, K.; Chen, Y.; Jin, Q.; Zhang, G.; LaBorde, L.F.; Macarisin, D. Prevalence and distribution of Listeria monocytogenes in three commercial tree fruit packinghouses. Front Microbiol 2021, 12, 652708. [Google Scholar] [CrossRef]
- Khan, R.; Petersen, F.C.; Shekhar, S. Commensal bacteria: an emerging player in defense against respiratory pathogens. Front Immunol 2019, 10, 1203. [Google Scholar] [CrossRef]
- Gibson, G.R.; Probert, H.M.; Van Loo, J.; Rastall, R.A.; Roberfroid, M.B. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev 2004, 17, 259–275. [Google Scholar] [CrossRef]
- Roberfroid, M.; Gibson, G.R.; Hoyles, L.; McCartney, A.L.; Rastall, R.; Rowland, I.; Wolvers, D.; Watzl, B.; Szajewska, H.; Stahl, B.; et al. Prebiotic effects: metabolic and health benefits. Br. J. Nutr. 2010, 104 Suppl. 2, 1–63. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: the International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 2017, 14, 491–502. [Google Scholar] [CrossRef]
- Charalampopoulos, D.; Rastall, R.A. Prebiotics in foods. Curr Opin Biotechnol 2012, 23, 187–191. [Google Scholar] [CrossRef]
- Musilova, S.; Rada, V.; Vlkova, E.; Bunesova, V. Beneficial effects of human milk oligosaccharides on gut microbiota. Benef Microbes 2014, 5, 273–283. [Google Scholar] [CrossRef]
- Rada, V.; Nevoral, J.; Trojanová, I.; Tománková, E.; Šmehilová, M.; Killer, J. Growth of infant faecal bifidobacteria and clostridia on prebiotic oligosaccharides in in vitro conditions. Anaerobe 2008, 14, 205–208. [Google Scholar] [CrossRef]
- Bunešová, V.; Vlková, E.; Rada, V.; Kňazovická, V.; Ročková, Š.; Geigerová, M.; Božik, M. Growth of infant fecal bacteria on commercial prebiotics. Folia Microbiol 2012, 57, 273–275. [Google Scholar] [CrossRef]
- Sauer, J.-D.; Herskovits, A.A.; O’Riordan, M.X.D. Metabolism of the Gram-positive bacterial pathogen Listeria monocytogenes. Microbiol Spectr 2019, 7, 1–12. [Google Scholar] [CrossRef]
- Friedman, M.E.; Roessler, W.G. Growth of Listeria monocytogenes in defined media. J Bacteriol 1961, 82, 528–533. [Google Scholar] [CrossRef]
- Balay, D.R.; Gänzle, M.G.; McMullen, L.M. The effect of carbohydrates and bacteriocins on the growth kinetics and resistance of Listeria monocytogenes. Front Microbiol 2018, 9, 347. [Google Scholar] [CrossRef]
- Gopal, S.; Berg, D.; Hagen, N.; Schriefer, E.M.; Stoll, R.; Goebel, W.; Kreft, J. Maltose and maltodextrin utilization by Listeria monocytogenes depend on an inducible ABC transporter which is repressed by glucose. PLoS One 2010, 5, 10349. [Google Scholar] [CrossRef]
- Pine, L.; Malcolm, G.B.; Brooks, J.B.; Daneshvar, M.I. Physiological studies on the growth and utilization of sugars by Listeria species. Can J Microbiol 1989, 35, 245–254. [Google Scholar] [CrossRef]
- Schardt, J.; Jones, G.; Müller-Herbst, S.; Schauer, K.; D’Orazio, S.E.F.; Fuchs, T.M. Comparison between Listeria sensu stricto and Listeria sensu lato strains identifies novel determinants involved in infection. Sci Rep 2017, 7, 17821. [Google Scholar] [CrossRef]
- Paspaliari, D.K.; Loose, J.S.M.; Larsen, M.H.; Vaaje-Kolstad, G. Listeria monocytogenes has a functional chitinolytic system and an active lytic polysaccharide monooxygenase. FEBS J 2015, 282, 921–936. [Google Scholar] [CrossRef]
- Bae, D.; Seo, K.S.; Zhang, T.; Wang, C. Characterization of a potential Listeria monocytogenes virulence factor associated with attachment to fresh produce. Appl Environ Microbiol 2013, 79, 6855–6861. [Google Scholar] [CrossRef]
- Salmonová, H.; Salmonová, S.; Killer, J.; Bunešová, V.; Bunešová, B.; Geigerová, M.; Geigerová, G.; Vlková, E.; Vlková, V. Cultivable bacteria from Pectinatella magnifica and the surrounding water in South Bohemia indicate potential new Gammaproteobacterial, Betaproteobacterial and Firmicutes taxa. FEMS Microbiol Lett 2018, 365. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: an important software for molecular biology. GERF Bull Biosci 2011, 2, 60–61. [Google Scholar]
- Liu, D.; Ainsworth, A.J.; Austin, F.W.; Lawrence, M.L. Characterization of virulent and avirulent Listeria monocytogenes strains by PCR amplification of putative transcriptional regulator and internalin genes. J Med Microbiol 2003, 52, 1065–1070. [Google Scholar] [CrossRef]
- Hungate, R.E. A roll tube method for cultivation of strict anaerobes. In Methods in Microbiology. 1st ed.; Norris, J.R.; Ribbons, D.W., Eds.; Academic Press, San Diego, US, 1969; Volume 3, pp. 117–132. [CrossRef]
- Rockova, S.; Nevoral, J.; Rada, V.; Marsik, P.; Sklenar, J.; Hinkova, A.; Vlkova, E.; Marounek, M. factors affecting the growth of bifidobacteria in human milk. Int Dairy J 2011, 21, 504–508. [Google Scholar] [CrossRef]
- Bai, Y.P.; Zhou, H.M.; Zhu, K.R.; Li, Q. Effect of thermal processing on the molecular, structural, and antioxidant characteristics of highland barley β-glucan. Carbohydr Polym 2021, 271, 118416. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhou, H.M.; Huang, Z.H.; Zhao, R.Y. Different aggregation states of barley β-glucan molecules affects their solution behavior: a comparative analysis. Food Hydrocoll 2020, 101, 105543. [Google Scholar] [CrossRef]
- Jones, G.S.; D’Orazio, S.E.F. Listeria monocytogenes: cultivation and laboratory. Curr Protoc Microbiol 2013, 31, 9B.2.1–9B.2.7. [Google Scholar] [CrossRef]
- Durica-Mitic, S.; Göpel, Y.; Görke, B. Carbohydrate utilization in bacteria: making the most out of sugars with the help of small regulatory RNAs. Microbiol Spectr 2018, 6. [Google Scholar] [CrossRef]
- Gahan, C.G.M.; Hill, C. Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol 2014, 5, 1–7. [Google Scholar] [CrossRef]
- Lockyer, S.; Stanner, S. Prebiotics – an added benefit of some fibre types. Nutr Bull 2019, 44, 74–91. [Google Scholar] [CrossRef]
- Ibrahim, O.O. Functional oligosaccharides: chemicals structure, manufacturing, health benefits, applications and regulations. J Food Chem Nanotechnol 2018, 4, 65–76. [Google Scholar] [CrossRef]
- Mudgil, D. The interaction between insoluble and soluble fiber. In Dietary fiber for the prevention of cardiovascular disease: fiber’s interaction between gut micoflora, sugar metabolism, weight control and cardiovascular health., 1st ed.; Samaan, R.A., Ed.; Academic Press: San Diego, US, 2017; pp. 35–59. [Google Scholar] [CrossRef]
- Stone, B.A. Chemistry of β-glucans. In Chemistry, biochemistry, and biology of 1-3 beta glucans and related polysaccharides, 1st ed.; Bacic, A., Fincher, G.B., Stone, B.A., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 5–46. [Google Scholar] [CrossRef]
- Kim, K.S.; Yun, H.S. Production of soluble β-glucan from the cell wall of Saccharomyces cerevisiae. Enzyme Microb Technol 2006, 39, 496–500. [Google Scholar] [CrossRef]
- Synytsya, A.; Novak, M. Structural analysis of glucans. Ann Transl Med 2014, 2, 17. [Google Scholar] [CrossRef]
- Jayachandran, M.; Chen, J.; Chung, S.S.M.; Xu, B. A critical review on the impacts of β-glucans on gut microbiota and human health. J Nutr Biochem 2018, 61, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Shokri, H.; Asadi, F.; Khosravi, A.R.; Shokriy, H.; Khosraviy, A.R. Isolation of β -glucan from the cell wall of Saccharomyces cerevisiae. Nat Pro Res 2009, 22, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Cheung, P.C.K. Fermentation of β-glucans derived from different sources by bifidobacteria: evaluation of their bifidogenic effect. J Agric Food Chem 2011, 59, 5986–5992. [Google Scholar] [CrossRef]
- Buddington, K.K.; Donahoo, J.B.; Buddington, R.K. Dietary oligofructose and inulin protect mice from enteric and systemic pathogens and tumor inducers. J Nutr 2002, 132, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Zheng, J.; Lin, A.; Xia, H.; Zhang, Z.; Gao, Q.; Lv, W.; Liu, H. A review: roles of carbohydrates in human diseases through regulation of imbalanced intestinal microbiota. J Funct Foods 2020, 74, 104197. [Google Scholar] [CrossRef]
- Chen, P.; Reiter, T.; Huang, B.; Kong, N.; Weimer, B.C. Prebiotic oligosaccharides potentiate host protective responses against L. monocytogenes infection. Pathogens 2017, 6, 68. [Google Scholar] [CrossRef] [PubMed]
- Karakan, T.; Tuohy, K.M.; Janssen-van Solingen, G. Low-dose lactulose as a prebiotic for improved gut health and enhanced mineral absorption. Front Nutr 2021, 8, 672925. [Google Scholar] [CrossRef] [PubMed]
- Sangwan, V.; Tomar, S.K.; Ali, B.; Singh, R.R.B.; Singh, A.K. Galactooligosaccharides reduce infection caused by Listeria monocytogenes and modulate IgG and IgA levels in mice. Int Dairy J 2015, 41, 58–63. [Google Scholar] [CrossRef]
- Ebersbach, T.; Jørgensen, J.B.; Heegaard, P.M.; Lahtinen, S.J.; Ouwehand, A.C.; Poulsen, M.; Frøkiær, H.; Licht, T.R. Certain dietary carbohydrates promote Listeria infection in a guinea pig model, while others prevent it. Int J Food Microbiol 2010, 140, 218–224. [Google Scholar] [CrossRef]
- Kupfahl, C.; Geginat, G.; Hof, H. Lentinan has a stimulatory effect on innate and adaptive immunity against murine Listeria monocytogenes infection. Int Immunopharmacol 2006, 6, 686–696. [Google Scholar] [CrossRef]
- Li, W.; Yajima, T.; Saito, K.; Nishimura, H.; Fushimi, T.; Ohshima, Y.; Tsukamoto, Y.; Yoshikai, Y. Immunostimulating properties of intragastrically administered Acetobacter-derived soluble branched (1,4)-β-d-glucans decrease murine susceptibility to Listeria monocytogenes. Infect Immun 2004, 72, 7005–7011. [Google Scholar] [CrossRef] [PubMed]
- Torello, C.O.; De Souza Queiroz, J.; Oliveira, S.C.; Queiroz, M.L.S. Immunohematopoietic modulation by oral β-1,3-glucan in mice infected with Listeria monocytogenes. Int Immunopharmacol 2010, 10, 1573–1579. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Fang, K.; Medina, D.; Wan, J.; Lee, J.L.; Hong, S.H. The probiotic Leuconostoc mesenteroides inhibits Listeria monocytogenes biofilm formation. J Food Saf 2020, 40, e12750. [Google Scholar] [CrossRef]
- Corr, S.C.; Gahan, C.G.M.; Hill, C. Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunol Med Microbiol 2007, 50, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Amézquita, A.; Brashears, M.M. Competitive inhibition of Listeria monocytogenes in ready-to-eat meat products by lactic acid bacteria. J Food Prot 2002, 65, 316–325. [Google Scholar] [CrossRef] [PubMed]
- García, M.J.; Ruíz, F.; Asurmendi, P.; Pascual, L.; Barberis, L. Searching potential candidates for development of protective cultures: evaluation of two Lactobacillus strains to reduce Listeria monocytogenes in artificially contaminated milk. J Food Saf 2020, 40, e12723. [Google Scholar] [CrossRef]
- Hascoët, A.S.; Ripolles-avila, C.; Cervantes-huamán, B.R.H.; Rodríguez-jerez, J.J. In vitro preformed biofilms of Bacillus safensis inhibit the adhesion and subsequent development of Listeria monocytogenes on stainless-steel surfaces. Biomolecules 2021, 11, 1–16. [Google Scholar] [CrossRef]
- Tran, T.D.; Del Cid, C.; Hnasko, R.; Gorski, L.; McGarvey, J.A. Bacillus amyloliquefaciens ALB65 inhibits the growth of Listeria monocytogenes on Cantaloupe melons. Appl Environ Microbiol 2020, 87, 1–10. [Google Scholar] [CrossRef]
- Alonso, V.P.P.; Harada, A.M.M.; Kabuki, D.Y. competitive and/or cooperative interactions of Listeria monocytogenes with Bacillus cereus in dual-species biofilm formation. Front Microbiol 2020, 11, 177. [Google Scholar] [CrossRef]
- da Silva Sabo, S.; Converti, A.; Todorov, S.D.; Domínguez, J.M.; de Souza Oliveira, R.P. effect of inulin on growth and bacteriocin production by Lactobacillus plantarum in stationary and shaken cultures. Int J Food Sci Technol 2015, 50, 864–870. [Google Scholar] [CrossRef]
- Kunová, G.; Rada, V.; Lisová, I.; Ročková, Š.; Vlková, E. In vitro fermentability of prebiotic oligosaccharides by lactobacilli. Czech J. Food Sci 2011, 29, 49–54. [Google Scholar] [CrossRef]
- Jaradat, Z.W.; Bhunia, A.K. Glucose and nutrient concentrations affect the expression of a 104-kilodalton listeria adhesion protein in Listeria monocytogenes. Appl Environ Microbiol 2002, 68, 4876–4883. [Google Scholar] [CrossRef] [PubMed]
- Aké, F.M.D.; Joyet, P.; Deutscher, J.; Milohanic, E. mutational analysis of glucose transport regulation and glucose-mediated virulence gene repression in Listeria monocytogenes. Mol Microbiol 2011, 81, 274–293. [Google Scholar] [CrossRef] [PubMed]
- Park, S.F.; Kroll, R.G. Expression of listeriolysin and phosphatidylinositol-specific phospholipase c is repressed by the plant-derived molecule cellobiose in Listeria monocytogenes. Mol Microbiol 1993, 8, 653–661. [Google Scholar] [CrossRef]
- Crespo Tapia, N.; Dorey, A.L.; Gahan, C.G.M.; den Besten, H.M.W.; O’Byrne, C.P.; Abee, T. different carbon sources result in differential activation of sigma B and stress resistance in Listeria monocytogenes. Int J Food Microbiol 2020, 320, 108504. [Google Scholar] [CrossRef]

| Strain code | LM1 | LM11 | LM56 | LM79 | Average ± SD | |
|---|---|---|---|---|---|---|
| Serotype | 4b | 1/2b | 1/2a | 1/2c | - | |
| Positive control (2 g/L glucose) | 0.156 ± 0.036Aa | 0.140 ± 0.027Aa | 0.127 ± 0.032Aa | 0.130 ± 0.030Aa | 0.138 ± 0.032A | |
| Residual control (0,1 g/L glucose) | 0.028 ± 0.002Ba | 0.029 ± 0.001BCa | 0.028 ± 0.001BCDab | 0.026 ± 0.001BCb | 0.028 ± 0.002BC | |
| Negative control (no saccharides) | 0.016 ± 0.015Ba | -0.006 ± 0.026Db | 0.008 ± 0.008Deab | 0.005 ± 0.005Dab | 0.006 ± 0.017EF | |
| Inulin | 0.027 ± 0.006Bb | 0.028 ± 0.004BCb | 0.047 ± 0.013Ba | 0.043 ± 0.005Ba | 0.036 ± 0.012B | |
| Fructooligosaccharides | 0.026 ± 0.031Ba | 0.029 ± 0.008Bca | 0.034 ± 0.011Bca | 0.016 ± 0.005Cda | 0.027 ± 0.016BCD | |
| Galactooligosaccharides | 0.008 ± 0.018Bb | 0.046 ± 0.006Ba | 0.018 ± 0.002CDEb | 0.011 ± 0.007CDb | 0.021 ± 0.018BCD | |
| Lactulose | 0.014 ± 0.040Ba | 0.010 ± 0.014Cda | -0.001 ± 0.005Ea | -0.005 ± 0.001Da | -0.001 ± 0.010F | |
| Raffinose | 0.004 ± 0.006Ba | 0.006 ± 0,002Cda | 0.004 ± 0.006DEa | -0.003 ± 0.007Da | 0.003 ± 0.006EF | |
| Stachyose | 0.032 ± 0.005Ba | 0.011 ± 0.009CDbc | 0.015 ± 0.005CDEb | 0.003 ± 0.005CDc | 0.015 ± 0.012CDE | |
| 2´-fucosyllactose | 0.019 ± 0.002Ba | 0.013 ± 0.003Cdab | 0.015 ± 0.004CDEab | 0.011 ± 0.006CDb | 0.014 ± 0.005CDE | |
| Mixture of HMOs | 0.028 ± 0.014Ba | 0.010 ± 0.007Cdab | 0.016 ± 0.007CDEa | -0.006 ± 0.010Db | 0.012 ± 0.015DEF | |
| Strain code | LM1 | LM11 | LM56 | LM79 | Average ± SD |
|---|---|---|---|---|---|
| Serotype | 4b | 1/2b | 1/2a | 1/2c | - |
| Positive control (2 g/L glucose) | 1.56 ± 0.72Aa | 1.2 ± 0.04Aba | 0.95 ± 0.01Aa | 1.24 ± 0.15Aa | 1.24 ± 0.39A |
| Negative control (no saccharides) | 0.33 ± 0.06Ba | 0.49 ± 0.12Ba | 0.16 ± 0.02Ba | 0.24 ± 0.11Ba | 0.31 ± 0.15B |
| Beta(1-3)-D-glucan | 1.05 ± 0.09Aba | 1.43 ± 0.67Aa | 0.96 ± 0.07Aa | 1.48 ± 0.72Aa | 1.23 ± 0.48A |
| Strain code | LM1 | LM11 | LM56 | LM79 | Average ± SD |
|---|---|---|---|---|---|
| Serotype | 4b | 1/2b | 1/2a | 1/2c | - |
| Positive control (2 g/L glucose) | 0.42 ± 0.21Aa | 0.63 ± 0.04Aa | 0.49 ± 0.02Ba | 0.64 ± 0.10Aa | 0.55 ± 0.14A |
| Residual control (0,1 g/L glucose) | 0.48 ± 0.07Aab | 0.32 ± 0.03Bb | 0.40 ± 0.08Bab | 0.55 ± 0.20ABCa | 0.44 ± 0.14A |
| Inulin | 0.35 ± 0.02ABc | 0.63 ± 0.12Aab | 0.68 ± 0.03Aa | 0.51 ± 0.04ABb | 0.54 ± 0.11A |
| Fructooligosaccharides | 0.38 ± 0.10ABb | 0.67 ± 0.12Aa | 0.70 ± 0.09Aa | 0.47 ± 0.09BCb | 0.55 ± 0.20A |
| Galactooligosaccharides | 0.19 ± 0.05Bb | 0.66 ± 0.03Aa | 0.73 ± 0.18Aa | 0.28 ± 0.13Cab | 0.46 ± 0.19A |
| Strain code | LM1 | LM11 | LM56 | LM79 | Average ± SD |
|---|---|---|---|---|---|
| Serotype | 4b | 1/2b | 1/2a | 1/2c | - |
| Positive control (2 g/L glucose) | 0.53 ± 0.01 | 0.53 ± 0.01 | 0.52 ± 0.01 | 0.47 ± 0.16 | 0.51 ± 0.07 |
| Beta(1,3)-D-glucan | 0.53 ± 0.01 | 0.54 ± 0.01 | 0.53 ± 0.01 | 0.52 ± 0.01 | 0.53 ± 0.01 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





