Submitted:
28 May 2024
Posted:
28 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Selection of Probiotic Candidate Strain
2.1.1. Preparation of Lactic Acid Bacteria Isolates
2.1.2. Hemolytic Analysis
2.1.3. Analysis of β-Glucosidase and β-Glucuronidase Activities
2.1.4. Analysis of Acid and Bile Salt Tolerance
2.1.5. Determination of ABTS-Scavenging Activity
2.2. Preparation of Artemisia herba-alba Extracts
2.2.1. Minimum Bactericidal Concentration of Artemisia herba-alba Extracts against Periodontal Pathogens
2.2.2. Growth of Probiotic Candidate Strains in the Presence of Artemisia herba-alba Extracts
2.2.3. Antimicrobial Effects of Artemisia herba-alba Cocultured Broths against Periodontal Pathogens
2.2.4. α-Glucosidase Inhibitory Activity of Artemisia herba-alba Cocultured Broths
2.3. Preparation of Bioconverted Milk
2.3.1. Antimicrobial Effects of Bioconverted Milk against Periodontal Pathogens
2.3.2. Analysis of the α-Glucosidase Inhibitory Activity of Bioconverted Milk
2.4. Whole-Genome Analysis of Novel Probiotics
2.4.1. DNA Extraction and Library Preparation
2.4.2. De Novo Sequencing
2.4.3. Comparison with Other Lactic Acid Bacteria
3. Results and Discussion
3.1. Determination of A. herba-alba Extract Concentration
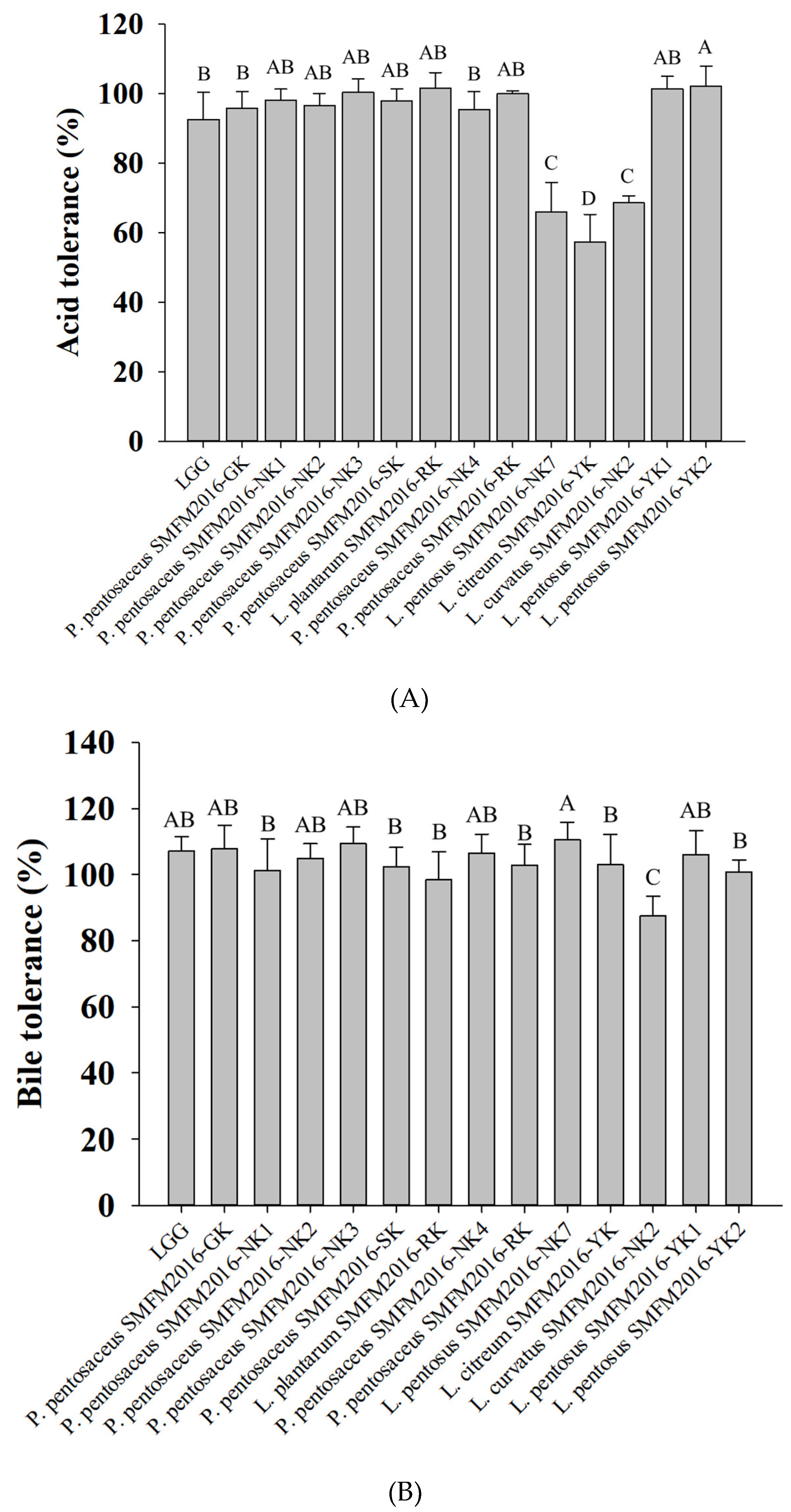
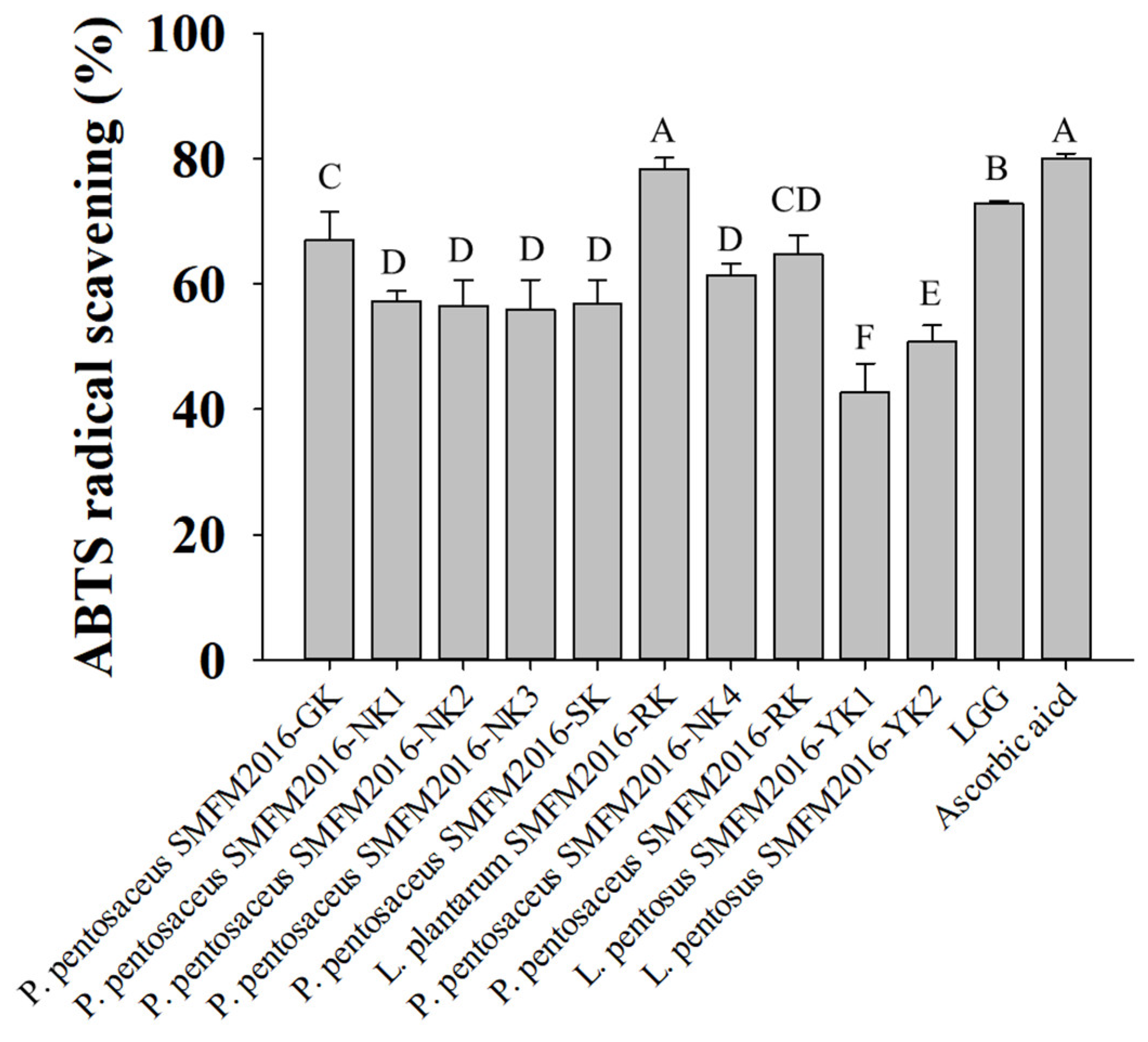
3.2. Selection of Probiotic Candidate Strain
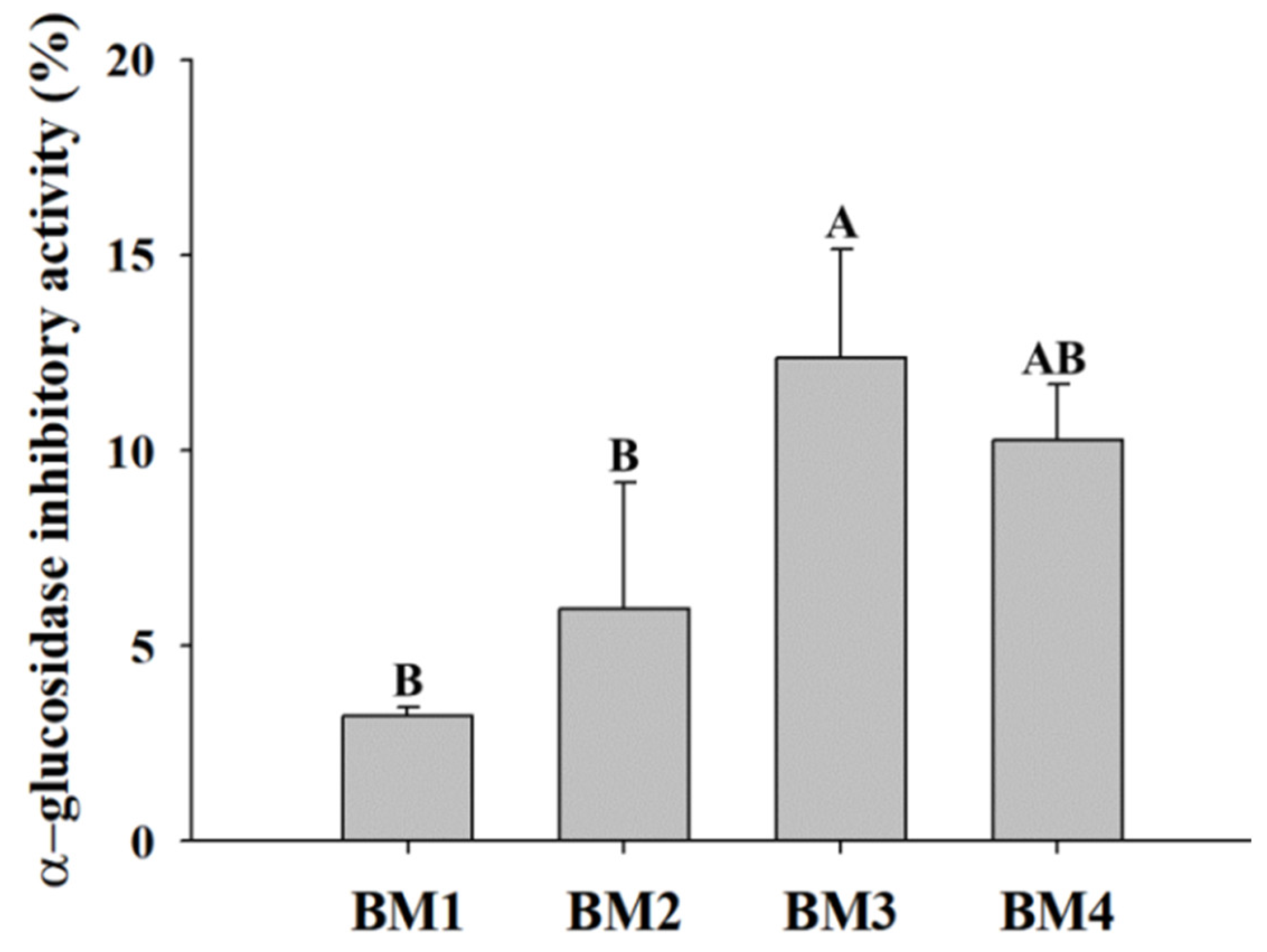
3.3. Efficacy Evaluation of Bioconverted Milk
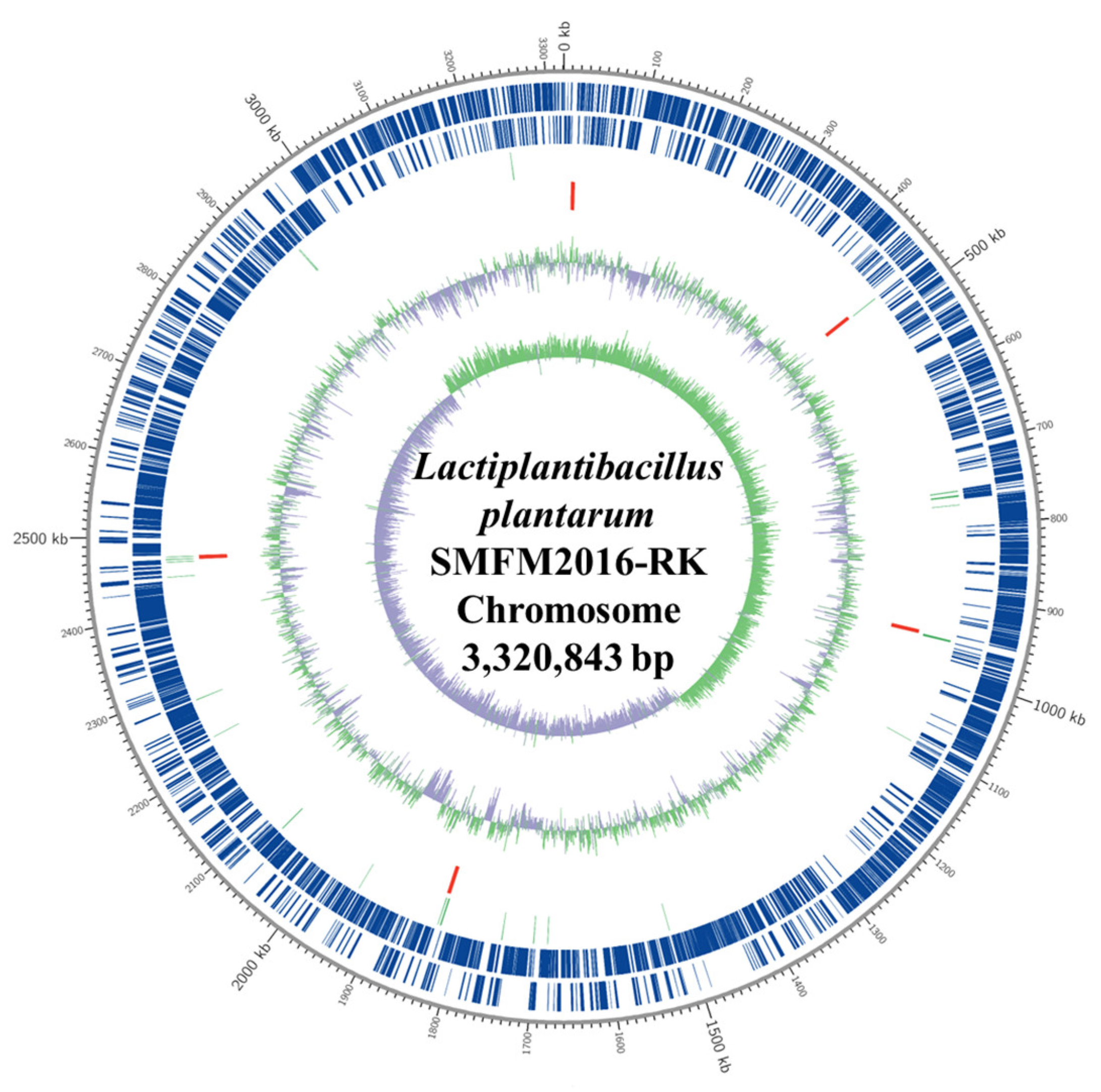
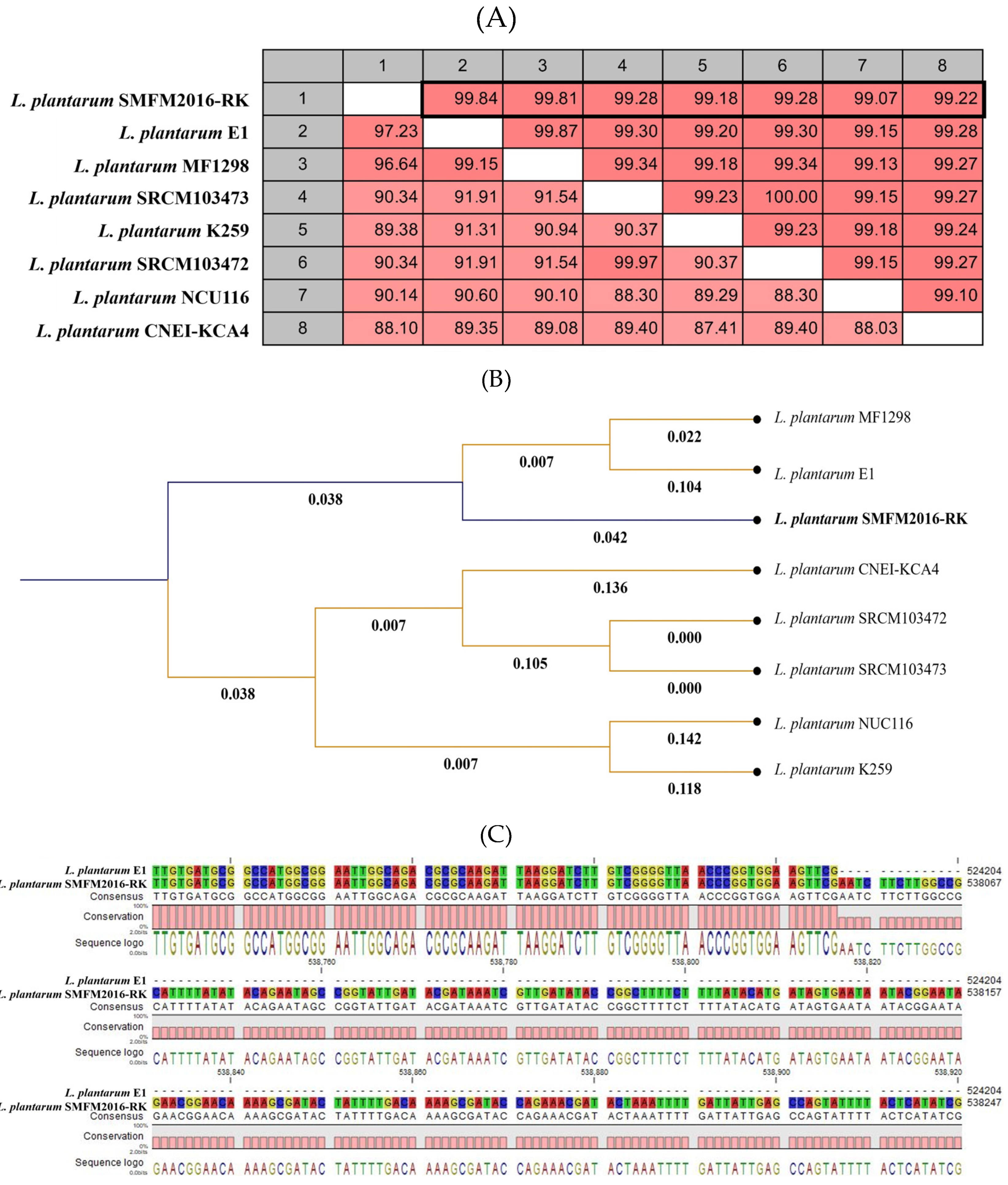
3.4. Whole-Genome Analysis of Novel Probiotics
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- WHO (World Health Organization). WHO highlights oral health neglect affecting nearly half of the world’s population. Available online: https://www.who.int/news/item/18-11-2022-who-highlights-oral-health-neglect-affecting-nearly-half-of-the-world-s-population (accessed on 10 August 2022).
- Borgnakke, W.S. IDF Diabetes Atlas: Diabetes and oral health – A two-way relationship of clinical importance. Diabetes Res. Clin. Pr. 2019, 157, 107839. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Haque, M. Oral Health Messiers: Diabetes Mellitus Relevance. Diabetes, Metab. Syndr. Obesity: Targets Ther. 3001; 14. [Google Scholar] [CrossRef]
- Van de Laar, F. A.; Lucassen, P. L.; Akkermans, R. P.; van de Lisdonk, E. H.; Rutten, G. E.; van Weel, C. α-Glucosidase inhibitors for patients with type 2 diabetes: results from a Cochrane systematic review and meta-analysis. Diabetes care, 2005, 28, 154–163. [Google Scholar] [CrossRef] [PubMed]
- Basri, R.; Ullah, S.; Halim, S. A.; Alharthy, R. D.; Rauf, U.; Khan, A. ; .. & Shafiq, Z. Synthesis, biological evaluation, and molecular docking study of chromen-linked hydrazine carbothioamides as potent α-glucosidase inhibitors. Drug Dev Res, 2023, 84, 962–974. [Google Scholar] [PubMed]
- Forozan, R.; Ghomi, M. K.; Iraji, A.; Montazer, M. N.; Noori, M.; Dastyafteh, N. ; .. ; Mahdavi, M. Synthesis, in vitro inhibitor screening, structure–activity relationship, and molecular dynamic simulation studies of novel thioquinoline derivatives as potent α-glucosidase inhibitors. Sci. Rep. 2023, 13, 7819. [Google Scholar] [PubMed]
- Ullah, S.; Waqas, M.; Halim, S. A.; Khan, I.; Khalid, A.; Abdalla, A. N. ; ..; Al-Harrasi, A. Triazolothiadiazoles and triazolothiadiazines as potent α-glucosidase inhibitors: Mechanistic insights from kinetics studies, molecular docking and dynamics simulations. Int. J. Biol. Macromol. 2023, 250, 126227. [Google Scholar]
- Huligere, S.S.; Kumari, V.B.C.; Alqadi, T.; Kumar, S.; Cull, C.A.; Amachawadi, R.G.; Ramu, R. Isolation and characterization of lactic acid bacteria with potential probiotic activity and further investigation of their activity by α-amylase and α-glucosidase inhibitions of fermented batters. Front. Microbiol. 2023, 13, 1042263. [Google Scholar] [CrossRef]
- Haque, M.; Yerex, K.; Kelekis-Cholakis, A.; Duan, K. Advances in novel therapeutic approaches for periodontal diseases. BMC Oral Heal. 2022, 22, 1–23. [Google Scholar] [CrossRef]
- Rosas-Val, P.; Adhami, M.; Brotons-Canto, A.; Gamazo, C.; Irache, J. M.; Larrañeta, E. 3D printing of microencapsulated Lactobacillus rhamnosus for oral delivery. Int. J. Pharm. 2023, 641123058. [Google Scholar] [CrossRef]
- Mahasneh, S.A.; Mahasneh, A.M. Probiotics: A Promising Role in Dental Health. Dent. J. 2017, 5, 26. [Google Scholar] [CrossRef]
- Zhu, Y.-F.; Chen, J.-J.; Ji, X.-M.; Hu, X.; Ling, T.-J.; Zhang, Z.-Z.; Bao, G.-H.; Wan, X.-C. Changes of major tea polyphenols and production of four new B-ring fission metabolites of catechins from post-fermented Jing-Wei Fu brick tea. Food Chem. 2015, 170, 110–117. [Google Scholar] [CrossRef]
- Luo, J.; Liu, S.; Lu, H.; Chen, Q.; Shi, Y. Microbial Community Variations and Bioconversion Improvements during Soybean-Based Fermentation by Kefir Grains. Foods 2023, 12, 1588. [Google Scholar] [CrossRef] [PubMed]
- Cosier, D.; Lambert, K.; Batterham, M.; Sanderson-Smith, M.; Mansfield, K.J.; Charlton, K. The INHABIT (synergIstic effect of aNtHocyAnin and proBIoTics in) Inflammatory Bowel Disease trial: a study protocol for a double-blind, randomised, controlled, multi-arm trial. J. Nutr. Sci. 2024, 13, e1. [Google Scholar] [CrossRef] [PubMed]
- Moufid, A.; Eddouks, M. Artemisia herba alba: A Popular Plant with Potential Medicinal Properties. Pak. J. Biol. Sci. 2012, 15, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Jang, H. J. Potential Use of Lactic acid Bacteria Isolated from Kimchi as Probiotics. 2018.
- Choi, Y.; Park, E.; Kim, S.; Ha, J.; Oh, H.; Kim, Y. ; ..; Choi, K. H. Alleviation of periodontal disease using Lactobacillus curvatus SMFM2016-NK. J. Funct. Foods 2021, 83, 104531. [Google Scholar]
- Hunt, R. Plant growth curves. The functional approach to plant growth analysis. Edward Arnold Ltd. 1982.
- Gallego, R.; Montero, L.; Cifuentes, A.; Ibáñez, E.; Herrero, M. Green Extraction of Bioactive Compounds from Microalgae. J. Anal. Test. 2018, 2, 109–123. [Google Scholar] [CrossRef]
- Mangia, N.P.; Saliba, L.; Deiana, P. Functional and safety characterization of autochthonous Lactobacillus paracasei FS103 isolated from sheep cheese and its survival in sheep and cow fermented milks during cold storage. Ann. Microbiol. 2018, 69, 161–170. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.-S.; Kim, Y.; Jeong, Y.; Kim, J.-E.; Paek, N.-S.; Kang, C.-H. Antioxidant and Probiotic Properties of Lactobacilli and Bifidobacteria of Human Origins. Biotechnol. Bioprocess Eng. 2020, 25, 421–430. [Google Scholar] [CrossRef]
- Nanno, M.; Morotomi, H.; Takayama, H.; Kuroshima, T.; Tanaka, R.; Mutai, M. Mutagenic activation of biliary metabolites of benzo (a) pyrene by β-glucuronidase-positive bacteria in human faeces. J.Med.Microbiol. 1986, 22, 351–5. [Google Scholar] [CrossRef]
- Rafter, J. Lactic acid bacteria and cancer: mechanistic perspective. Br. J. Nutr. 2002, 88, S89–S94. [Google Scholar] [CrossRef]
- Monteagudo-Mera, A.; Caro, I.; Rodríguez-Aparicio, L.B.; Rúa, J.; Ferrero, M.A.; García-Armesto, M.R. Characterization of Certain Bacterial Strains for Potential Use as Starter or Probiotic Cultures in Dairy Products. J. Food Prot. 2011, 74, 1379–1386. [Google Scholar] [CrossRef] [PubMed]
- Rice-Evans, C.; Miller, N.; Paganga, G. Antioxidant properties of phenolic compounds. Trends Plant Sci. 1997, 2, 152–159. [Google Scholar] [CrossRef]
- Ebringer, L.; Ferenčík, M.; Krajčovič, J. Beneficial health effects of milk and fermented dairy products — Review. Folia Microbiol. 2008, 53, 378–394. [Google Scholar] [CrossRef] [PubMed]
- Ostaff, M. J.; Stange, E. F.; Wehkamp, J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol. Med. 2013, 5, 1465–1483. [Google Scholar] [CrossRef] [PubMed]
- Oboh, G.; Ogunsuyi, O.B.; Ogunbadejo, M.D.; Adefegha, S.A. Influence of gallic acid on α-amylase and α-glucosidase inhibitory properties of acarbose. J. Food Drug Anal. 2016, 24, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Ramchandran, L.; Shah, N. Proteolytic Profiles and Angiotensin-I Converting Enzyme and α-Glucosidase Inhibitory Activities of Selected Lactic Acid Bacteria. J. Food Sci. 2008, 73, M75–M81. [Google Scholar] [CrossRef] [PubMed]
- Kwun, S.Y.; Bae, Y.W.; Yoon, J.A.; Park, E.H.; Kim, M.D. Isolation of acid tolerant lactic acid bacteria and evaluation of α-glucosidase inhibitory activity. Food Sci. Biotechnol. 2020, 29, 1125–1130. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M. J.; Anand, U.; Altemimi, A. B.; Tripathi, V.; Guo, Y.; Pratap-Singh, A. Phenolic composition, antioxidant capacity and antibacterial activity of white wormwood (Artemisia herba-alba). Plants. 2021, 10, 164. [Google Scholar] [CrossRef] [PubMed]
- Younsi, F.; Trimech, R.; Boulila, A.; Ezzine, O.; Dhahri, S.; Boussaid, M.; Messaoud, C. Essential oil and phenolic compounds of Artemisia herba-alba (Asso. ): Composition, antioxidant, antiacetylcholinesterase, and antibacterial activities. Int. J. Food Prop. 2016, 19, 1425–1438. [Google Scholar]
- Loo, Y.T.; Howell, K.; Chan, M.; Zhang, P.; Ng, K. Modulation of the human gut microbiota by phenolics and phenolic fiber-rich foods. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1268–1298. [Google Scholar] [CrossRef]
- Leonard, W.; Zhang, P.; Ying, D.; Fang, Z. Hydroxycinnamic acids on gut microbiota and health. Compr. Rev. Food Sci. Food Saf. 2021, 20, 710–737. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Miao, M. Dietary polyphenols modulate starch digestion and glycaemic level: a review. Crit. Rev. Food Sci. Nutr. 2020, 60, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Varghese, N.J.; Mukherjee, S.; Ivanova, N.; Konstantinidis, K.T.; Mavrommatis, K.; Kyrpides, N.C.; Pati, A. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015, 43, 6761–6771. [Google Scholar] [CrossRef] [PubMed]
| Artermisia herba-alba extracts |
F. nucleatum ATCC43718 | A. actinomycetemcomitans ATCC10953 | P. gingivalis ATCC33277 | Total average |
|---|---|---|---|---|
| Ethanol extracts | 4.3±1.7C | 5.3±1.7C | 1.4±0.4C | 4.0±2.1 |
| Hot-water extracts | 5.9±1.3C | 10.7±3.3B | 26.5±2.0A | 13.6±9.1 |
| Artermisia herba-alba extracts |
F. nucleatum ATCC43718 | A. actinomycetemcomitans ATCC10953 | P. gingivalis ATCC33277 | Total average |
|---|---|---|---|---|
| Ethanol extracts | 4.3±1.7C | 5.3±1.7C | 1.4±0.4C | 4.0±2.1 |
| Hot-water extracts | 5.9±1.3C | 10.7±3.3B | 26.5±2.0A | 13.6±9.1 |
| Lactic acid bacteria strain | MRS broth | MRS broth+ A. herba-alba ethanol extract (5 mg/mL) |
MRS broth+ A. herba-alba hot-water extract (25 mg/mL) |
|---|---|---|---|
| Pediococcus pentosaceus SMFM2016-GK | 0.054±0.002Aa* | 0.052±0.002Aab | 0.052±0.002Aab |
| P. pentosaceus SMFM2016-NK1 | 0.052±0.002Aab | 0.051±0.003Aab | 0.050±0.001Aab |
| P. pentosaceus SMFM2016-NK2 | 0.052±0.001Aab | 0.052±0.004Aab | 0.052±0.003Aab |
| P. pentosaceus SMFM2016-NK3 | 0.053±0.002Aab | 0.051±0.002Aab | 0.051±0.002Aab |
| P. pentosaceus SMFM2016-SK | 0.053±0.001Aa | 0.053±0.004Aab | 0.052±0.002Aab |
| Lactiplantibacillus plantarum SMFM2016-RK | 0.049±0.001Ab | 0.048±0.002Ab | 0.049±0.002Ab |
| P. pentosaceus SMFM2016-NK4 | 0.053±0.002Aa | 0.052±0.002Aab | 0.052±0.002Aab |
| P. pentosaceus SMFM2016-RK | 0.053±0.002Aab | 0.050±0.002Aab | 0.050±0.002Aab |
| Lactobacillus curvatus SMFM2016-NK | 0.050±0.002Ab | 0.050±0.001Ab | 0.049±0.002Ab |
| Lactobacillus rhamnosus GG | 0.042±0.004Ac | 0.028±0.006Bc | 0.44±0.004Ac |
| Lactic acid bacteria strain | MRS broth | MRS broth+ A. herba-alba ethanol extract (5 mg/mL) |
MRS broth+ A. herba-alba hot-water extract (25 mg/mL) |
| Pediococcus pentosaceus SMFM2016-GK | 12.9±0.6Ac | 13.5±0.4Ab | 13.3±0.5Ab |
| P. pentosaceus SMFM2016-NK1 | 13.3±0.4Aab | 13.6±0.8Ab | 13.9±0.2Ab |
| P. pentosaceus SMFM2016-NK2 | 13.2±0.3Aab | 13.3±0.9Ab | 13.3±0.7Ab |
| P. pentosaceus SMFM2016-NK3 | 13.2±0.4Aab | 13.5±0.6Ab | 13.7±0.6Ab |
| P. pentosaceus SMFM2016-SK | 13.1±0.4Aab | 13.2±0.9Ab | 13.3±0.5Ab |
| Lactiplantibacillus plantarum SMFM2016-RK | 14.3±0.3Ab | 14.5±0.6Ab | 14.2±0.5Ab |
| P. pentosaceus SMFM2016-NK4 | 13.0±0.5Aab | 13.4±0.4Ab | 13.2±0.6Ab |
| P. pentosaceus SMFM2016-RK | 13.1±0.5Aab | 13.9±0.5Ab | 13.8±0.7Ab |
| Lactobacillus curvatus SMFM2016-NK | 13.8±0.5Aab | 3.9±0.4Ab | 14.3±0.5Ab |
| Lactobacillus rhamnosus GG | 16.7±1.6Ba | 25.6±6.3Aa | 15.7±1.4Ba |
| Lactic acid bacteria strain | MRS broth | MRS broth + A. herba-alba ethanol extract (5 mg/mL) | MRS broth + A. herba-alba hot-water extract (25 mg/mL) | |||
|---|---|---|---|---|---|---|
| Aggregatibacter actinomycetemcomitans ATCC43718 | Fusobacterium nucleatum ATCC10953 | A. actinomycetemcomitans ATCC43718 | F. nucleatum ATCC10953 | A. actinomycetemcomitans ATCC43718 | F. nucleatum ATCC10953 | |
| Pediococcus pentosaceus SMFM2016-GK | 1.0±0.0Ab | 1.5±0.6ABbc | 0.5±0.6Bd | 1.8±0.3Ac | 1.1±0.3Ab | 0.7±0.6Bc |
| P. pentosaceus SMFM2016-NK1 | 1.0±0.0Ab | 1.3±0.5Abc | 1.0±0.0Ac | 1.4±0.5Ac | 1.1±0.3Ab | 0.3±0.5Bc |
| P. pentosaceus SMFM2016-NK2 | 1.3±0.3Ab | 1.4±0.5ABbc | 1.0±0.0Ac | 1.8±0.3Ac | 1.1±0.3Ab | 0.7±0.6Bc |
| P. pentosaceus SMFM2016-NK3 | 1.0±0.0Ab | 1.5±0.9Abc | 1.0±0.0Ac | 1.2±1.0Ac | 1.1±0.3Ab | 0.0±0.0Bc |
| P. pentosaceus SMFM2016-SK | 1.0±0.0Ab | 2.0±0.5Ab | 1.0±0.0Ac | 1.6±0.5Ac | 0.8±0.8Ab | 0.3±0.5Bc |
| Lactiplantibacillus plantarum SMFM2016-RK | 1.4±0.5Ca | 3.1±0.3Aa | 2.1±0.3Aa | 2.8±0.9Ab | 2.0±0.0Ba | 3.1±0.3Aa |
| P. pentosaceus SMFM2016-NK4 | 1.0±0.4Ab | 0.9±0.6Ac | 1.0±0.0Ac | 1.1±0.3Ac | 0.9±0.3Ab | 0.8±0.3Ac |
| P. pentosaceus SMFM2016-RK | 1.0±0.0Ab | 0.8±0.5Ac | 1.1±0.3Abc | 1.0±0.0Ac | 1.0±0.0Ab | 1.0±0.0Abc |
| Lactobacillus curvatus SMFM2016-NK | 1.0±0.0Bb | 3.1±1.0Ba | 1.9±0.3Aab | 4.0±0.9Aa | 1.6±0.5Aa | 2.6±0.9Ba |
| Lactobacillus rhamnosus GG | 1.0±0.0Bb | 1.5±0.6Abc* | 1.5±0.6Ab | 2.1±0.9Abc | 1.8±0.5Aa | 1.8±0.5Ab |
| Lactic acid bacteria strain | MRS broth | MRS broth+ A. herba-alba ethanol extract (5 mg/mL) |
MRS broth+ A. herba-alba hot-water extract (25 mg/mL) |
|---|---|---|---|
| Positive control (Lactobacillus rhamnosus GG) |
83.2±1.7Aa* | 60.0±5.5Ba | 26.2±0.9Cb |
| Lactiplantibacillus plantarum SMFM2016-RK | 85.2±0.3Aa | 60.8±5.0Ba | 35.0±0.2Ca |
| Lactobacillus curvatus SMFM2016-NK | 0.0±0.0Bb | 2.7±1.2Ab | 5.9±2.6Ac |
| Sample | 0 h | 6 h | 24 h | 37 h | ||||
|---|---|---|---|---|---|---|---|---|
| pH | cell counts (Log CFU/mL) | pH | cell counts (Log CFU/mL) | pH | cell counts (Log CFU/mL) | pH | cell counts (Log CFU/mL) | |
| BM1 | 6.57±0.08 | 8.3±0.0 | - | - | 4.42±0.57 | 8.8±0.2 | - | - |
| BM2 | 6.39±0.07 | 8.3±0.0 | - | - | - | - | 4.58±0.50 | 8.8±0.3 |
| BM3 | 6.20±0.05 | 8.3±0.2 | 4.62±0.30 | 9.2±0.1 | - | - | - | - |
| BM4 | 6.11±0.05 | 8.2±0.1 | 4.58±0.25 | 9.2±0.1 | - | - | - | - |
| Sample | Periodontal pathogen | Total average | ||
|---|---|---|---|---|
| Aggregatibacter actinomycetemcomitans ATCC43718 | Fusobacterium nucleatum ATCC10953 | Porphyromonas gingivalis ATCC33277 | ||
| 10% skim milk |
0.0±0.0D | 0.0±0.0D | 0.0±0.0D | 0.0±0.0 |
| BM1 | 2.6±1.1B | 1.6±0.5C | 1.9±0.3BC | 2.0±0.8 |
| BM2 | 3.0±0.8AB | 2.5±0.6BC | 1.8±0.5BC | 2.4±0.8 |
| BM3 | 3.6±0.8A | 1.9±1.0BC | 1.9±0.3BC | 2.5±1.1 |
| BM4 | 3.3±1.5AB | 2.9±0.6AB | 1.6±0.5C | 2.6±1.1 |
| Category | Gene ontology | Number of transcripts |
|---|---|---|
| Cellular component | Cell part | 263 |
| Cell | 263 | |
| Protein-containing complex | 120 | |
| Organelle | 61 | |
| Extracellular region | 5 | |
| Membrane | 397 | |
| Membrane part | 249 | |
| Extracellular region part | 4 | |
| Organelle part | 16 | |
| Molecular function | Catalytic activity | 1,087 |
| Binding | 805 | |
| Molecular carrier activity | 2 | |
| Transport activity | 219 | |
| Antioxidant activity | 8 | |
| Transcription regulator activity | 105 | |
| Molecular function regulator | 1 | |
| Structural molecule activity | 51 | |
| Molecular transducer activity | 2 | |
| Biological process | Localization | 317 |
| Response to stimulus | 104 | |
| Metabolic process | 1,089 | |
| Cellular process | 845 | |
| Biological regulation | 223 | |
| Regulation of biological process | 217 | |
| Cellular component organization or biogenesis | 56 | |
| Negative regulation of biological process | 9 | |
| Multi-organism process | 9 | |
| Signaling | 22 | |
| Developmental process | 9 | |
| Immune system process | 1 | |
| Biological adhesion | 8 | |
| Detoxification | 2 | |
| Carbon utilization | 1 |
| Description | Number of ORFs | Ratio (%) |
|---|---|---|
| Translation, ribosomal structure, and biogenesis | 150 | 4.7847 |
| Transcription | 258 | 8.2297 |
| Replication, recombination, and repair | 196 | 6.2520 |
| Cell cycle control, cell division, chromosome partitioning | 26 | 0.8293 |
| Defense mechanisms | 66 | 2.1053 |
| Signal transduction mechanisms | 70 | 2.2329 |
| Cell wall/membrane/envelope biogenesis | 179 | 5.7097 |
| Cell motility | 4 | 0.1276 |
| Intracellular trafficking, secretion, and vesicular transport | 25 | 0.7974 |
| Posttranslational modification, protein turnover, chaperones | 69 | 2.2010 |
| Energy production and conversion | 111 | 3.5407 |
| Carbohydrate transport and metabolism | 290 | 9.2504 |
| Amino acid transport and metabolism | 206 | 6.5710 |
| Nucleotide transport and metabolism | 86 | 2.7432 |
| Coenzyme transport and metabolism | 63 | 2.0096 |
| Lipid transport and metabolism | 62 | 1.9777 |
| Inorganic ion transport and metabolism | 125 | 3.9872 |
| Secondary metabolites biosynthesis, transport, and catabolism | 20 | 0.6380 |
| General function prediction only | 349 | 11.1324 |
| Function unknown | 780 | 24.8804 |
| Total | 3,135 | 100 |
| Start | End | Product | Gene | Identity | e-value | Bit score |
|---|---|---|---|---|---|---|
| 1849164 | 1850129 | Oligopeptide transport ATP-binding protein OppF | oppF | 77.636 | 0.0 | 518 |
| 1850136 | 1851215 | Oligopeptide transport ATP-binding protein OppD | oppD | 79.883 | 0.0 | 570 |
| 2470957 | 2472429 | Di-/tripeptide transporter | dtpT | 66.189 | 0.0 | 648 |
| 962965 | 964776 | Oligoendopeptidase F, plasmid | pepF1 | 57.333 | 0.0 | 728 |
| 3234367 | 3236283 | Neutral endopeptidase | pepO | 59.528 | 0.0 | 791 |
| 504486 | 505796 | D-serine dehydratase | dsdA | 57.619 | 9.40e-175 | 501 |
| 2530776 | 2531678 | L-serine dehydratase, alpha chain | sdhA | 72.069 | 5.73e-138 | 397 |
| 820964 | 822202 | Serine hydroxymethyltransferase | glyA | 70.270 | 0.0 | 607 |
| 1173945 | 1175741 | Aspartate--tRNA ligase | aspS | 74.617 | 0.0 | 917 |
| 2291674 | 2292909 | Argininosuccinate synthase | argG | 73.350 | 0.0 | 634 |
| 2290271 | 2291674 | Argininosuccinate lyase | argH | 71.024 | 0.0 | 699 |
| Start | End | Product | Gene | Identity | e-value | Bit score |
|---|---|---|---|---|---|---|
| 335672 | 337144 | Gallate decarboxylase | lpdC | 82.857 | 0 | 841 |
| 3008485 | 3009021 | Phenolic acid decarboxylase PadC | padC | 87.64 | 3.70E-116 | 332 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).





