Submitted:
21 May 2024
Posted:
24 May 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Network Pharmacology
Target Identification
Gene Ontology
String
Cytoscape
Materials and Methods
Text Mining
GO Enrichment Analysis
PPI Network Analysis
Lead Identification
Molecular Docking
Results
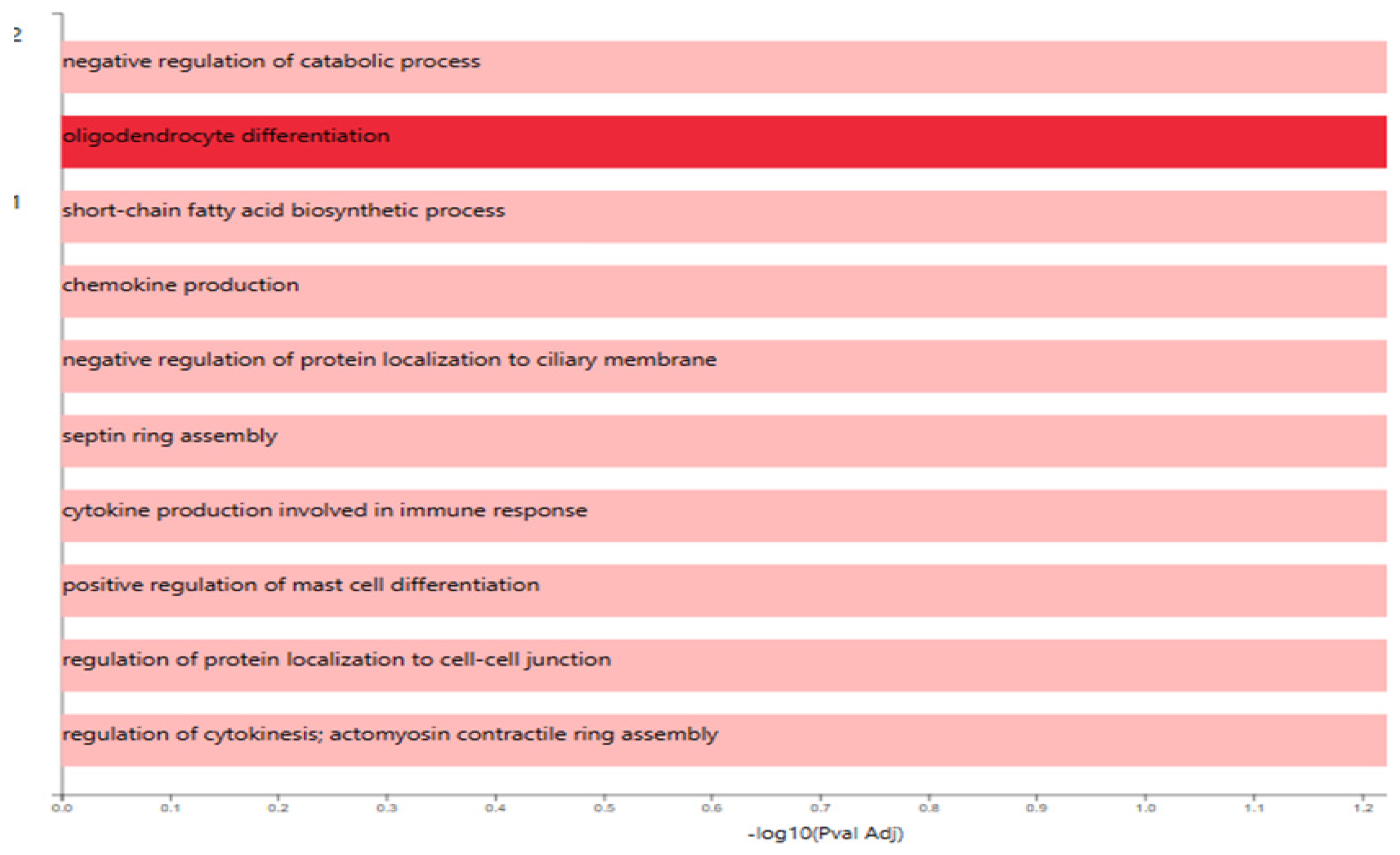
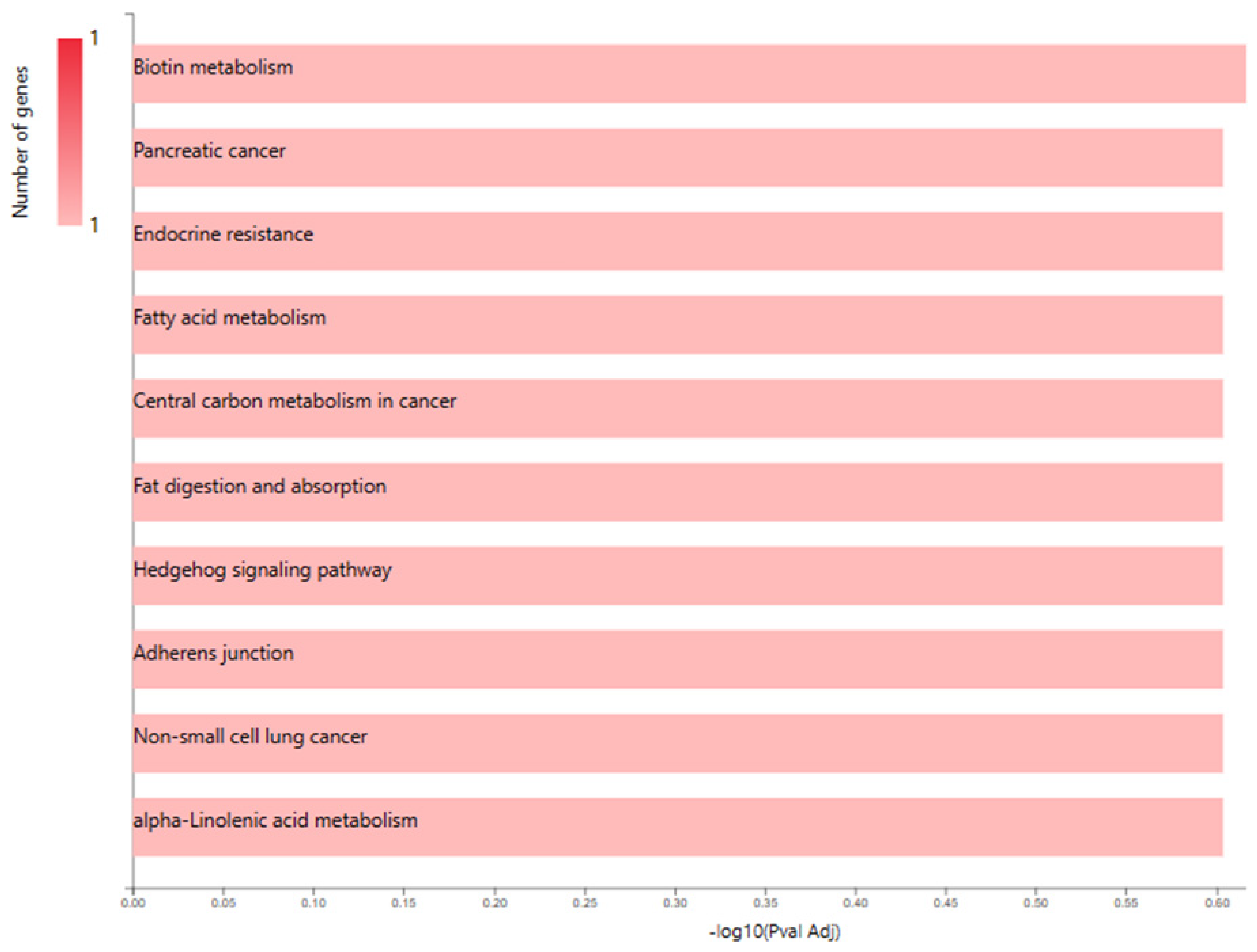
Go Enrichment Analysis
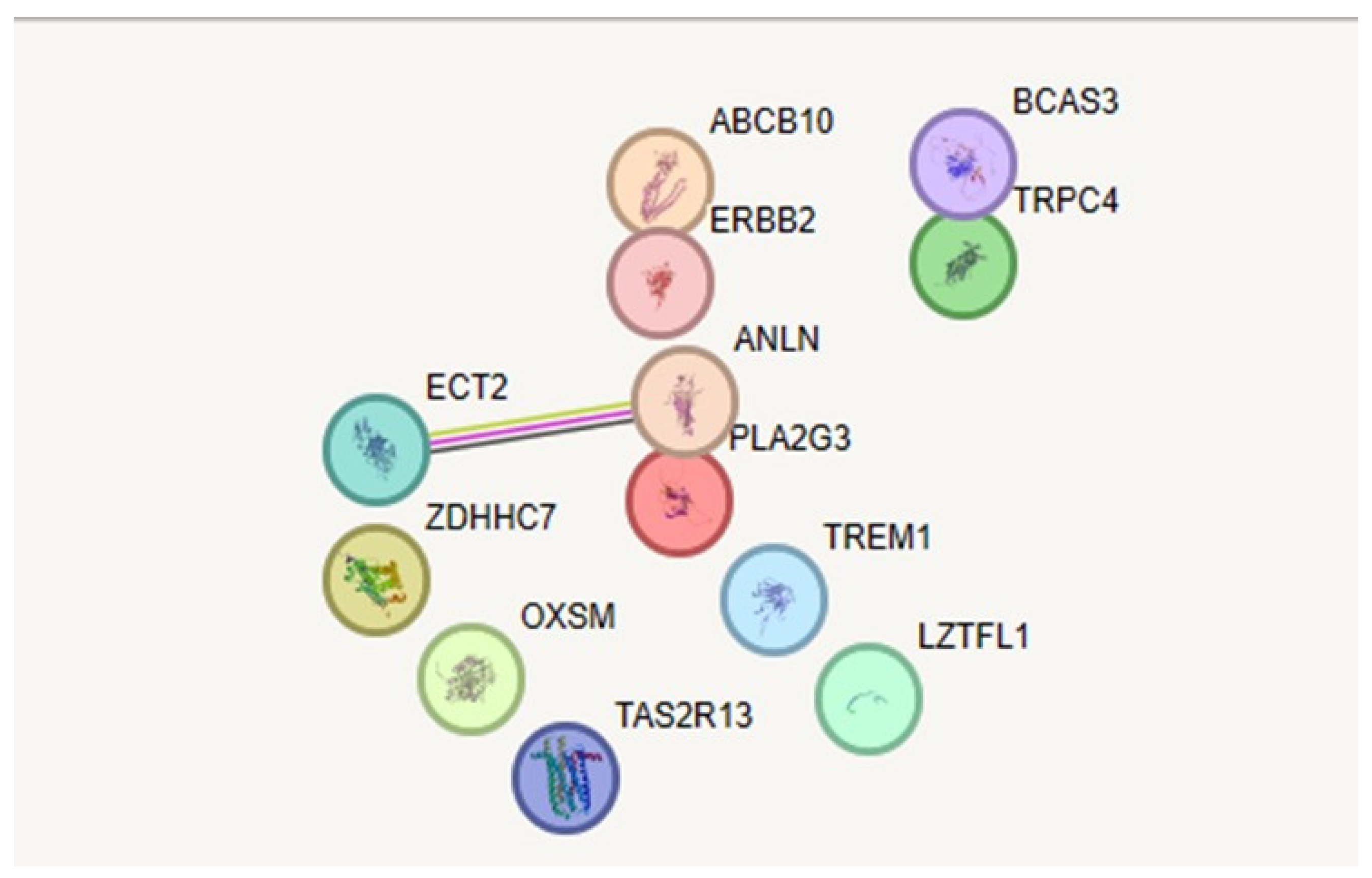
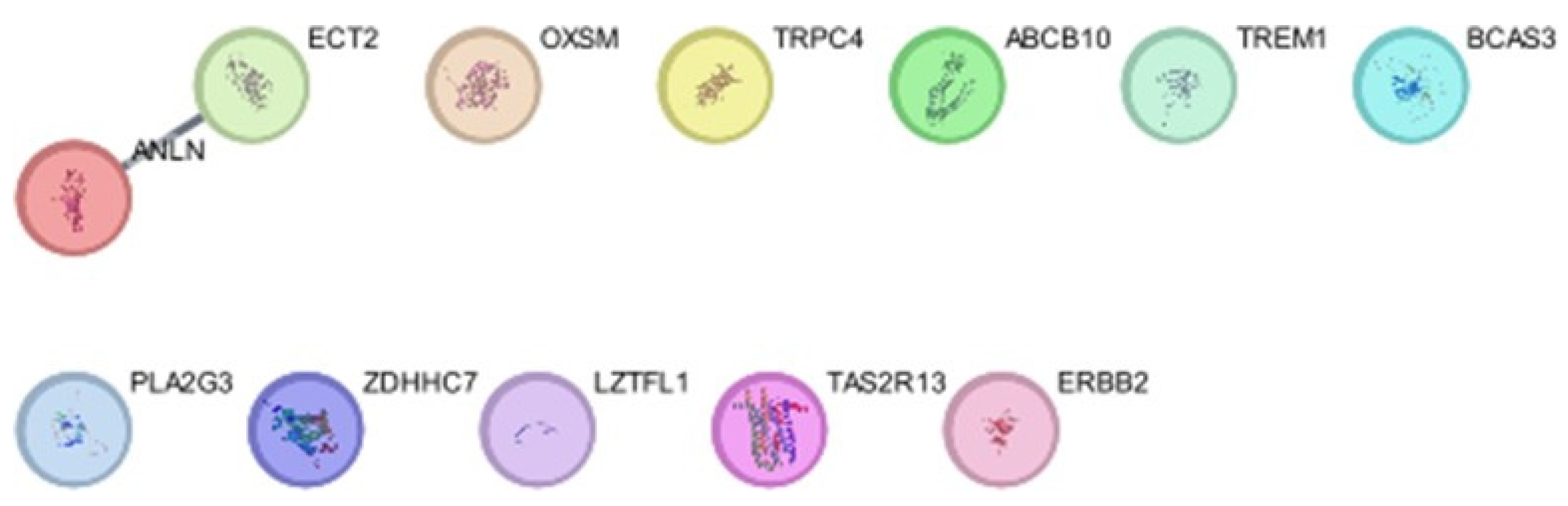
PPI Network Analysis
Degree and betweenness Analysis
Molecular Docking
|
Entry |
Gene name |
|
Q9H2R5 |
KLK15 |
|
Q9H2X6 |
HIPK2 |
|
Q9H2X9 |
SLC12A5 KCC2 KIAA1176 |
|
Q9H3D4 |
TP63 KET P63 P73H P73L TP73L |
|
Q9H3H5 |
DPAGT1 DPAGT2 |
|
Q9H3M9 |
ATXN3L ATX3L MJDL |
|
Q9H3R5 |
CENPH ICEN35 |
|
Q9H3T3 |
SEMA6B SEMAN SEMAZ UNQ1907/PRO4353 |
|
Q9H3V2 |
MS4A5 CD20L2 TETM4 |
|
Q9H3Y6 |
SRMS C20orf148 |
|
Q9H467 |
CUEDC2 C10orf66 HOYS6 |
|
Q9H4A3 |
WNK1 HSN2 KDP KIAA0344 PRKWNK1 |
|
Q9H4B4 |
PLK3 CNK FNK PRK |
|
Q9H4D0 |
CLSTN2 CS2 |
|
Q9H4H8 |
FAM83D C20orf129 |
|
Q9H4L2 |
BRCA2 |
|
Q9H4L3 |
BRCA2 |
|
Q9H4T2 |
ZSCAN16 ZNF392 ZNF435 |
|
Q9H4X1 |
RGCC C13orf15 RGC32 |
|
Q9H582 |
ZNF644 KIAA1221 ZEP2 |
|
Q9H596 |
DUSP21 LMWDSP21 |
|
Q9H5I1 |
SUV39H2 KMT1B |
|
Q9H5J0 |
ZBTB3 |
|
Q9H5V8 |
CDCP1 TRASK UNQ2486/PRO5773 |
|
Q9H6B1 |
ZNF385D ZNF659 |
|
Q9H6R7 |
WDCP C2orf44 PP384 |
|
Q9H6U6 |
BCAS3 |
|
Q9H7D7 |
WDR26 CDW2 MIP2 PRO0852 |
|
Q9H7L9 |
SUDS3 SAP45 SDS3 |
|
Q9H7N4 |
SCAF1 SFRS19 SRA1 |
|
Q9H7R0 |
ZNF442 |
|
Q9H7S9 |
ZNF703 ZEPPO1 ZPO1 |
|
Q9H813 |
PACC1 C1orf75 TMEM206 |
|
Q9H8L6 |
MMRN2 EMILIN3 |
|
Q9H8V3 |
ECT2 |
|
Q9H8Y1 |
VRTN C14orf115 |
|
Q9H910 |
JPT2 C16orf34 HN1L L11 |
|
Q9H9B1 |
EHMT1 EUHMTASE1 GLP KIAA1876 KMT1D |
|
Q9HAU4 |
SMURF2 |
|
Q9HAU8 |
RNPEPL1 |
|
Q9HAW4 |
CLSPN |
|
Q9HAX1 |
DSC3 |
|
Q9HB09 |
BCL2L12 BPR |
|
Q9HB58 |
SP110 |
|
Q9HBL0 |
TNS1 TNS |
|
Q9HBT6 |
CDH20 CDH7L3 |
|
Q9HBW1 |
LRRC4 BAG NAG14 UNQ554/PRO1111 |
|
Q9HBX8 |
LGR6 UNQ6427/PRO21331 VTS20631 |
|
Q9HC10 |
OTOF FER1L2 |
|
Q9HC57 |
WFDC1 PS20 |
|
Q9HC96 |
CAPN10 KIAA1845 |
|
Q9HCE0 |
EPG5 KIAA1632 |
|
Q9HCE3 |
ZNF532 KIAA1629 |
|
Q9HCE6 |
ARHGEF10L GRINCHGEF KIAA1626 |
|
Q9HCH5 |
SYTL2 KIAA1597 SGA72M SLP2 SLP2A |
|
Q9HCI5 |
MAGEE1 HCA1 KIAA1587 |
|
Q9HCS4 |
TCF7L1 TCF3 |
|
Q9HCU0 |
CD248 CD164L1 TEM1 |
|
Q9HCU9 |
BRMS1 |
|
Q9HD15 |
SRA1 PP7684 |
|
Q9HD64 |
XAGE1A GAGED2 XAGE1; XAGE1B XAGE1C XAGE1D XAGE1E |
|
Q9HDB8 |
ERVK-5 ERVK5 |
|
Q9NP09 |
ERBB2 |
|
Q9NP58 |
ABCB6 MTABC3 PRP UMAT |
|
Q9NP60 |
IL1RAPL2 IL1R9 |
|
Q9NP61 |
ARFGAP3 ARFGAP1 |
|
Q9NP99 |
TREM1 |
|
Q9NPC2 |
KCNK9 TASK3 |
|
Q9NPD5 |
SLCO1B3 LST2 OATP1B3 OATP8 SLC21A8 |
|
Q9NPD8 |
UBE2T HSPC150 PIG50 |
|
Q9NPG8 |
ZDHHC4 ZNF374 DC1 UNQ5787/PRO19576 |
|
Q9NPY3 |
CD93 C1QR1 MXRA4 |
|
Q9NQ31 |
AKIP1 BCA3 C11orf17 |
|
Q9NQ48 |
LZTFL1 |
|
Q9NQ66 |
PLCB1 KIAA0581 |
|
Q9NQ88 |
TIGAR C12orf5 |
|
Q9NQB0 |
TCF7L2 TCF4 |
|
Q9NQC3 |
RTN4 KIAA0886 NOGO My043 SP1507 |
|
Q9NQG5 |
RPRD1B C20orf77 CREPT |
|
Q9NQR3 |
BRCA1 |
|
Q9NQU5 |
PAK6 PAK5 |
|
Q9NQW6 |
ANLN |
|
Q9NQX1 |
PRDM5 PFM2 |
|
Q9NR12 |
PDLIM7 ENIGMA |
|
Q9NR30 |
DDX21 |
|
Q9NR80 |
ARHGEF4 KIAA1112 |
|
Q9NR82 |
KCNQ5 |
|
Q9NR96 |
TLR9 UNQ5798/PRO19605 |
|
Q9NRA2 |
SLC17A5 |
|
Q9NRC1 |
ST7 FAM4A1 HELG RAY1 |
|
Q9NRD0 |
FBXO8 FBS FBX8 DC10 UNQ1877/PRO4320 |
|
Q9NRH2 |
SNRK KIAA0096 SNFRK |
|
Q9NRJ1 |
C8orf17 |
|
Q9NRK6 |
ABCB10 |
|
Q9NRM2 |
ZNF277 NRIF4 ZNF277P |
|
Q9NRN9 |
METTL5 DC3 HSPC133 |
|
Q9NRP0 |
OSTC DC2 HDCMD45P HSPC307 |
|
Q9NRP7 |
STK36 KIAA1278 |
|
Q9NRY4 |
ARHGAP35 GRF1 GRLF1 KIAA1722 P190A p190ARHOGAP |
|
Q9NS23 |
RASSF1 RDA32 |
|
Q9NS39 |
ADARB2 ADAR3 RED2 |
|
Q9NS71 |
GKN1 AMP18 CA11 UNQ489/PRO1005 |
|
Q9NS87 |
KIF15 KLP2 KNSL7 |
|
Q9NSC7 |
ST6GALNAC1 SIAT7A UNQ543/PRO848 |
|
Q9NTF0 |
DKFZp727E011 |
|
Q9NTG1 |
PKDREJ |
|
Q9NTK1 |
DEPP1 C10orf10 DEPP FIG |
|
Q9NTK5 |
OLA1 GTPBP9 PRO2455 PTD004 |
|
Q9NTM9 |
CUTC CGI-32 |
|
Q9NTW7 |
ZFP64 ZNF338 |
|
Q9NTX7 |
RNF146 |
|
Q9NTX9 |
FAM217B C20orf177 |
|
Q9NU02 |
ANKEF1 ANKRD5 |
|
Q9NUM3 |
SLC39A9 ZIP9 UNQ714/PRO1377 |
|
Q9NUQ7 |
UFSP2 C4orf20 |
|
Q9NUT2 |
ABCB8 MABC1 MITOSUR |
|
Q9NV12 |
TMEM140 |
|
Q9NV23 |
OLAH THEDC1 |
|
Q9NV64 |
TMEM39A SUSR2 |
|
Q9NVA1 |
UQCC1 BZFB C20orf44 UQCC |
|
Q9NVD3 |
SETD4 C21orf18 C21orf27 |
|
Q9NVE7 |
PANK4 |
|
Q9NVM9 |
INTS13 ASUN C12orf11 GCT1 |
|
Q9NVP1 |
DDX18 cPERP-D |
|
Q9NVW2 |
RLIM RNF12 |
|
Q9NVX2 |
NLE1 HUSSY-07 |
|
Q9NW75 |
GPATCH2 GPATC2 |
|
Q9NWB6 |
ARGLU1 |
|
Q9NWK9 |
ZNHIT6 BCD1 C1orf181 |
|
Q9NWM0 |
SMOX C20orf16 SMO UNQ3039/PRO9854 |
|
Q9NWU1 |
OXSM |
|
Q9NX61 |
TMEM161A UNQ582/PRO1152 |
|
Q9NXF8 |
ZDHHC7 |
|
Q9NXL2 |
ARHGEF38 |
|
Q9NY72 |
SCN3B KIAA1158 |
|
Q9NY74 |
ETAA1 ETAA16 |
|
Q9NYC9 |
DNAH9 DNAH17L DNEL1 KIAA0357 |
|
Q9NYF0 |
DACT1 DPR1 HNG3 |
|
Q9NYV9 |
TAS2R13 |
|
Q9NZ20 |
PLA2G3 |
|
Q9NZ52 |
GGA3 KIAA0154 |
|
Q9NZC7 |
WWOX FOR SDR41C1 WOX1 |
|
Q9NZC9 |
SMARCAL1 HARP |
|
Q9NZJ4 |
SACS KIAA0730 |
|
Q9NZJ5 |
EIF2AK3 PEK PERK |
|
Q9NZL6 |
RGL1 KIAA0959 RGL |
|
Q9NZP6 |
NPAP1 C15orf2 |
|
Q9P032 |
NDUFAF4 C6orf66 HRPAP20 HSPC125 My013 |
|
Q9P0G3 |
KLK14 KLKL6 |
|
Q9P253 |
VPS18 KIAA1475 |
|
Q9P260 |
RELCH KIAA1468 |
|
Q9P283 |
SEMA5B KIAA1445 SEMAG UNQ5867/PRO34001 |
|
Q9P287 |
BCCIP TOK1 |
|
Q9P2J8 |
ZNF624 KIAA1349 |
|
Q9P2U8 |
SLC17A6 DNPI VGLUT2 |
|
Q9T3Q5 |
ND2 NADH2 |
|
Q9UBB6 |
NCDN KIAA0607 |
|
Q9UBF1 |
MAGEC2 HCA587 MAGEE1 |
|
Q9UBF9 |
MYOT TTID |
|
Q9UBK9 |
UXT HSPC024 |
|
Q9UBN4 |
TRPC4 |
|
Q9UBN7 |
HDAC6 KIAA0901 JM21 |
|
Q9UBP0 |
SPAST ADPSP FSP2 KIAA1083 SPG4 |
|
Q9UBS9 |
SUCO C1orf9 CH1 OPT SLP1 |
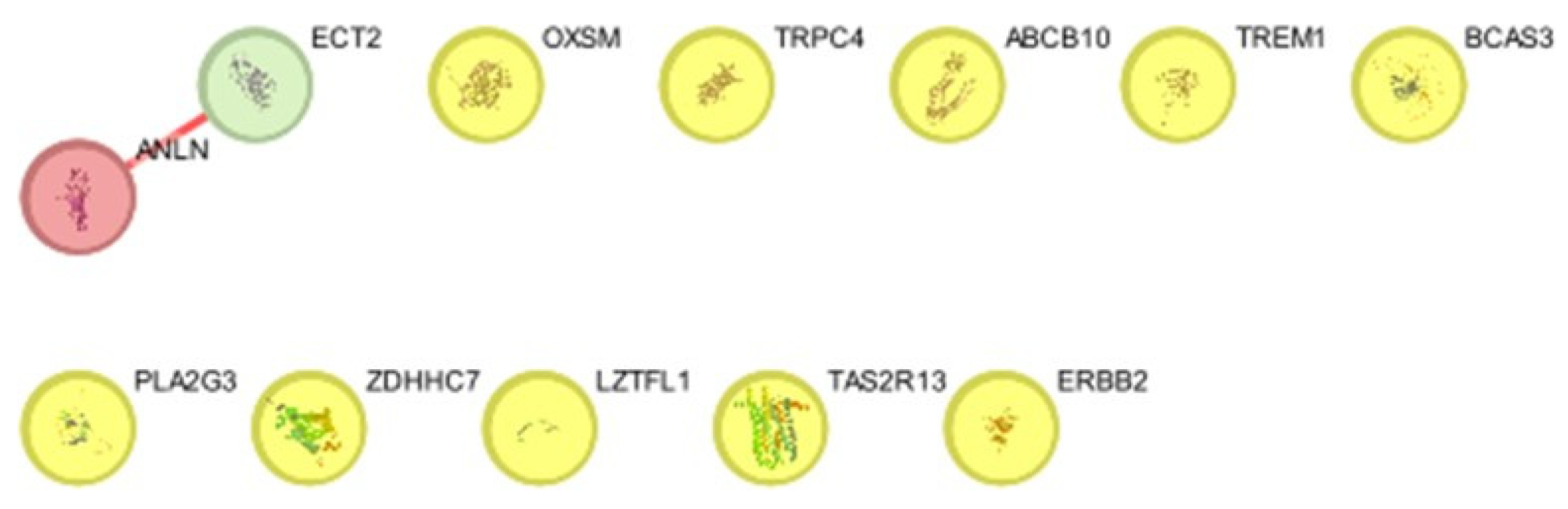
| GENES | DEGREE | BETWEENES |
|
ERBB2 |
0 |
0 |
|
LZTFL1 |
0 |
0 |
|
ZDHHC7 |
0 |
0 |
|
PLA2G3 |
0 |
0 |
|
BCAS3 |
0 |
0 |
|
TREM1 |
0 |
0 |
|
ABCB10 |
0 |
0 |
|
ECT2 |
1 |
0 |
| TRPC4 | 0 | 0 |
| OXSM | 0 | 0 |
|
ANLN |
1 |
0 |
| Epigallocatechin | Gallocatechol |
| Epicatechin gallate | Gallic acid |
| Catechin | Theaflavin |
| Theanine | Chlorogenic acid |
| Theobromine | Epicatechin 3 gallate |
| Catechol | epigallocatechin |
| Gallocatechin | linolool |
| Quinic acid |
|
Ligand |
Binding Affinity |
Ligand |
Binding Affinity |
|
ABCB10_catechin |
-6.6 |
ERBB2_catechin |
-7.5 |
|
ABCB10_CATECHOL |
-4.4 |
ERBB2_CATECHOL | -5.2 |
|
ABCB10_chlorogenic_acid |
-6.4 |
ERBB2_chlorogenic_acid |
-7 |
|
ABCB10_epicatechin_3_gallate |
-7.3 |
ERBB2_epicatechin_3_galla te |
-8.8 |
|
ABCB10_epicatechin_gallate |
-7.2 |
ERBB2_epicatechin_gallate | -7.6 |
|
ABCB10_epigallocatechin |
-7.2 |
ERBB2_epigallocatechin |
-8.9 |
|
ABCB10_epigallocatechol |
-6.7 |
ERBB2_epigallocatechol |
-7.5 |
|
ABCB10_gallic_acid |
-5 |
ERBB2_gallic_acid |
-6.3 |
|
ABCB10_gallocatechin |
-6.8 |
ERBB2_gallocatechin |
-7.2 |
|
ABCB10_gallocatechol |
-6.8 |
ERBB2_gallocatechol |
-7.2 |
|
ABCB10_linalool |
-5 |
ERBB2_linalool |
-5 |
|
ABCB10_quinic_acid |
-4.9 |
ERBB2_quinic_acid | -5.9 |
|
ABCB10_theaflavin |
-7.9 |
ERBB2_theaflavin |
-9.4 |
|
ABCB10_theanine |
-4.4 |
ERBB2_theanine | -4.6 |
|
ABCB10_theobromine |
-5.1 |
ERBB2_theobromine |
-5.3 |
|
TREM1_catechin |
-6.5 |
TREM1_linalool |
-4.3 |
|
TREM1_CATECHOL |
-5.2 |
TREM1_quinic_acid |
-5.1 |
|
TREM1_chlorogenic_acid |
-7.1 |
TREM1_theaflavin |
-7.7 |
|
TREM1_epicatechin_3_gallate |
-7.3 |
TREM1_theanine |
-4.4 |
|
TREM1_epicatechin_gallate |
-6.9 |
TREM1_theobromine |
-4.6 |
| TREM1_epigallocatechin | -6.9 | TREM1_gallocatechin | -6.6 |
|
TREM1_epigallocatechol |
-6.5 |
TREM1_gallocatechol |
-6.6 |
|
TREM1_gallic_acid |
-5.6 |
||
Discussion
Conclusion
References
- ZJinrui, Huang. (2022). Research on the Growth of Breast Tumor. Advances in social science, education and humanities research. [CrossRef]
- Mr., Jayantkumar, Rathod., Pushvin, Gowda, M, R., Preethi, M., Manila, S, Koddaddi., Bindhu, R. (2023). A Review Paper on Brief Study on Breast Cancer Classification using Deep Learning. International Journal of Advanced Research in Science, Communication and Technology, 18-20. [CrossRef]
- Dong, Y., Tao, B., Xue, X. et al. Molecular mechanism of Epicedium treatment for depression based on network pharmacology and molecular docking technology. BMC Complement Med Ther 21, 222 (2021). [CrossRef]
- Chandran, U., Mehendale, N. E. E. L. A. Y., Tillu, G. I. R. I. S. H., & Patwardhan, B. H. U. S. H. A. N. (2015, June). Network pharmacology: an emerging technique for natural product drug discovery and scientific research on ayurveda. In Proc Indian Natn Sci Acad (Vol. 81, No. 3, pp. 561-8).
- Vijayarani, S., Ilamathi, M. J., & Nithya, M. (2015). Preprocessing techniques for text mining-an overview. International Journal of Computer Science & Communication Networks, 5(1), 7-16.
- Charlie, Mayor., Lyn, Robinson. (2014). Ontological realism and classification: Structures and concepts in the Gene Ontology. 65(4):686-697. [CrossRef]
- Doncheva, N. T., Morris, J. H., Gorodkin, J., & Jensen, L. J. (2018). Cytoscape StringApp: network analysis and visualization of proteomics data. Journal of proteome research, 18(2), 623-632. [CrossRef]
- Wu, G., Dawson, E., Duong, A., Haw, R., & Stein, L. (2014). ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Research, 3. [CrossRef]
- Yuyan, Pan., Yong, Zhang., Jiaqi, Liu. (2018). Text mining-based drug discovery in cutaneous squamous cell carcinoma. Oncology Reports, 40(6):3830-3842. [CrossRef]
- Noor, F., Tahir ul Qamar, M., Ashfaq, U. A., Albutti, A., Alwashmi, A. S., & Aljasir, M. A. (2022). Network pharmacology approach for medicinal plants: review and assessment. Pharmaceuticals, 15(5), 572. [CrossRef]
- S Azmi, A. (2013). Adopting network pharmacology for cancer drug discovery. Current drug discovery technologies, 10(2), 95-105.
- Pei, C., Yang, K., Chen, Y., Dong, Y., Meng, X., & Song, N. (2024). Mechanisms of action of Qinghao in treating breast cancer and doxorubicin-induced cardiotoxicity based on network pharmacology and molecular docking. [CrossRef]
- Hopkins, A. L. (2008). Network pharmacology: the next paradigm in drug discovery. Nature chemical biology, 4(11), 682-690. [CrossRef]
- Li, Z., Han, P., You, Z. H., Li, X., Zhang, Y., Yu, H., ... & Chen, X. (2017). In silico prediction of drug-target interaction networks based on drug chemical structure and protein sequences. Scientific reports, 7(1), 11174. [CrossRef]
- Robert, C., Goldman. (2020). Target Discovery for New Antitubercular Drugs Using a Large Dataset of Growth Inhibitors from PubChem.. Infectious disorders drug targets. [CrossRef]
- Rupali, Patil, Bhagat., Mitali, B, Ghormade. (2020). Docking of rigid macromolecules using autodok. Journal of emerging technologies and innovative research.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




