Submitted:
18 May 2024
Posted:
21 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Sample Collection and Cryopreservation
Cloning and Expression Clones
Protein Expression
Protein Purification
Indirect Enzyme Linked Immunosorbent Assay (iELISA)
Statistical Analysis
3. Results
3.1. Sample Collection and Cryopreservation
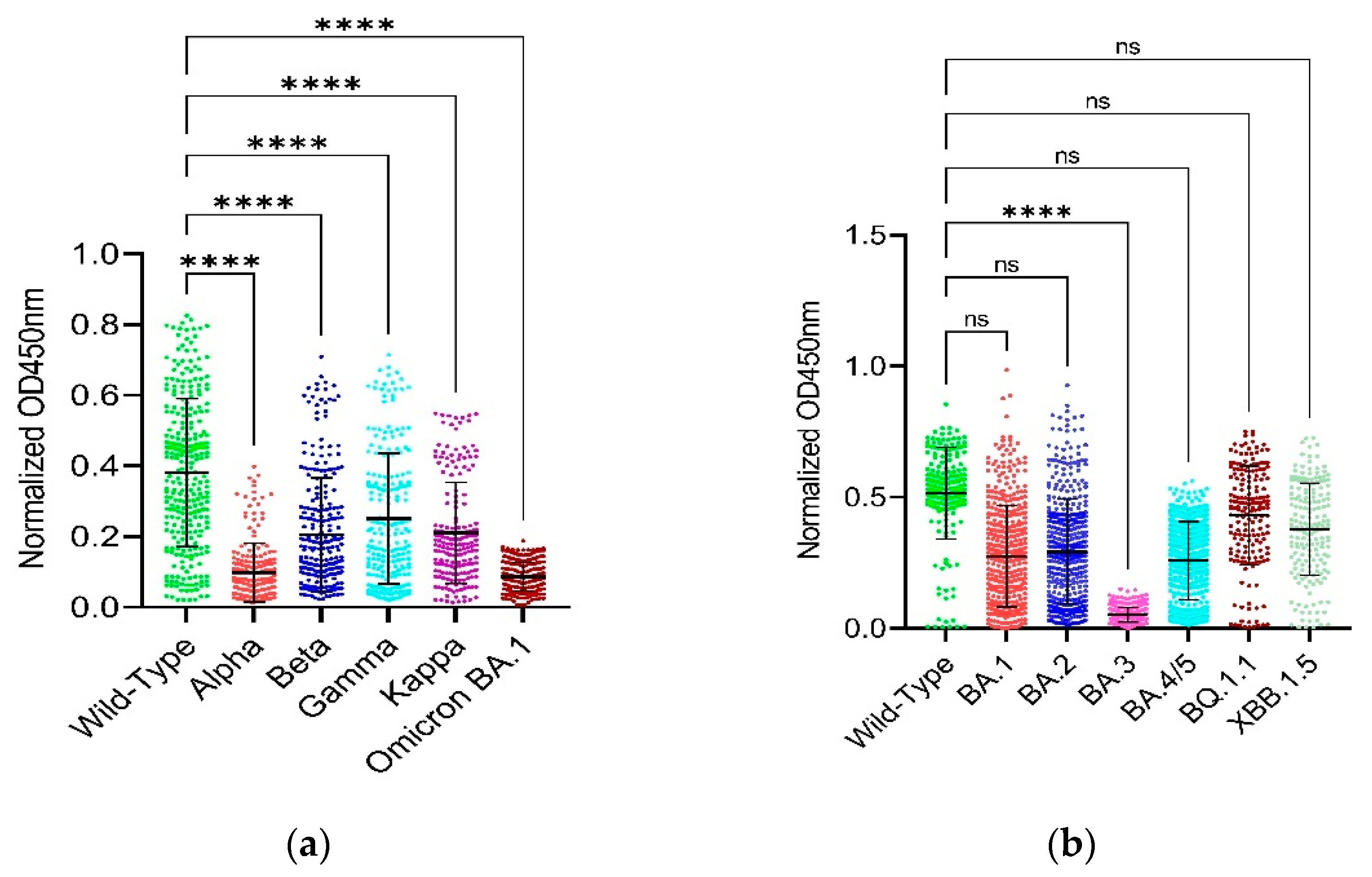
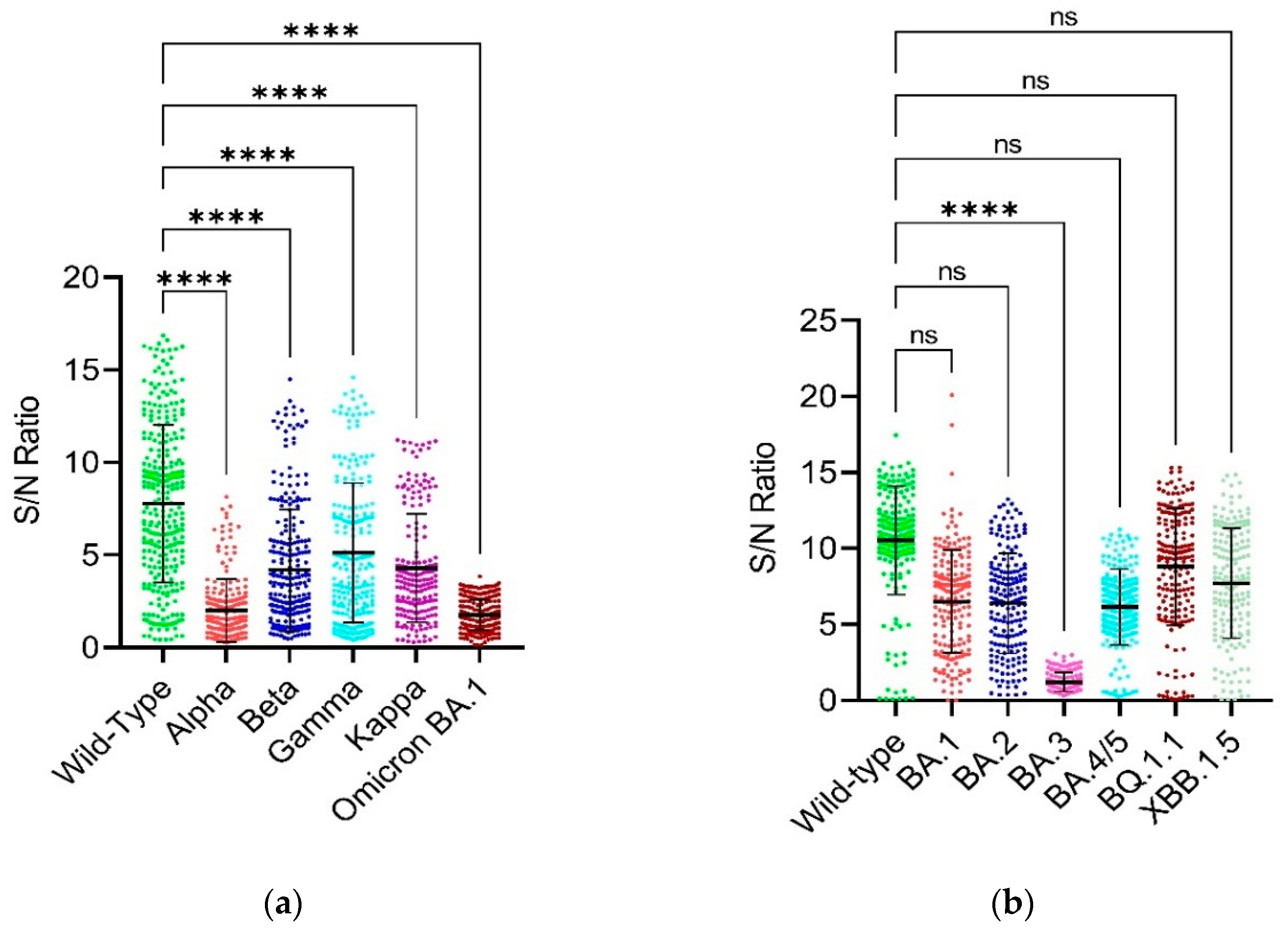
3.2. Binding Capability of RBD and Full-Length Spike
| Variant Name (As per WHO) | Other Names | Key RBD/Spike/other mutation (s) | First time detected in | First time detected from | Reference |
|---|---|---|---|---|---|
| Alpha | B.1.1.7, 20I/501Y.V1, GRY, VOC 202012/01 | N501Y, A570D, P681H, T716I, S982A and D1118H | 2020 | United Kingdom | [29] |
| Beta | B.1.351, GH, 20H/501Y.V2, | RBD: K417N, E484K and N501Y | 2020 | South Africa | [29] |
| Gamma | P.1, B.1.1.28.1, GR, 20J/501Y.V3 | K417T, E484K and N501Y | 2020 | Brazil | [30] |
| Kappa | B.1.617.1, 21B, G/452R.V3, 20A/S:154K | L452R and E484Q | 2021 | India | [30] |
| Omicron | B.1.1529, 21K, 21L, 21M, GR/484A | G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y and Y505H | 2021 | Botswana | [31,32] |
| Omicron BA.1 | B.1.1.529.1 | T547K | 2021 | South Africa | [33] |
| Omicron BA.2 | B.1.1.529.2 | V213G | 2021 | South Africa | [33] |
| Omicron BA.3 | B.1.1.529.3 | No unique RBD or spike mutation. Others: C832T and C11235T |
2021 | South Africa | [33] |
| Omicron BA.4/5 | B.1.1.529.4/5 | 69-70del, L452R, and F486V; Others: N: P151S, ORF7b: L11F, and NSP1:141-143del |
2022 | South Africa | [34,35] |
| Omicron BQ.1.1 | B.1.1.529.5.3.1.1.1.1.1.1 | ORF1b: N1191S and S: R346T, N460K | 2022 | Nigeria | [36] |
| Omicron XBB.1.5 | 23A | S: F486P | 2022 | United States of America | [37] |
| Sample ID | Wild-type | Alpha | Beta | Gamma | Kappa | Omicron BA.1 | Alpha | Beta | Gamma | Kappa | Omicron BA.1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Normalized OD450nm | % Reduction rate | ||||||||||
| Y1 | 0.5906 | 0.0217 | 96.3241 | ||||||||
| Y3 | 0.5516 | 0.0327 | 94.0699 | ||||||||
| Y6 | 0.6169 | 0.0260 | 95.7786 | ||||||||
| Y11 | 0.6216 | 0.0387 | 93.7724 | ||||||||
| Y14 | 0.0772 | 0.0080 | 89.5870 | ||||||||
| Y17 | 0.0712 | 0.0185 | 0.0429 | 0.0633 | 0.0499 | 0.0767 | 74.0250 | 39.7036 | 11.0764 | 29.8947 | -7.7028 |
| Y18 | 0.6452 | 0.0244 | 96.2223 | ||||||||
| Y20 | 0.4822 | 0.0825 | 0.2151 | 0.2907 | 0.2029 | 0.1307 | 82.8917 | 55.3917 | 39.7235 | 57.9176 | 72.8946 |
| 434 | 0.0743 | 0.0365 | 0.0627 | 0.0598 | 0.0564 | 0.0697 | 50.8754 | 15.7011 | 19.5872 | 24.1833 | 6.2286 |
| A20 | 0.0372 | 0.0200 | 0.0358 | 0.0320 | 0.0307 | 0.0364 | 46.3758 | 3.8943 | 14.0421 | 17.5490 | 2.2900 |
| 32 | 0.4472 | 0.0708 | 0.2168 | 0.2827 | 0.1998 | 84.1615 | 51.5280 | 36.7950 | 55.3323 | ||
| 22 | 0.3422 | 0.1392 | 0.1734 | 0.2278 | 0.1814 | 0.0809 | 59.3188 | 49.3190 | 33.4264 | 46.9854 | 76.3518 |
| 1048 | 0.3469 | 0.0649 | 0.1329 | 0.1356 | 0.1050 | 0.0818 | 81.3050 | 61.6918 | 60.9231 | 69.7234 | 76.4137 |
| 1090 | 0.5393 | 0.1511 | 0.4273 | 0.3681 | 0.2831 | 0.1146 | 71.9890 | 20.7672 | 31.7477 | 47.5026 | 78.7523 |
| 1092 | 0.6090 | 0.2219 | 0.4743 | 0.3924 | 0.3439 | 0.1232 | 63.5714 | 22.1134 | 35.5598 | 43.5281 | 79.7780 |
| 1094 | 0.3238 | 0.0967 | 0.1162 | 0.1201 | 0.1000 | 0.0527 | 70.1217 | 64.1049 | 62.9038 | 69.1065 | 83.7211 |
| 1096 | 0.1120 | 0.0302 | 0.0557 | 0.0780 | 0.0618 | 0.0675 | 73.0502 | 50.3000 | 30.3604 | 44.8563 | 39.7474 |
| 1435 | 0.6398 | 0.2261 | 0.5523 | 0.5703 | 0.4649 | 0.1504 | 64.6668 | 13.6773 | 10.8552 | 27.3300 | 76.4959 |
| 1443 | 0.4051 | 0.0876 | 0.1973 | 0.2662 | 0.1593 | 0.0928 | 78.3876 | 51.3034 | 34.2987 | 60.6869 | 77.0882 |
| 1444 | 0.6337 | 0.1072 | 0.2221 | 0.2755 | 0.1893 | 0.1025 | 83.0806 | 64.9513 | 56.5267 | 70.1346 | 83.8257 |
| 2234 | 0.2787 | 0.0811 | 0.1219 | 0.1707 | 0.1423 | 0.0763 | 70.9189 | 56.2687 | 38.7682 | 48.9585 | 72.6131 |
| 2237 | 0.7146 | 0.2917 | 0.4901 | 0.5675 | 0.4804 | 0.1592 | 59.1743 | 31.4104 | 20.5800 | 32.7651 | 77.7251 |
| 2255 | 0.5131 | 0.1106 | 0.1944 | 0.2577 | 0.2131 | 0.0825 | 78.4515 | 62.1007 | 49.7780 | 58.4651 | 83.9226 |
| 2256 | 0.2202 | 0.0345 | 0.0851 | 0.1022 | 0.0756 | 0.0652 | 84.3262 | 61.3536 | 53.6091 | 65.6735 | 70.4161 |
| 2494 | 0.3659 | 0.0601 | 0.1266 | 0.1683 | 0.1196 | 0.0843 | 83.5864 | 65.3963 | 53.9933 | 67.3132 | 76.9511 |
| 2497 | 0.7069 | 0.1897 | 0.4673 | 0.5163 | 0.4076 | 0.1543 | 73.1610 | 33.8966 | 26.9569 | 42.3393 | 78.1672 |
| 2506 | 0.4711 | 0.0977 | 0.2163 | 0.2747 | 0.1899 | 0.0740 | 79.2546 | 54.0866 | 41.6912 | 59.6798 | 84.2906 |
| 2601 | 0.2567 | 0.0452 | 0.0868 | 0.1167 | 0.0849 | 0.0612 | 82.3848 | 66.1978 | 54.5553 | 66.9173 | 76.1740 |
| 2602 | 0.3916 | 0.0919 | 0.1969 | 0.2517 | 0.1704 | 0.1588 | 76.5323 | 49.7020 | 35.7264 | 56.4735 | 59.4353 |
| 2604 | 0.5237 | 0.1294 | 0.3768 | 0.4072 | 0.3023 | 0.1483 | 75.2944 | 28.0577 | 22.2552 | 42.2855 | 71.6771 |
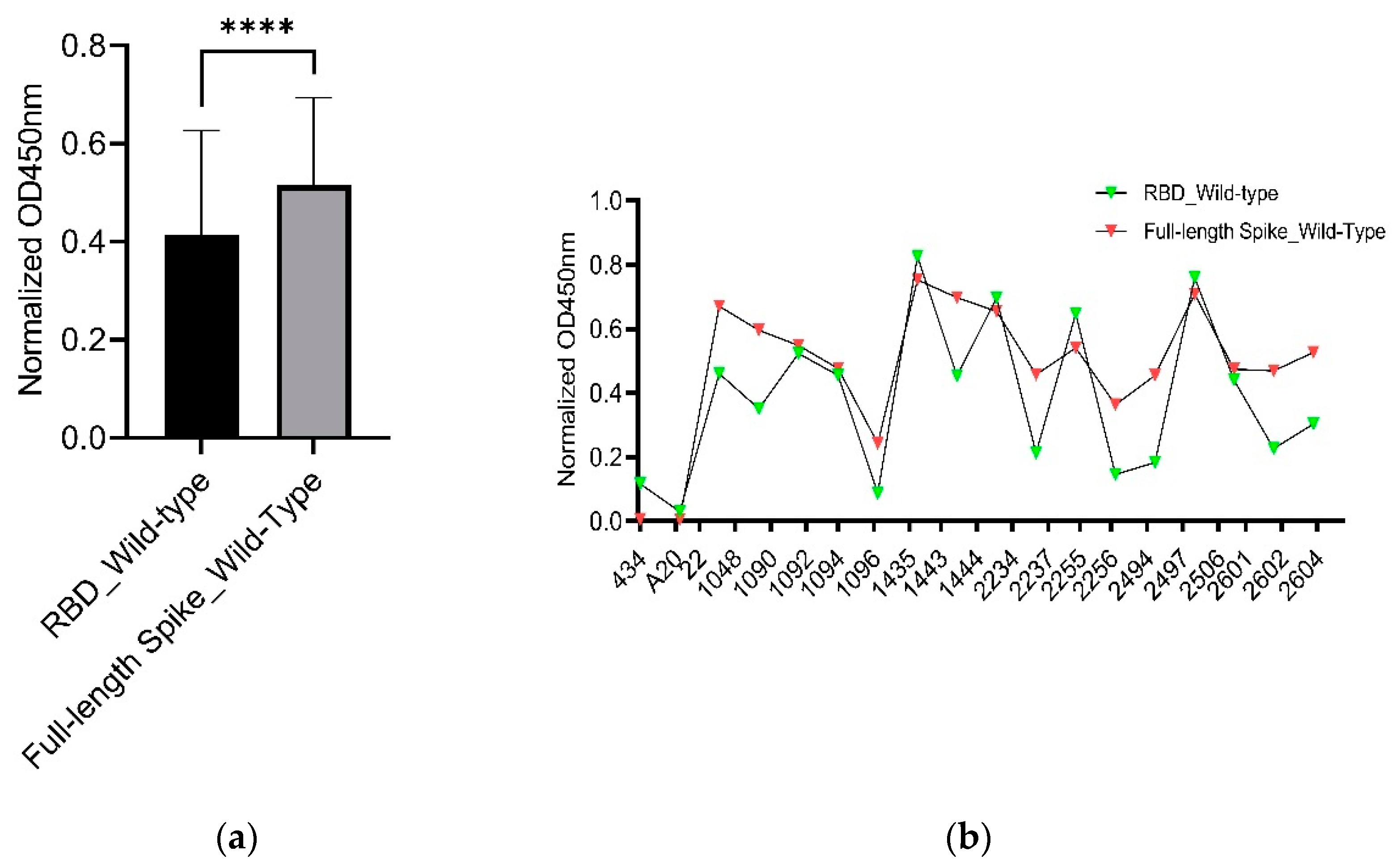
3.3. Comparison of Binding Capability between the RBD and Full-Length Spike of SARS-CoV-2 Wild-Type
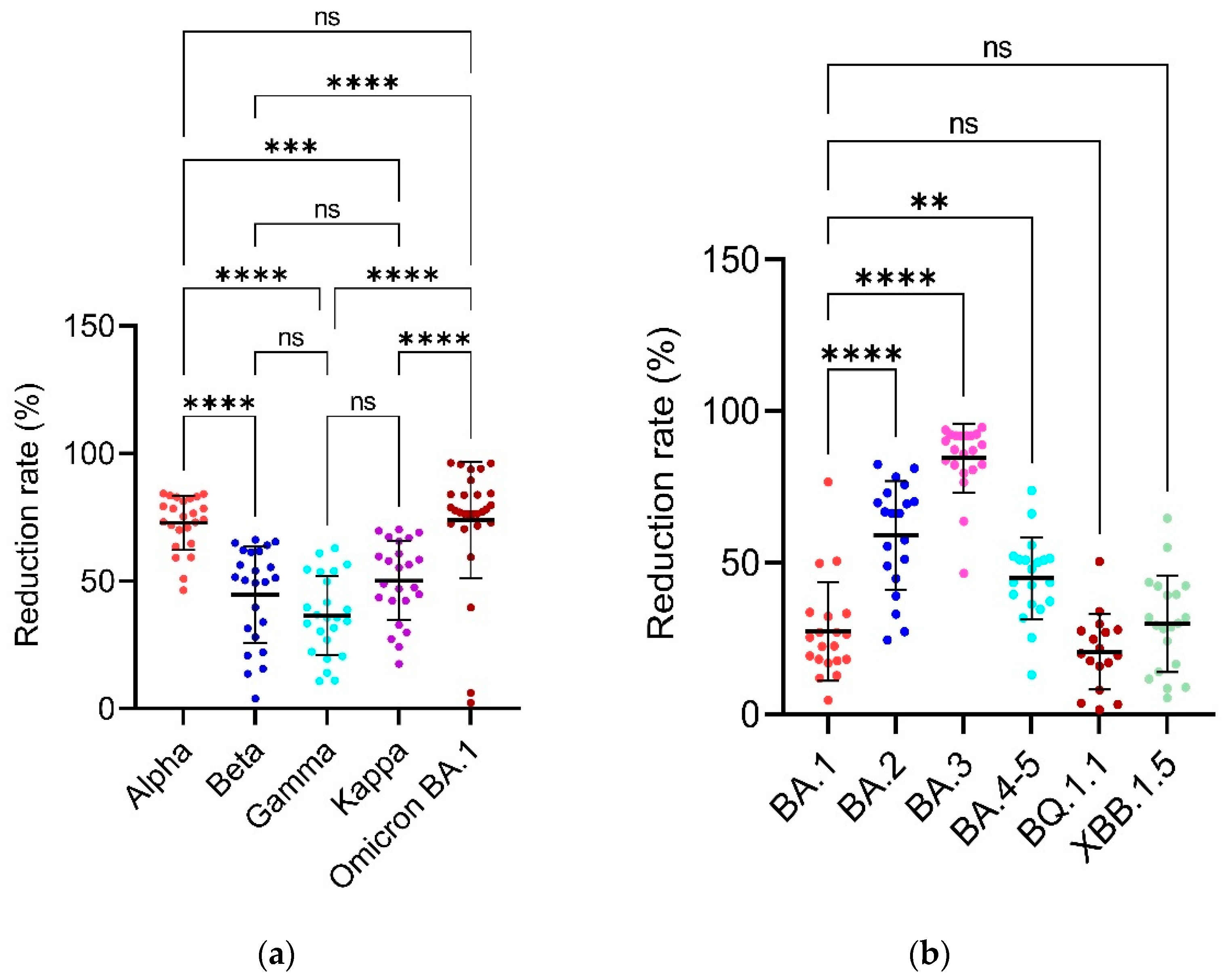
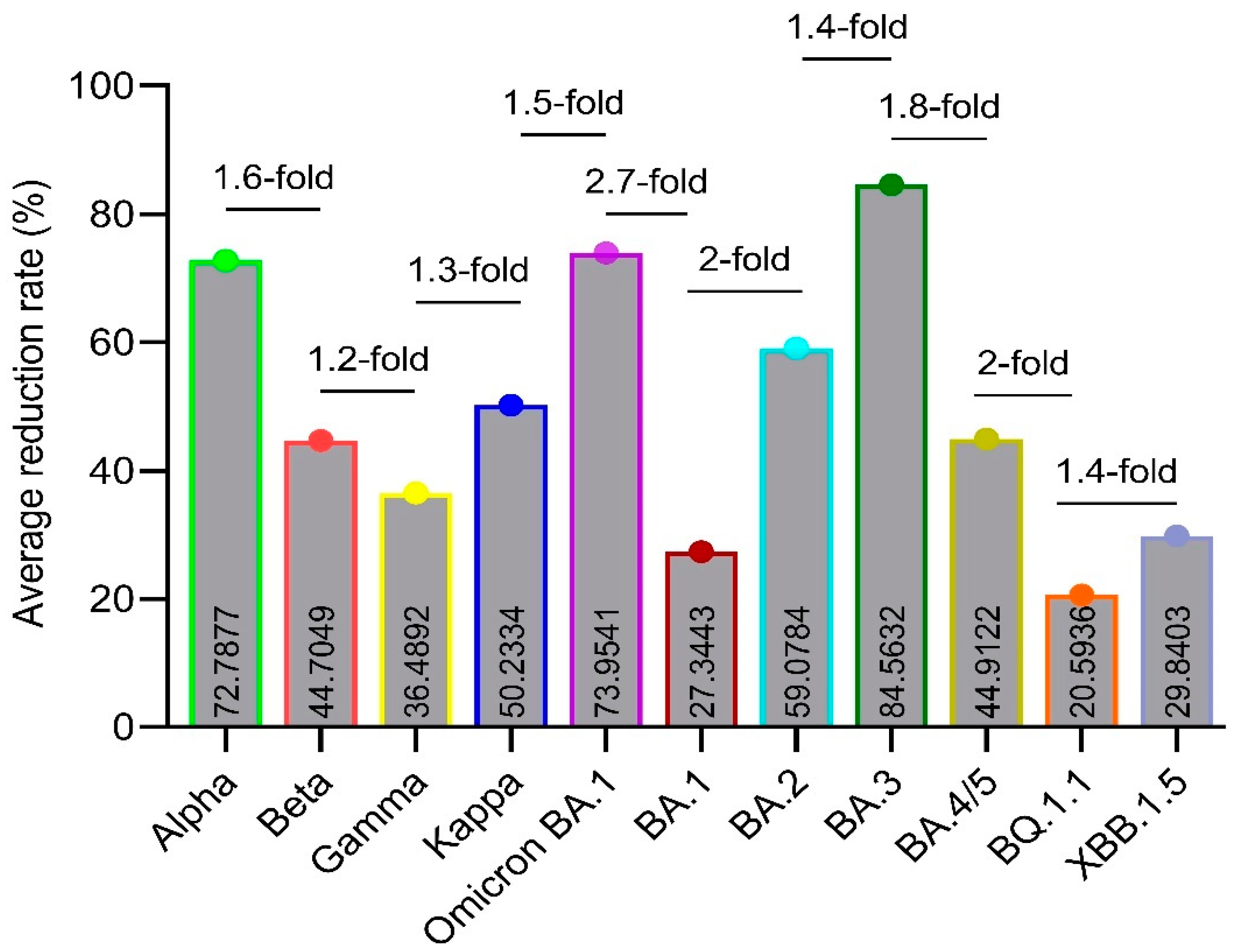
3.4. Reduction Rates
3.5. Figures and Tables
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Cucinotta, D.; Vanelli, M.J.A.b.m.A.p. WHO declares COVID-19 a pandemic. 2020, 91, 157.
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z.J.N.R.I. Immunological considerations for COVID-19 vaccine strategies. 2020, 20, 615-632. [CrossRef]
- Hoffmann, M.; Arora, P.; Groß, R.; Seidel, A.; Hörnich, B.F.; Hahn, A.S.; Krüger, N.; Graichen, L.; Hofmann-Winkler, H.; Kempf, A.J.C. SARS-CoV-2 variants B. 1.351 and P. 1 escape from neutralizing antibodies. 2021, 184, 2384–2393.e2312. [Google Scholar] [CrossRef] [PubMed]
- Norman, M.; Gilboa, T.; Ogata, A.F.; Maley, A.M.; Cohen, L.; Busch, E.L.; Lazarovits, R.; Mao, C.-P.; Cai, Y.; Zhang, J.J.N.B.E. Ultrasensitive high-resolution profiling of early seroconversion in patients with COVID-19. 2020, 4, 1180-1187. [CrossRef]
- Krammer, F.; Simon, V.J.S. Serology assays to manage COVID-19. 2020, 368, 1060-1061. [CrossRef]
- Esposito, D.; Mehalko, J.; Drew, M.; Snead, K.; Wall, V.; Taylor, T.; Frank, P.; Denson, J.-P.; Hong, M.; Gulten, G.J.P.e.; et al. Optimizing high-yield production of SARS-CoV-2 soluble spike trimers for serology assays. 2020, 174, 105686.
- Gribble, J.; Stevens, L.J.; Agostini, M.L.; Anderson-Daniels, J.; Chappell, J.D.; Lu, X.; Pruijssers, A.J.; Routh, A.L.; Denison, M.R.J.P.p. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. 2021, 17, e1009226. [CrossRef]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.J.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. 2020, 182, 812-827. e819.
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.J.V.e. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. 2021, 7, veab064. [CrossRef]
- Control, C.f.D. Prevention. SARS-CoV-2 variant classifications and definitions. 2021.
- SARS-CoV, C. Variant classifications and definitions.
- Evans, J.P.; Qu, P.; Zeng, C.; Zheng, Y.-M.; Carlin, C.; Bednash, J.S.; Lozanski, G.; Mallampalli, R.K.; Saif, L.J.; Oltz, E.M.J.N.E.J.o.M. Neutralization of the SARS-CoV-2 deltacron and BA. 3 variants. 2022, 386, 2340–2342. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Microbiology, C.-G.U.C.J.N.R. SARS-CoV-2 variants, spike mutations and immune escape. 2021, 19, 409-424.
- Chi, X.; Guo, Y.; Zhang, G.; Sun, H.; Zhang, J.; Li, M.; Chen, Z.; Han, J.; Zhang, Y.; Zhang, X.J.S.T.; et al. Broadly neutralizing antibodies against Omicron-included SARS-CoV-2 variants induced by vaccination. 2022, 7, 139.
- Maeda, K.; Amano, M.; Uemura, Y.; Tsuchiya, K.; Matsushima, T.; Noda, K.; Shimizu, Y.; Fujiwara, A.; Takamatsu, Y.; Ichikawa, Y.J.S.R. Correlates of neutralizing/SARS-CoV-2-S1-binding antibody response with adverse effects and immune kinetics in BNT162b2-vaccinated individuals. 2021, 11, 22848.
- Tsuchiya, K.; Maeda, K.; Matsuda, K.; Takamatsu, Y.; Kinoshita, N.; Kutsuna, S.; Hayashida, T.; Gatanaga, H.; Ohmagari, N.; Oka, S.J.S.R. Neutralization activity of IgG antibody in COVID-19-convalescent plasma against SARS-CoV-2 variants. 2023, 13, 1263.
- Wall, E.C.; Wu, M.; Harvey, R.; Kelly, G.; Warchal, S.; Sawyer, C.; Daniels, R.; Hobson, P.; Hatipoglu, E.; Ngai, Y.J.T.L. Neutralising antibody activity against SARS-CoV-2 VOCs B. 1.617. 2 and B. 1.351 by BNT162b2 vaccination. 2021, 397, 2331–2333. [Google Scholar] [PubMed]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.d.S.; Mishra, S.; Crispim, M.A.; Sales, F.C.; Hawryluk, I.; McCrone, J.T.J.S. Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil. 2021, 372, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Casner, R.G.; Nair, M.S.; Wang, M.; Yu, J.; Cerutti, G.; Liu, L.; Kwong, P.D.; Huang, Y.; Shapiro, L.J.C.h.; et al. Increased resistance of SARS-CoV-2 variant P. 1 to antibody neutralization. 2021, 29, 747–751 e744. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Lam, E.C.; Denis, K.S.; Nitido, A.D.; Garcia, Z.H.; Hauser, B.M.; Feldman, J.; Pavlovic, M.N.; Gregory, D.J.; Poznansky, M.C.J.C. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. 2021, 184, 2372-2383. e2379.
- Kurhade, C.; Zou, J.; Xia, H.; Liu, M.; Chang, H.C.; Ren, P.; Xie, X.; Shi, P.Y.J.N.m. Low neutralization of SARS-CoV-2 Omicron BA. 2.75. 2, BQ. 1.1 and XBB. 1 by parental mRNA vaccine or a BA. 5 bivalent booster. 2023, 29, 344–347. [Google Scholar] [PubMed]
- Wang, Q.; Iketani, S.; Li, Z.; Liu, L.; Guo, Y.; Huang, Y.; Bowen, A.D.; Liu, M.; Wang, M.; Yu, J.J.C. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. 2023, 186, 279-286. e278.
- Yue, C.; Song, W.; Wang, L.; Jian, F.; Chen, X.; Gao, F.; Shen, Z.; Wang, Y.; Wang, X.; Cao, Y.J.T.L.I.D. ACE2 binding and antibody evasion in enhanced transmissibility of XBB. 1.5. 2023, 23, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Collier, A.-r.Y.; Rowe, M.; Mardas, F.; Ventura, J.D.; Wan, H.; Miller, J.; Powers, O.; Chung, B.; Siamatu, M.J.N.E.J.o.M. Neutralization of the SARS-CoV-2 Omicron BA. 1 and BA. 2 variants. 2022, 386, 1579–1580. [Google Scholar] [CrossRef]
- Caniels, T.G.; Bontjer, I.; van der Straten, K.; Poniman, M.; Burger, J.A.; Appelman, B.; Lavell, A.A.; Oomen, M.; Godeke, G.-J.; Valle, C.J.S.A. Emerging SARS-CoV-2 variants of concern evade humoral immune responses from infection and vaccination. 2021, 7, eabj5365.
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.J.N. Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7. 2021, 593, 130–135. [Google Scholar]
- Adams, O.; Andrée, M.; Hermsen, D.; Lübke, N.; Timm, J.; Schaal, H.; Müller, L.J.J.o.V.M. Comparison of commercial SARS-CoV-2 surrogate neutralization assays with a full virus endpoint dilution neutralization test in two different cohorts. 2022, 307, 114569.
- Vilibic-Cavlek, T.; Bogdanic, M.; Borko, E.; Hruskar, Z.; Zilic, D.; Ferenc, T.; Tabain, I.; Barbic, L.; Vujica Ferenc, M.; Ferencak, I.J.A. Detection of SARS-CoV-2 Antibodies: Comparison of Enzyme Immunoassay, Surrogate Neutralization and Virus Neutralization Test. 2023, 12, 35.
- O’Toole, Á.; Hill, V.; Pybus, O.G.; Watts, A.; Bogoch, I.I.; Khan, K.; Messina, J.P.; COVID, T.; Network, B.-U.C.G.; Tegally, H.J.W.O.R. Tracking the international spread of SARS-CoV-2 lineages B. 1.1. 7 and B. 1.351/501Y-V2 with grinch. 2021, 6.
- Tulimilli, S.V.; Dallavalasa, S.; Basavaraju, C.G.; Kumar Rao, V.; Chikkahonnaiah, P.; Madhunapantula, S.V.; Veeranna, R.P.J.V. Variants of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and Vaccine Effectiveness. 2022, 10, 1751.
- Saxena, S.K.; Kumar, S.; Ansari, S.; Paweska, J.T.; Maurya, V.K.; Tripathi, A.K.; Abdel-Moneim, A.S.J.J.o.m.v. Characterization of the novel SARS-CoV-2 Omicron (B. 1.1. 529) variant of concern and its global perspective. 2022, 94, 1738–1744. [Google Scholar]
- Karim, S.S.A.; Karim, Q.A.J.T.l. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. 2021, 398, 2126-2128.
- Viana, R.; Moyo, S.; Amoako, D.G.; Tegally, H.; Scheepers, C.; Althaus, C.L.; Anyaneji, U.J.; Bester, P.A.; Boni, M.F.; Chand, M.J.N. Rapid epidemic expansion of the SARS-CoV-2 Omicron variant in southern Africa. 2022, 603, 679-686.
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D.G.J.N.m. Emergence of SARS-CoV-2 omicron lineages BA. 4 and BA. 5 in South Africa. 2022, 28, 1785–1790. [Google Scholar]
- Islam, M.R.; Shahriar, M.; Bhuiyan, M.A.J.H.s.r. The latest Omicron BA. 4 and BA. 5 lineages are frowning toward COVID-19 preventive measures: a threat to global public health. 2022, 5.
- Qu, P.; Evans, J.P.; Faraone, J.N.; Zheng, Y.-M.; Carlin, C.; Anghelina, M.; Stevens, P.; Fernandez, S.; Jones, D.; Lozanski, G.J.C.h.; et al. Enhanced neutralization resistance of SARS-CoV-2 omicron subvariants BQ. 1, BQ. 1.1, BA. 4.6, BF. 7, and BA. 2.75. 2. 2023, 31, 9–17.e13. [Google Scholar] [PubMed]
- Kamp, J.; Abbott, B.J.W.S.J.-O.E. New Covid-19 Subvariant, XBB. 1.5, Takes Over in Parts of US. 2023, N. PAG-N. PAG.
| Sr. No. | Sample ID | Sample Collection Date | Demographic Location (City) | Blood type | Gender | Age (years) |
PCR* status (-ve or +ve) |
|---|---|---|---|---|---|---|---|
| 1 | Y1 | 18-02-2020 | Yuncheng | O | Male | 27 | +ve |
| 2 | Y2 | 11-05-2020 | Yuncheng | B | Female | 24 | +ve |
| 3 | Y3 | 26-04-2020 | Yuncheng | O | Male | 27 | +ve |
| 4 | Y4 | 29-01-2020 | Yuncheng | A | Male | 23 | +ve |
| 5 | Y5 | 22-02-2020 | Yuncheng | A | Male | 32 | +ve |
| 6 | Y6 | 03-03-2020 | Yuncheng | O | Male | 27 | +ve |
| 7 | Y7 | 19-03-2020 | Yuncheng | B | Female | 22 | +ve |
| 8 | Y8 | 19-03-2020 | Yuncheng | A | Male | 32 | +ve |
| 9 | Y9 | 23-01-2020 | Yuncheng | B | Female | 22 | +ve |
| 10 | Y10 | 02-05-2020 | Yuncheng | AB | Male | 35 | +ve |
| 11 | Y11 | 19-03-2020 | Yuncheng | O | Male | 27 | +ve |
| 12 | Y12 | 19-03-2020 | Yuncheng | B | Male | 38 | +ve |
| 13 | Y13 | 09-02-2020 | Yuncheng | B | Male | 28 | +ve |
| 14 | Y14 | 18-02-2020 | Yuncheng | AB | Male | 38 | +ve |
| 15 | Y15 | 03-05-2020 | Yuncheng | O | Male | 27 | +ve |
| 16 | Y16 | 19-03-2020 | Yuncheng | O | Female | 26 | +ve |
| 17 | Y17 | 19-03-2020 | Yuncheng | O | Male | 32 | +ve |
| 18 | Y18 | 18-02-2020 | Yuncheng | A | Male | 32 | +ve |
| 19 | Y19 | 20-02-2020 | Yuncheng | O | Male | 27 | +ve |
| 20 | Y20 | 19-03-2020 | Yuncheng | A | Male | 45 | +ve |
| 21 | Y21 | 10-04-2020 | Yuncheng | O | Male | 27 | +ve |
| 22 | Y22 | 02-05-2020 | Yuncheng | A | Male | 23 | +ve |
| 23 | Y23 | 10-02-2020 | Yuncheng | A | Male | 45 | +ve |
| 24 | Y24 | 19-03-2020 | Yuncheng | B | Male | 43 | +ve |
| 25 | Y25 | 19-03-2020 | Yuncheng | O | Male | 39 | +ve |
| 26 | Y26 | 19-03-2020 | Yuncheng | A | Male | 37 | +ve |
| 27 | Y27 | 19-03-2020 | Yuncheng | O | Female | 26 | +ve |
| 28 | Y28 | 19-03-2020 | Yuncheng | AB | Male | 26 | +ve |
| 29 | 22 | 02-05-2020 | Datong | B | Male | 25 | +ve |
| 30 | 32 | 02-05-2020 | Yuncheng | A | Male | 45 | +ve |
| 31 | 34 | 02-05-2020 | Luliang | A | Male | 28 | +ve |
| 32 | 434 | 02-05-2020 | Jinzhong | AB | Male | 32 | +ve |
| 33 | A20 | 02-05-2020 | Taiyuan | O | Female | 40 | +ve |
| 34 | 1048 | 02-05-2020 | Xinzhou | A | Female | 38 | +ve |
| 35 | 1090 | 02-05-2020 | Xinzhou | B | Male | 43 | +ve |
| 36 | 1092 | 02-05-2020 | Pingyao | AB | Male | 26 | +ve |
| 37 | 1094 | 02-05-2020 | Jinzhong | O | Male | 32 | +ve |
| 38 | 1096 | 02-05-2020 | Yuncheng | AB | Male | 35 | +ve |
| 39 | 1435 | 02-05-2020 | Jinzhong | O | Male | 21 | +ve |
| 40 | 1443 | 02-05-2020 | Datong | B | Male | 25 | +ve |
| 41 | 1444 | 02-05-2020 | Jinzhong | O | Male | 27 | +ve |
| 42 | 2234 | 02-05-2020 | Changzhi | B | Male | 27 | +ve |
| 43 | 2237 | 02-05-2020 | Yuncheng | AB | Male | 26 | +ve |
| 44 | 2255 | 02-05-2020 | Jincheng | A | Female | 44 | +ve |
| 45 | 2256 | 02-05-2020 | Shuozhou | A | Male | 30 | +ve |
| 46 | 2494 | 02-05-2020 | Yuncheng | O | Male | 39 | +ve |
| 47 | 2497 | 02-05-2020 | Taiyuan | B | Male | 42 | +ve |
| 48 | 2506 | 02-05-2020 | Jinzhong | O | Male | 49 | +ve |
| 49 | 2601 | 02-05-2020 | Luliang | A | Male | 28 | +ve |
| 50 | 2602 | 02-05-2020 | Changzhi | AB | Female | 23 | +ve |
| 51 | 2604 | 02-05-2020 | Taiyuan | A | Male | 35 | +ve |
| 52 | 2080 | 02-05-2020 | Taiyuan | O | Male | 31 | -ve |
| 53 | 2081 | 02-05-2020 | Taiyuan | O | Male | 35 | -ve |
| 54 | 2082 | 02-05-2020 | Taiyuan | B | Male | 28 | -ve |
| 55 | 2083 | 02-05-2020 | Taiyuan | AB | Male | 38 | -ve |
| 56 | 2084 | 02-05-2020 | Taiyuan | B | Male | 30 | -ve |
| 57 | 2085 | 02-05-2020 | Taiyuan | A | Male | 44 | -ve |
| 58 | 2086 | 02-05-2020 | Taiyuan | A | Male | 27 | -ve |
| 59 | 2087 | 02-05-2020 | Taiyuan | O | Male | 40 | -ve |
| 60 | 2088 | 02-05-2020 | Taiyuan | AB | Male | 37 | -ve |
| 61 | 2089 | 02-05-2020 | Taiyuan | AB | Male | 26 | -ve |
| 62 | 2090 | 02-05-2020 | Taiyuan | O | Male | 28 | -ve |
| 63 | 2092 | 02-05-2020 | Yangquan | B | Male | 38 | -ve |
| 64 | 2093 | 02-05-2020 | Jinzhong | O | Male | 49 | -ve |
| 65 | 2094 | 02-05-2020 | Jincheng | A | Female | 28 | -ve |
| Sample ID | Wild-type | Alpha | Beta | Gamma | Kappa | Omicron BA.1 | Alpha | Beta | Gamma | Kappa | Omicron BA.1 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Normalized OD450nm | % Reduction rate | ||||||||||
| Y1 | 0.5906 | 0.0217 | 96.3241 | ||||||||
| Y3 | 0.5516 | 0.0327 | 94.0699 | ||||||||
| Y6 | 0.6169 | 0.0260 | 95.7786 | ||||||||
| Y11 | 0.6216 | 0.0387 | 93.7724 | ||||||||
| Y14 | 0.0772 | 0.0080 | 89.5870 | ||||||||
| Y17 | 0.0712 | 0.0185 | 0.0429 | 0.0633 | 0.0499 | 0.0767 | 74.0250 | 39.7036 | 11.0764 | 29.8947 | -7.7028 |
| Y18 | 0.6452 | 0.0244 | 96.2223 | ||||||||
| Y20 | 0.4822 | 0.0825 | 0.2151 | 0.2907 | 0.2029 | 0.1307 | 82.8917 | 55.3917 | 39.7235 | 57.9176 | 72.8946 |
| 434 | 0.0743 | 0.0365 | 0.0627 | 0.0598 | 0.0564 | 0.0697 | 50.8754 | 15.7011 | 19.5872 | 24.1833 | 6.2286 |
| A20 | 0.0372 | 0.0200 | 0.0358 | 0.0320 | 0.0307 | 0.0364 | 46.3758 | 3.8943 | 14.0421 | 17.5490 | 2.2900 |
| 32 | 0.4472 | 0.0708 | 0.2168 | 0.2827 | 0.1998 | 84.1615 | 51.5280 | 36.7950 | 55.3323 | ||
| 22 | 0.3422 | 0.1392 | 0.1734 | 0.2278 | 0.1814 | 0.0809 | 59.3188 | 49.3190 | 33.4264 | 46.9854 | 76.3518 |
| 1048 | 0.3469 | 0.0649 | 0.1329 | 0.1356 | 0.1050 | 0.0818 | 81.3050 | 61.6918 | 60.9231 | 69.7234 | 76.4137 |
| 1090 | 0.5393 | 0.1511 | 0.4273 | 0.3681 | 0.2831 | 0.1146 | 71.9890 | 20.7672 | 31.7477 | 47.5026 | 78.7523 |
| 1092 | 0.6090 | 0.2219 | 0.4743 | 0.3924 | 0.3439 | 0.1232 | 63.5714 | 22.1134 | 35.5598 | 43.5281 | 79.7780 |
| 1094 | 0.3238 | 0.0967 | 0.1162 | 0.1201 | 0.1000 | 0.0527 | 70.1217 | 64.1049 | 62.9038 | 69.1065 | 83.7211 |
| 1096 | 0.1120 | 0.0302 | 0.0557 | 0.0780 | 0.0618 | 0.0675 | 73.0502 | 50.3000 | 30.3604 | 44.8563 | 39.7474 |
| 1435 | 0.6398 | 0.2261 | 0.5523 | 0.5703 | 0.4649 | 0.1504 | 64.6668 | 13.6773 | 10.8552 | 27.3300 | 76.4959 |
| 1443 | 0.4051 | 0.0876 | 0.1973 | 0.2662 | 0.1593 | 0.0928 | 78.3876 | 51.3034 | 34.2987 | 60.6869 | 77.0882 |
| 1444 | 0.6337 | 0.1072 | 0.2221 | 0.2755 | 0.1893 | 0.1025 | 83.0806 | 64.9513 | 56.5267 | 70.1346 | 83.8257 |
| 2234 | 0.2787 | 0.0811 | 0.1219 | 0.1707 | 0.1423 | 0.0763 | 70.9189 | 56.2687 | 38.7682 | 48.9585 | 72.6131 |
| 2237 | 0.7146 | 0.2917 | 0.4901 | 0.5675 | 0.4804 | 0.1592 | 59.1743 | 31.4104 | 20.5800 | 32.7651 | 77.7251 |
| 2255 | 0.5131 | 0.1106 | 0.1944 | 0.2577 | 0.2131 | 0.0825 | 78.4515 | 62.1007 | 49.7780 | 58.4651 | 83.9226 |
| 2256 | 0.2202 | 0.0345 | 0.0851 | 0.1022 | 0.0756 | 0.0652 | 84.3262 | 61.3536 | 53.6091 | 65.6735 | 70.4161 |
| 2494 | 0.3659 | 0.0601 | 0.1266 | 0.1683 | 0.1196 | 0.0843 | 83.5864 | 65.3963 | 53.9933 | 67.3132 | 76.9511 |
| 2497 | 0.7069 | 0.1897 | 0.4673 | 0.5163 | 0.4076 | 0.1543 | 73.1610 | 33.8966 | 26.9569 | 42.3393 | 78.1672 |
| 2506 | 0.4711 | 0.0977 | 0.2163 | 0.2747 | 0.1899 | 0.0740 | 79.2546 | 54.0866 | 41.6912 | 59.6798 | 84.2906 |
| 2601 | 0.2567 | 0.0452 | 0.0868 | 0.1167 | 0.0849 | 0.0612 | 82.3848 | 66.1978 | 54.5553 | 66.9173 | 76.1740 |
| 2602 | 0.3916 | 0.0919 | 0.1969 | 0.2517 | 0.1704 | 0.1588 | 76.5323 | 49.7020 | 35.7264 | 56.4735 | 59.4353 |
| 2604 | 0.5237 | 0.1294 | 0.3768 | 0.4072 | 0.3023 | 0.1483 | 75.2944 | 28.0577 | 22.2552 | 42.2855 | 71.6771 |
| Sample ID | Wild-type | BA.1 | BA.2 | BA.3 | BA.4/5 | BQ.1.1 | XBB.1.5 | BA.1 | BA.2 | BA.3 | BA.4/5 | BQ.1.1 | XBB.1.5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Normalized OD450nm | % Reduction rate | ||||||||||||
| 434 | 0.0543 | 0.05933 | 0.0623 | 0.0290 | 0.2110 | 0.0694 | 0.0453 | -9.2062 | -14.7278 | 46.6239 | -288.3568 | -27.7722 | 16.6151 |
| A20 | 0.0570 | 0.03800 | 0.0430 | 0.0207 | 0.0280 | 0.0675 | 0.0432 | 33.3312 | 24.5590 | 63.7415 | 50.8756 | -18.4805 | 24.2201 |
| 22 | 0.5859 | 0.13633 | 0.1580 | 0.0363 | 0.5093 | 0.4823 | 0.4137 | 76.7304 | 73.0323 | 93.7986 | 13.0663 | 17.6788 | 29.3808 |
| 1048 | 0.6076 | 0.44333 | 0.2013 | 0.0987 | 0.3160 | 0.4568 | 0.3500 | 27.0298 | 66.8616 | 83.7600 | 47.9881 | 24.8208 | 42.3967 |
| 1090 | 0.5864 | 0.51633 | 0.3577 | 0.0760 | 0.3997 | 0.5772 | 0.5364 | 11.9550 | 39.0108 | 87.0405 | 31.8490 | 1.5763 | 8.5306 |
| 1092 | 0.6096 | 0.50633 | 0.4430 | 0.1180 | 0.3980 | 0.5870 | 0.5385 | 16.9338 | 27.3239 | 80.6416 | 34.7063 | 3.7040 | 11.6523 |
| 1094 | 0.5342 | 0.35433 | 0.2383 | 0.0410 | 0.2363 | 0.3852 | 0.3230 | 33.6728 | 55.3867 | 92.3253 | 55.7611 | 27.8952 | 39.5435 |
| 1096 | 0.2888 | 0.14500 | 0.1227 | 0.0367 | 0.0757 | 0.1432 | 0.1019 | 49.7881 | 57.5219 | 87.3027 | 73.7974 | 50.4120 | 64.7270 |
| 1435 | 0.6120 | 0.50367 | 0.2990 | 0.0677 | 0.3897 | 0.6219 | 0.5784 | 17.7013 | 51.1436 | 88.9433 | 36.3288 | -1.6122 | 5.4874 |
| 1443 | 0.6080 | 0.49733 | 0.2050 | 0.0493 | 0.3433 | 0.5112 | 0.4326 | 18.2015 | 66.2828 | 91.8859 | 43.5305 | 15.9211 | 28.8423 |
| 1444 | 0.6041 | 0.45067 | 0.2033 | 0.0483 | 0.3410 | 0.4869 | 0.4327 | 25.3998 | 66.3416 | 91.9992 | 43.5533 | 19.4078 | 28.3658 |
| 2234 | 0.5033 | 0.37033 | 0.1093 | 0.0493 | 0.2447 | 0.3648 | 0.2902 | 26.4236 | 78.2781 | 90.1986 | 51.3905 | 27.5321 | 42.3455 |
| 2237 | 0.5961 | 0.52000 | 0.3990 | 0.1047 | 0.3733 | 0.5481 | 0.5126 | 12.7677 | 33.0660 | 82.4417 | 37.3717 | 8.0559 | 14.0028 |
| 2255 | 0.5719 | 0.44367 | 0.1707 | 0.0807 | 0.2847 | 0.4572 | 0.4003 | 22.4206 | 70.1573 | 85.8947 | 50.2233 | 20.0544 | 30.0029 |
| 2256 | 0.5073 | 0.25100 | 0.0953 | 0.0410 | 0.1717 | 0.3351 | 0.2279 | 50.5254 | 81.2089 | 91.9185 | 66.1628 | 33.9510 | 55.0866 |
| 2494 | 0.5384 | 0.39300 | 0.1647 | 0.0407 | 0.2643 | 0.4463 | 0.3662 | 27.0117 | 69.4180 | 92.4474 | 50.9078 | 17.1110 | 31.9903 |
| 2497 | 0.6054 | 0.48833 | 0.3090 | 0.1420 | 0.3660 | 0.5858 | 0.5516 | 19.3428 | 48.9630 | 76.5461 | 39.5484 | 3.2520 | 8.8869 |
| 2506 | 0.5287 | 0.35800 | 0.1277 | 0.0433 | 0.2527 | 0.3858 | 0.3211 | 32.2822 | 75.8511 | 91.8032 | 52.2066 | 27.0324 | 39.2655 |
| 2601 | 0.4848 | 0.37567 | 0.0850 | 0.0260 | 0.2373 | 0.3401 | 0.2736 | 22.5072 | 82.4661 | 94.6367 | 51.0427 | 29.8465 | 43.5538 |
| 2602 | 0.5938 | 0.48567 | 0.1790 | 0.1050 | 0.3403 | 0.4641 | 0.4043 | 18.2071 | 69.8539 | 82.3166 | 42.6832 | 21.8414 | 31.9096 |
| 2604 | 0.6279 | 0.59867 | 0.3463 | 0.1283 | 0.4693 | 0.6625 | 0.6641 | 4.6538 | 44.8415 | 79.5611 | 25.2519 | -5.5177 | -5.7646 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





