Submitted:
14 May 2024
Posted:
15 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Respiratory Proteases
3. Serine Proteases
3.1. Furin
3.2. Coagulation Cascade Proteases (Plasmin, Thrombin, Factor Xa)
3.4. Elastase
3.3. Type II Transmembrane Serine Proteases (TTSP):
| Type II Transmembrane Serine Proteases (TTSPs) | Cleavage Preferences | Exploiting Respiratory Viruses |
|---|---|---|
| DESC1 | R-R/A/L-L-A↓ [70] | Influenza virus [71], MERS-CoV [71], SARS-CoV-1 [71] |
| Human Airway Trypsin-like Protease | R/K↓ [72,73] | Influenza virus [74], HCoV-229E [75], SARS-CoV-1 [76], Mammalian Orthoreovirus [6] |
| Matriptase | Minimum: R/K [22]↓ Preferred: R-X(non-basic)-S-R↓ [70] |
Influenza A virus [77,78] |
| TMPRSS11a | Unconfirmed, Putative R/K↓ | SARS-CoV [32], Influenza virus [79], MERS-CoV [79] |
| TMPRSS13/MSPL | R/K↓, Preference for dibasic P2-P1 [80] | Influenza virus [71], SARS-CoV-1 [71], MERS-CoV [71], SARS-CoV-2 [81] |
| TMPRSS2 | R/K↓ [82] | Influenza A + B virus [74], SARS-CoV-2 [83], HMPV [84], HCoV-229E [75], MERS-CoV [85], SARS-CoV [85], HPIV [86], Mammalian Orthoreovirus [6] |
| TMPRSS4 | Unconfirmed, Putative R/K↓[87] | Influenza A virus [88], SARS-CoV-2 [89] |
3.3. TMPRSS2
4. Cysteine Proteases - Cathepsins
| Cysteine Proteases | Cleavage Determinants | Exploiting Respiratory Viruses |
|---|---|---|
| Cathepsin L | Prefers aromatic or aliphatic residues in P2 [108] | HCoV-229E [75], SARS-CoV-1 [109,110,111], SARS-CoV-2 [112], Mammalian Orthoreovirus [113], MERS-CoV [114], Nipah [115], RSV [107], Hendra [115] |
| Cathepsin B | Prefers an aromatic or aliphatic residue and tolerates a basic P2, an aromatic residue in P1’ and a P3' G [108] | Influenza A virus [116], Mammalian Orthoreovirus [113], Nipah [105], SARS-CoV-2 [117] |
| Cathepsin S | Prefers aliphatic residues in P2, G/E in P1 [108] | Mammalian Orthoreovirus [118], SARS-CoV-2, SARS-CoV-1, RSV [40] |
| Cathepsin W | W/F–L/V–G/A/R↓V–D/N/E/Q (Suggested [119]] | Influenza A virus [120], RSV [40] |
| Cathepsin K | Prefers non-aromatic hydrophobic residues in P2 [121] | SARS-CoV-2 [117] |
| Cathepsin V | Prefers hydrophobic residues in P2, P in P3 [122] | SARS-CoV-2 [117] |
4. Metalloproteases
4.1. ACE2
| Metalloprotease | Exploiting Respiratory Viruses | Role |
|---|---|---|
| MT-MMP | SARS-CoV-2 [130] | Proteolytic cleavage |
| ADAM | SARS-CoV-2 [130] | Proteolytic cleavage |
| ACE2 | SARS-CoV-2 [135], SARS-CoV [136], HcoV NL63 [137] | Influenza A virus [134], MERS-CoV [134] | Receptor| Regulation of TMPRSS2 |
| APN | HCoV-229E [138] | Receptor |
5. Protease Inhibitors –
6. Current Therapeutic Strategies Targeting Coronaviruses, Influenza viruses and Para-Myxoviruses
7. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schechter I, Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967 Apr 20;27(2):157–62.
- McKelvey MC, Brown R, Ryan S, Mall MA, Weldon S, Taggart CC. Proteases, Mucus, and Mucosal Immunity in Chronic Lung Disease. Int J Mol Sci. 2021 May 9;22(9):5018. [CrossRef]
- Majchrzak M, Poręba M. The roles of cellular protease interactions in viral infections and programmed cell death: a lesson learned from the SARS-CoV-2 outbreak and COVID-19 pandemic. Pharmacol Rep. 2022 Dec 1;74(6):1149–65.
- Rawlings ND, Barrett AJ. Chapter 559 - Introduction: Serine Peptidases and Their Clans. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes (Third Edition) [Internet]. Academic Press; 2013 [cited 2023 Jul 13]. p. 2491–523. Available from: https://www.sciencedirect.com/science/article/pii/B9780123822192005597.
- Andersson MK, Enoksson M, Gallwitz M, Hellman L. The extended substrate specificity of the human mast cell chymase reveals a serine protease with well-defined substrate recognition profile. Int Immunol. 2009 Jan;21(1):95–104. [CrossRef]
- Nygaard RM, Golden JW, Schiff LA. Impact of Host Proteases on Reovirus Infection in the Respiratory Tract. Journal of Virology. 2012 Jan 15;86(2):1238–43. [CrossRef]
- Brown MA, Stenberg LM, Stenflo J. Coagulation Factor Xa. Handbook of Proteolytic Enzymes. 2013;2908–15.
- Du L, Kao RY, Zhou Y, He Y, Zhao G, Wong C, et al. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochemical and Biophysical Research Communications. 2007 Jul 20;359(1):174–9. [CrossRef]
- Kastenhuber ER, Mercadante M, Nilsson-Payant B, Johnson JL, Jaimes JA, Muecksch F, et al. Coagulation factors directly cleave SARS-CoV-2 spike and enhance viral entry. van der Meer JW, editor. eLife. 2022 Mar 16;11:e77444.
- Hosaka M, Nagahama M, Kim WS, Watanabe T, Hatsuzawa K, Ikemizu J, et al. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. Journal of Biological Chemistry. 1991 Jul 5;266(19):12127–30. [CrossRef]
- Kawaoka Y, Webster RG. Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proceedings of the National Academy of Sciences. 1988 Jan;85(2):324–8. [CrossRef]
- Peacock TP, Goldhill DH, Zhou J, Baillon L, Frise R, Swann OC, et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021 Jul;6(7):899–909.
- Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):15214–9. [CrossRef]
- Collins PL, Huang YT, Wertz GW. Nucleotide sequence of the gene encoding the fusion (F) glycoprotein of human respiratory syncytial virus. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7683–7. [CrossRef]
- Ortmann D, Ohuchi M, Angliker H, Shaw E, Garten W, Klenk HD. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J Virol. 1994 Apr;68(4):2772–6. [CrossRef]
- Watanabe M, Hirano A, Stenglein S, Nelson J, Thomas G, Wong TC. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J Virol. 1995 May;69(5):3206–10. [CrossRef]
- Ueo A, Kubota M, Shirogane Y, Ohno S, Hashiguchi T, Yanagi Y. Lysosome-Associated Membrane Proteins Support the Furin-Mediated Processing of the Mumps Virus Fusion Protein. J Virol. 2020 Jun 1;94(12):e00050-20. [CrossRef]
- Borgoño CA, Gavigan JA, Alves J, Bowles B, Harris JL, Sotiropoulou G, et al. Defining the extended substrate specificity of kallikrein 1-related peptidases. 2007 Nov 1;388(11):1215–25. [CrossRef]
- Milewska A, Falkowski K, Kulczycka M, Bielecka E, Naskalska A, Mak P, et al. Kallikrein 13 serves as a priming protease during infection by the human coronavirus HKU1. Science Signaling. 2020 Nov 24;13(659):eaba9902. [CrossRef]
- Li HX, Hwang BY, Laxmikanthan G, Blaber SI, Blaber M, Golubkov PA, et al. Substrate specificity of human kallikreins 1 and 6 determined by phage display. Protein Sci. 2008 Apr;17(4):664–72. [CrossRef]
- Hamilton BS, Whittaker GR. Cleavage Activation of Human-adapted Influenza Virus Subtypes by Kallikrein-related Peptidases 5 and 12 *. Journal of Biological Chemistry. 2013 Jun 14;288(24):17399–407. [CrossRef]
- Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc Natl Acad Sci U S A. 2000 Jul 5;97(14):7754–9. [CrossRef]
- Golden JW, Schiff LA. Neutrophil elastase, an acid-independent serine protease, facilitates reovirus uncoating and infection in U937 promonocyte cells. Virology Journal. 2005 May 31;2(1):48. [CrossRef]
- Belouzard S, Madu I, Whittaker GR. Elastase-mediated Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein at Discrete Sites within the S2 Domain *. Journal of Biological Chemistry. 2010 Jul 23;285(30):22758–63. [CrossRef]
- Matsuyama S, Ujike M, Morikawa S, Tashiro M, Taguchi F. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proceedings of the National Academy of Sciences. 2005 Aug 30;102(35):12543–7. [CrossRef]
- Seidah NG. Chapter 730 - Proprotein Convertase 5. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes (Third Edition) [Internet]. Academic Press; 2013 [cited 2024 May 4]. p. 3305–10. Available from: https://www.sciencedirect.com/science/article/pii/B9780123822192007304.
- Basak A, Zhong M, Munzer JS, Chrétien M, Seidah NG. Implication of the proprotein convertases furin, PC5 and PC7 in the cleavage of surface glycoproteins of Hong Kong, Ebola and respiratory syncytial viruses: a comparative analysis with fluorogenic peptides. Biochem J. 2001 Feb 1;353(Pt 3):537–45.
- Castellino FJ. Chapter 648 - Plasmin. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes (Third Edition) [Internet]. Academic Press; 2013 [cited 2023 Jul 13]. p. 2958–68. Available from: https://www.sciencedirect.com/science/article/pii/B9780123822192006487.
- Xue F, Seto CT. Selective Inhibitors of the Serine Protease Plasmin: Probing the S3 and S3‘ Subsites Using a Combinatorial Library. J Med Chem. 2005 Nov 1;48(22):6908–17. [CrossRef]
- Tse LV, Marcano VC, Huang W, Pocwierz MS, Whittaker GR. Plasmin-Mediated Activation of Pandemic H1N1 Influenza Virus Hemagglutinin Is Independent of the Viral Neuraminidase. J Virol. 2013 May;87(9):5161–9. [CrossRef]
- Dubovi EJ, Geratz JD, Tidwell RR. Enhancement of respiratory syncytial virus-induced cytopathology by trypsin, thrombin, and plasmin. Infect Immun. 1983 Apr;40(1):351–8. [CrossRef]
- Kam YW, Okumura Y, Kido H, Ng LFP, Bruzzone R, Altmeyer R. Cleavage of the SARS Coronavirus Spike Glycoprotein by Airway Proteases Enhances Virus Entry into Human Bronchial Epithelial Cells In Vitro. PLOS ONE. 2009 Nov 17;4(11):e7870. [CrossRef]
- Hou Y, Yu T, Wang T, Ding Y, Cui Y, Nie H. Competitive cleavage of SARS-CoV-2 spike protein and epithelial sodium channel by plasmin as a potential mechanism for COVID-19 infection. Am J Physiol Lung Cell Mol Physiol. 2022 Nov 1;323(5):L569–77.
- Gallwitz M, Enoksson M, Thorpe M, Hellman L. The Extended Cleavage Specificity of Human Thrombin. PLoS One. 2012 Feb 27;7(2):e31756. [CrossRef]
- Rodriguez T, Dobrovolny HM. Quantifying the effect of trypsin and elastase on in vitro SARS-CoV infections. Virus Research. 2021 Jul 2;299:198423.
- Kirchdoerfer RN, Wang N, Pallesen J, Wrapp D, Turner HL, Cottrell CA, et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep. 2018 Oct 24;8(1):15701.
- Schowalter RM, Smith SE, Dutch RE. Characterization of Human Metapneumovirus F Protein-Promoted Membrane Fusion: Critical Roles for Proteolytic Processing and Low pH. J Virol. 2006 Nov;80(22):10931–41. [CrossRef]
- Henrickson KJ. Parainfluenza Viruses. Clin Microbiol Rev. 2003 Apr;16(2):242–64.
- Thorpe M, Fu Z, Chahal G, Akula S, Kervinen J, de Garavilla L, et al. Extended cleavage specificity of human neutrophil cathepsin G: A low activity protease with dual chymase and tryptase-type specificities. PLoS One. 2018 Apr 13;13(4):e0195077. [CrossRef]
- Foronjy RF, Taggart CC, Dabo AJ, Weldon S, Cummins N, Geraghty P. Type-I interferons induce lung protease responses following respiratory syncytial virus infection via RIG-I-like receptors. Mucosal Immunology. 2015 Jan 1;8(1):161–75. [CrossRef]
- Tian S. A 20 Residues Motif Delineates the Furin Cleavage Site and its Physical Properties May Influence Viral Fusion. Biochem Insights. 2009 Jan 1;2:BCI.S2049. [CrossRef]
- Choi A, Kots ED, Singleton DT, Weinstein H, Whittaker GR. Analysis of the molecular determinants for furin cleavage of the spike protein S1/S2 site in defined strains of the prototype coronavirus murine hepatitis virus (MHV). Virus Res. 2024 Feb;340:199283.
- Molloy SS, Bresnahan PA, Leppla SH, Klimpel KR, Thomas G. Human furin is a calcium-dependent serine endoprotease that recognizes the sequence Arg-X-X-Arg and efficiently cleaves anthrax toxin protective antigen. Journal of Biological Chemistry. 1992 Aug 15;267(23):16396–402. [CrossRef]
- Böttcher-Friebertshäuser E, Klenk HD, Garten W. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathogens and Disease. 2013 Nov 1;69(2):87–100. [CrossRef]
- Lu X, Shi Y, Gao F, Xiao H, Wang M, Qi J, et al. Insights into Avian Influenza Virus Pathogenicity: the Hemagglutinin Precursor HA0 of Subtype H16 Has an Alpha-Helix Structure in Its Cleavage Site with Inefficient HA1/HA2 Cleavage. Journal of Virology. 2012 Dec;86(23):12861–70.
- Mayer G, Boileau G, Bendayan M. Sorting of Furin in Polarized Epithelial and Endothelial Cells: Expression Beyond the Golgi Apparatus. J Histochem Cytochem. 2004 May 1;52(5):567–79. [CrossRef]
- Krzyzaniak MA, Zumstein MT, Gerez JA, Picotti P, Helenius A. Host Cell Entry of Respiratory Syncytial Virus Involves Macropinocytosis Followed by Proteolytic Activation of the F Protein. PLOS Pathogens. 2013 Apr 11;9(4):e1003309. [CrossRef]
- Takeda M. Proteolytic activation of SARS-CoV-2 spike protein. Microbiol Immunol. 2022 Jan;66(1):15–23.
- Gobeil SMC, Janowska K, McDowell S, Mansouri K, Parks R, Manne K, et al. D614G Mutation Alters SARS-CoV-2 Spike Conformation and Enhances Protease Cleavage at the S1/S2 Junction. Cell Reports. 2021 Jan 12;34(2):108630.
- Sasaki M, Uemura K, Sato A, Toba S, Sanaki T, Maenaka K, et al. SARS-CoV-2 variants with mutations at the S1/S2 cleavage site are generated in vitro during propagation in TMPRSS2-deficient cells. PLOS Pathogens. 2021 Jan 21;17(1):e1009233.
- Akatsuki Saito, et al. Nature. 2021 [cited 2022 Sep 8]. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation | Nature. Available from: https://www.nature.com/articles/s41586-021-04266-9.
- Lubinski B, Frazier LE, Phan MVT, Bugembe DL, Cunningham JL, Tang T, et al. Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: a case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. bioRxiv. 2022 Mar 28;2021.06.30.450632.
- Lubinski B, Fernandes MHV, Frazier L, Tang T, Daniel S, Diel DG, et al. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience. 2022 Jan 21;25(1):103589.
- Liu Y, Liu J, Johnson BA, Xia H, Ku Z, Schindewolf C, et al. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Reports. 2022 May 17;39(7):110829.
- Gellenoncourt S, Saunders N, Robinot R, Auguste L, Rajah MM, Kervevan J, et al. The Spike-Stabilizing D614G Mutation Interacts with S1/S2 Cleavage Site Mutations To Promote the Infectious Potential of SARS-CoV-2 Variants. J Virol. 96(19):e01301-22.
- Lau SKP, Li KSM, Li X, Tsang KY, Sridhar S, Woo PCY. Fatal Pneumonia Associated With a Novel Genotype of Human Coronavirus OC43. Front Microbiol [Internet]. 2022 Jan 14 [cited 2024 May 7];12. [CrossRef]
- Stodola JK, Dubois G, Le Coupanec A, Desforges M, Talbot PJ. The OC43 human coronavirus envelope protein is critical for infectious virus production and propagation in neuronal cells and is a determinant of neurovirulence and CNS pathology. Virology. 2018 Feb;515:134–49. [CrossRef]
- Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor | Nature [Internet]. [cited 2024 Apr 9]. Available from: https://www.nature.com/articles/nature07082.
- Julie M. Murray, et al. Journal of General Virology. 1993 [cited 2022 Sep 8]. Processing of the dengue virus type 2 proteins prM and C-prM | Microbiology Society. [CrossRef]
- Dejnirattisai W, Jumnainsong A, Onsirisakul N, Fitton P, Vasanawathana S, Limpitikul W, et al. Cross-Reacting Antibodies Enhance Dengue Virus Infection in Humans. Science. 2010 May 7;328(5979):745–8. [CrossRef]
- Luo YY, Feng JJ, Zhou JM, Yu ZZ, Fang DY, Yan HJ, et al. Identification of a novel infection-enhancing epitope on dengue prM using a dengue cross-reacting monoclonal antibody. BMC Microbiology. 2013 Aug 29;13(1):194. [CrossRef]
- Schreuder H, Matter H. Serine Proteinases from the Blood Coagulation Cascade. In: Structural Biology in Drug Discovery [Internet]. John Wiley & Sons, Ltd; 2020 [cited 2024 Apr 9]. p. 395–422. [CrossRef]
- Franczuk P, Tkaczyszyn M, Kulak M, Domenico E, Ponikowski P, Jankowska EA. Cardiovascular Complications of Viral Respiratory Infections and COVID-19. Biomedicines. 2022 Dec 27;11(1):71. [CrossRef]
- Lazarowitz SG, Choppin PW. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology. 1975 Dec 1;68(2):440–54. [CrossRef]
- Sun X, Tse LV, Ferguson AD, Whittaker GR. Modifications to the Hemagglutinin Cleavage Site Control the Virulence of a Neurotropic H1N1 Influenza Virus. J Virol. 2010 Sep;84(17):8683–90. [CrossRef]
- Gotoh B, Ogasawara T, Toyoda T, Inocencio NM, Hamaguchi M, Nagai Y. An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J. 1990 Dec;9(12):4189–95. [CrossRef]
- Kummarapurugu AB, Hawkridge AM, Ma J, Osei S, Martin RK, Zheng S, et al. Neutrophil elastase decreases SARS-CoV-2 spike protein binding to human bronchial epithelia by clipping ACE-2 ectodomain from the epithelial surface. Journal of Biological Chemistry [Internet]. 2023 Jun 1 [cited 2024 May 1];299(6). Available from: https://www.jbc.org/article/S0021-9258(23)01848-3/abstract.
- Masic A, Booth JS, Mutwiri GK, Babiuk LA, Zhou Y. Elastase-Dependent Live Attenuated Swine Influenza A Viruses Are Immunogenic and Confer Protection against Swine Influenza A Virus Infection in Pigs. Journal of Virology. 2009 Oct;83(19):10198–210. [CrossRef]
- Bugge TH, Antalis TM, Wu Q. Type II Transmembrane Serine Proteases. J Biol Chem. 2009 Aug 28;284(35):23177–81.
- Béliveau F, Désilets A, Leduc R. Probing the substrate specificities of matriptase, matriptase-2, hepsin and DESC1 with internally quenched fluorescent peptides. The FEBS Journal. 2009;276(8):2213–26. [CrossRef]
- Zmora P, Blazejewska P, Moldenhauer AS, Welsch K, Nehlmeier I, Wu Q, et al. DESC1 and MSPL Activate Influenza A Viruses and Emerging Coronaviruses for Host Cell Entry. Journal of Virology. 2014 Oct 15;88(20):12087–97. [CrossRef]
- Yasuoka S, Ohnishi T, Kawano S, Tsuchihashi S, Ogawara M, Masuda K, et al. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. Am J Respir Cell Mol Biol. 1997 Mar;16(3):300–8. [CrossRef]
- Wysocka M, Spichalska B, Lesner A, Jaros M, Brzozowski K, Łęgowska A, et al. Substrate specificity and inhibitory study of human airway trypsin-like protease. Bioorganic & Medicinal Chemistry. 2010 Aug 1;18(15):5504–9. [CrossRef]
- Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. Proteolytic Activation of Influenza Viruses by Serine Proteases TMPRSS2 and HAT from Human Airway Epithelium. Journal of Virology. 2006 Oct;80(19):9896–8. [CrossRef]
- Bertram S, Dijkman R, Habjan M, Heurich A, Gierer S, Glowacka I, et al. TMPRSS2 Activates the Human Coronavirus 229E for Cathepsin-Independent Host Cell Entry and Is Expressed in Viral Target Cells in the Respiratory Epithelium. Journal of Virology. 2013 Jun;87(11):6150–60. [CrossRef]
- Bertram S, Glowacka I, Müller MA, Lavender H, Gnirss K, Nehlmeier I, et al. Cleavage and Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by Human Airway Trypsin-Like Protease. Journal of Virology. 2011 Dec 15;85(24):13363–72. [CrossRef]
- Beaulieu A, Gravel É, Cloutier A, Marois I, Colombo É, Désilets A, et al. Matriptase Proteolytically Activates Influenza Virus and Promotes Multicycle Replication in the Human Airway Epithelium. Journal of Virology. 2013 Apr 15;87(8):4237–51. [CrossRef]
- Whittaker GR, Straus MR. Human matriptase/ST 14 proteolytically cleaves H7N9 hemagglutinin and facilitates the activation of influenza A/Shanghai/2/2013 virus in cell culture. Influenza Other Respir Viruses. 2020 Mar;14(2):189–95.
- Zmora P, Hoffmann M, Kollmus H, Moldenhauer AS, Danov O, Braun A, et al. TMPRSS11A activates the influenza A virus hemagglutinin and the MERS coronavirus spike protein and is insensitive against blockade by HAI-1. Journal of Biological Chemistry. 2018 Sep 1;293(36):13863–73. [CrossRef]
- Kido H, Okumura Y. MSPL/TMPRSS13. FBL. 2008 Jan 1;13(2):754–8.
- Kishimoto M, Uemura K, Sanaki T, Sato A, Hall WW, Kariwa H, et al. TMPRSS11D and TMPRSS13 Activate the SARS-CoV-2 Spike Protein. Viruses. 2021 Mar;13(3):384.
- Yun Kim S, Park D, Oh M, Sellamuthu S, Park WJ. Detection of site-specific proteolysis in secretory pathways. Biochemical and Biophysical Research Communications. 2002 Aug 16;296(2):419–24.
- Mahmoud IS, Jarrar YB, Alshaer W, Ismail S. SARS-CoV-2 entry in host cells-multiple targets for treatment and prevention. Biochimie. 2020 Aug 1;175:93–8.
- Shirogane Y, Takeda M, Iwasaki M, Ishiguro N, Takeuchi H, Nakatsu Y, et al. Efficient Multiplication of Human Metapneumovirus in Vero Cells Expressing the Transmembrane Serine Protease TMPRSS2. Journal of Virology. 2008 Sep;82(17):8942–6. [CrossRef]
- Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J Virol. 2019 Mar 5;93(6):e01815-18. [CrossRef]
- Abe M, Tahara M, Sakai K, Yamaguchi H, Kanou K, Shirato K, et al. TMPRSS2 Is an Activating Protease for Respiratory Parainfluenza Viruses. J Virol. 2013 Nov;87(21):11930–5. [CrossRef]
- Esram P, Arumugam P. Development and Validation of an Enzymatic Assay for TMPRSS4: Evaluation of Molecular Inhibitors. Journal of Advanced Zoology. 2023 Oct 9;44(3):309–21. [CrossRef]
- Kühn N, Bergmann S, Kösterke N, Lambertz RLO, Keppner A, van den Brand JMA, et al. The Proteolytic Activation of (H3N2) Influenza A Virus Hemagglutinin Is Facilitated by Different Type II Transmembrane Serine Proteases. Journal of Virology. 2016 Apr 14;90(9):4298–307. [CrossRef]
- Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020 May 13;5(47):eabc3582.
- Epstein RJ. The secret identities of TMPRSS2: Fertility factor, virus trafficker, inflammation moderator, prostate protector and tumor suppressor. Tumor Biology. 2021 Jan 1;43(1):159–76. [CrossRef]
- Murza A, Dion SP, Boudreault PL, Désilets A, Leduc R, Marsault É. Inhibitors of type II transmembrane serine proteases in the treatment of diseases of the respiratory tract – A review of patent literature. Expert Opinion on Therapeutic Patents. 2020 Nov 1;30(11):807–24. [CrossRef]
- Heurich A, Hofmann-Winkler H, Gierer S, Liepold T, Jahn O, Pöhlmann S. TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. J Virol. 2014 Jan;88(2):1293–307. [CrossRef]
- Kirschke H. Chapter 410 - Cathepsin L. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes (Third Edition) [Internet]. Academic Press; 2013 [cited 2023 Jul 13]. p. 1808–17. Available from: https://www.sciencedirect.com/science/article/pii/B9780123822192004105.
- Yadati T, Houben T, Bitorina A, Shiri-Sverdlov R. The Ins and Outs of Cathepsins: Physiological Function and Role in Disease Management. Cells. 2020 Jul 13;9(7):1679. [CrossRef]
- Du X, Tang H, Gao L, Wu Z, Meng F, Yan R, et al. Omicron adopts a different strategy from Delta and other variants to adapt to host. Sig Transduct Target Ther. 2022 Feb 10;7(1):1–3. [CrossRef]
- Willett BJ, Grove J, MacLean OA, Wilkie C, De Lorenzo G, Furnon W, et al. SARS-CoV-2 Omicron is an immune escape variant with an altered cell entry pathway. Nat Microbiol. 2022 Aug;7(8):1161–79.
- Mykytyn AZ, Breugem TI, Geurts MH, Beumer J, Schipper D, van Acker R, et al. SARS-CoV-2 Omicron entry is type II transmembrane serine protease-mediated in human airway and intestinal organoid models. J Virol. 97(8):e00851-23.
- Scarcella M, d’Angelo D, Ciampa M, Tafuri S, Avallone L, Pavone LM, et al. The Key Role of Lysosomal Protease Cathepsins in Viral Infections. Int J Mol Sci. 2022 Aug 13;23(16):9089. [CrossRef]
- Mainou BA. The Orchestra of Reovirus Cell Entry. Curr Clin Micro Rpt. 2017 Sep 1;4(3):142–9. [CrossRef]
- Baer GS, Dermody TS. Mutations in reovirus outer-capsid protein sigma3 selected during persistent infections of L cells confer resistance to protease inhibitor E64. Journal of Virology. 1997 Jul;71(7):4921–8. [CrossRef]
- Wilson GJ, Nason EL, Hardy CS, Ebert DH, Wetzel JD, Venkataram Prasad BV, et al. A Single Mutation in the Carboxy Terminus of Reovirus Outer-Capsid Protein σ3 Confers Enhanced Kinetics of σ3 Proteolysis, Resistance to Inhibitors of Viral Disassembly, and Alterations in σ3 Structure. Journal of Virology. 2002 Oct;76(19):9832–43.
- Doyle JD, Danthi P, Kendall EA, Ooms LS, Wetzel JD, Dermody TS. Molecular Determinants of Proteolytic Disassembly of the Reovirus Outer Capsid *. Journal of Biological Chemistry. 2012 Mar 9;287(11):8029–38. [CrossRef]
- Mainou BA, Dermody TS. In Search of Cathepsins: How Reovirus Enters Host Cells. DNA Cell Biol. 2012 Dec;31(12):1646–9. [CrossRef]
- Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of Endosomal Cathepsins in Entry Mediated by the Ebola Virus Glycoprotein. Journal of Virology. 2006 Apr 15;80(8):4174–8. [CrossRef]
- Diederich S, Sauerhering L, Weis M, Altmeppen H, Schaschke N, Reinheckel T, et al. Activation of the Nipah virus fusion protein in MDCK cells is mediated by cathepsin B within the endosome-recycling compartment. J Virol. 2012 Apr;86(7):3736–45. [CrossRef]
- Vogt C, Eickmann M, Diederich S, Moll M, Maisner A. Endocytosis of the Nipah Virus Glycoproteins. J Virol. 2005 Mar;79(6):3865–72. [CrossRef]
- Corry J, Johnson SM, Cornwell J, Peeples ME. Preventing Cleavage of the Respiratory Syncytial Virus Attachment Protein in Vero Cells Rescues the Infectivity of Progeny Virus for Primary Human Airway Cultures. J Virol. 2016 Jan 15;90(3):1311–20. [CrossRef]
- Biniossek ML, Nägler DK, Becker-Pauly C, Schilling O. Proteomic Identification of Protease Cleavage Sites Characterizes Prime and Non-prime Specificity of Cysteine Cathepsins B, L, and S. J Proteome Res. 2011 Dec 2;10(12):5363–73.
- Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci USA. 2005 Aug 16;102(33):11876–81. [CrossRef]
- Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. Simultaneous Treatment of Human Bronchial Epithelial Cells with Serine and Cysteine Protease Inhibitors Prevents Severe Acute Respiratory Syndrome Coronavirus Entry. Journal of Virology. 2012 Jun 15;86(12):6537–45. [CrossRef]
- Belouzard S, Chu VC, Whittaker GR. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proceedings of the National Academy of Sciences. 2009 Apr 7;106(14):5871–6. [CrossRef]
- Zhao MM, Yang WL, Yang FY, Zhang L, Huang WJ, Hou W, et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Sig Transduct Target Ther. 2021 Mar 27;6(1):1–12.
- Ebert DH, Deussing J, Peters C, Dermody TS. Cathepsin L and Cathepsin B Mediate Reovirus Disassembly in Murine Fibroblast Cells *. Journal of Biological Chemistry. 2002 Jul 5;277(27):24609–17. [CrossRef]
- Kleine-Weber H, Elzayat MT, Hoffmann M, Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci Rep. 2018 Nov 9;8(1):16597. [CrossRef]
- Pager CT, Craft WW, Patch J, Dutch RE. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology. 2006 Mar 15;346(2):251–7. [CrossRef]
- Coleman MD, Ha SD, Haeryfar SMM, Barr SD, Kim SO. Cathepsin B plays a key role in optimal production of the influenza A virus. J Virol Antivir Res. 2018 Apr;7(1):1–20. [CrossRef]
- Bollavaram K, Leeman TH, Lee MW, Kulkarni A, Upshaw SG, Yang J, et al. Multiple sites on SARS-CoV-2 spike protein are susceptible to proteolysis by cathepsins B, K, L, S, and V. Protein Science. 2021;30(6):1131–43.
- Golden JW, Bahe JA, Lucas WT, Nibert ML, Schiff LA. Cathepsin S Supports Acid-independent Infection by Some Reoviruses *. Journal of Biological Chemistry. 2004 Mar 2;279(10):8547–57. [CrossRef]
- Brinkworth RI, Tort JF, Brindley PJ, Dalton JP. Phylogenetic relationships and theoretical model of human cathepsin W (lymphopain), a cysteine proteinase from cytotoxic T lymphocytes. The International Journal of Biochemistry & Cell Biology. 2000 Mar 1;32(3):373–84. [CrossRef]
- Edinger TO, Pohl MO, Yángüez E, Stertz S. Cathepsin W Is Required for Escape of Influenza A Virus from Late Endosomes. mBio. 2015 Jun 9;6(3):e00297.
- Brömme D. Chapter 409 - Cathepsin K. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes (Third Edition) [Internet]. Academic Press; 2013 [cited 2024 May 4]. p. 1801–7. Available from: https://www.sciencedirect.com/science/article/pii/B9780123822192004099.
- Brömme D. Chapter 414 - Cathepsin V. In: Rawlings ND, Salvesen G, editors. Handbook of Proteolytic Enzymes (Third Edition) [Internet]. Academic Press; 2013 [cited 2024 May 4]. p. 1831–4. Available from: https://www.sciencedirect.com/science/article/pii/B9780123822192004130.
- Cabral-Pacheco GA, Garza-Veloz I, Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA, Guerrero-Rodriguez JF, et al. The Roles of Matrix Metalloproteinases and Their Inhibitors in Human Diseases. Int J Mol Sci. 2020 Dec 20;21(24):9739. [CrossRef]
- Yeo SJ, Yun YJ, Lyu MA, Woo SY, Woo ER, Kim SJ, et al. Respiratory syncytial virus infection induces matrix metalloproteinase-9 expression in epithelial cells. Arch Virol. 2002 Feb 1;147(2):229–42. [CrossRef]
- Rojas-Quintero J, Wang X, Tipper J, Burkett PR, Zuñiga J, Ashtekar AR, et al. Matrix metalloproteinase-9 deficiency protects mice from severe influenza A viral infection. JCI Insight. 2018 Dec 20;3(24):e99022, 99022. [CrossRef]
- Tacon CE, Wiehler S, Holden NS, Newton R, Proud D, Leigh R. Human Rhinovirus Infection Up-Regulates MMP-9 Production in Airway Epithelial Cells via NF-κB. Am J Respir Cell Mol Biol. 2010 Aug;43(2):201–9.
- Elliott MB, Welliver RC, Laughlin TS, Pryharski KS, LaPierre NA, Chen T, et al. Matrix metalloproteinase-9 and tissue inhibitor of matrix metalloproteinase-1 in the respiratory tracts of human infants following paramyxovirus infection. J Med Virol. 2007 Apr;79(4):447–56. [CrossRef]
- Phillips JM, Gallagher T, Weiss SR. Neurovirulent Murine Coronavirus JHM.SD Uses Cellular Zinc Metalloproteases for Virus Entry and Cell-Cell Fusion. J Virol. 2017 Mar 29;91(8):e01564-16. [CrossRef]
- Yamamoto M, Gohda J, Kobayashi A, Tomita K, Hirayama Y, Koshikawa N, et al. Metalloproteinase-Dependent and TMPRSS2-Independent Cell Surface Entry Pathway of SARS-CoV-2 Requires the Furin Cleavage Site and the S2 Domain of Spike Protein. mBio. 2022 Aug 30;13(4):e0051922.
- Chan JFW, Huang X, Hu B, Chai Y, Shi H, Zhu T, et al. Altered host protease determinants for SARS-CoV-2 Omicron. Science Advances. 2023 Jan 20;9(3):eadd3867.
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar 27;367(6485):1444–8.
- Yu S, Zheng X, Zhou B, Li J, Chen M, Deng R, et al. SARS-CoV-2 spike engagement of ACE2 primes S2′ site cleavage and fusion initiation. Proceedings of the National Academy of Sciences. 2022 Jan 4;119(1):e2111199119.
- Zhang H, Lv P, Jiang J, Liu Y, Yan R, Shu S, et al. Advances in developing ACE2 derivatives against SARS-CoV-2. Lancet Microbe. 2023 May;4(5):e369–78.
- Heindl MR, Rupp AL, Schwerdtner M, Bestle D, Harbig A, De Rocher A, et al. ACE2 acts as a novel regulator of TMPRSS2-catalyzed proteolytic activation of influenza A virus in airway cells. Subbarao K, editor. J Virol. 2024 Mar 12;e00102-24. [CrossRef]
- Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol. 2020 Jun;17(6):613–20. [CrossRef]
- Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003 Nov 27;426(6965):450–4. [CrossRef]
- Hofmann H, Pyrc K, van der Hoek L, Geier M, Berkhout B, Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci U S A. 2005 May 31;102(22):7988–93. [CrossRef]
- Funk CJ, Wang J, Ito Y, Travanty EA, Voelker DR, Holmes KV, et al. Infection of human alveolar macrophages by human coronavirus strain 229E. J Gen Virol. 2012 Mar;93(Pt 3):494–503. [CrossRef]
- Hajizadeh M, Moosavi-Movahedi Z, Sheibani N, Moosavi-Movahedi AA. An outlook on suicide enzyme inhibition and drug design. Journal of the Iranian Chemical Society. 2022;19(5):1575–92. [CrossRef]
- Ben-Tal AK Nir. Introduction to Proteins: Structure, Function, and Motion, Second Edition. 2nd ed. New York: Chapman and Hall/CRC; 2018. 988 p.
- Rajendran L, Knölker HJ, Simons K. Subcellular targeting strategies for drug design and delivery. Nat Rev Drug Discov. 2010 Jan;9(1):29–42. [CrossRef]
- Donahue ND, Acar H, Wilhelm S. Concepts of nanoparticle cellular uptake, intracellular trafficking, and kinetics in nanomedicine. Advanced Drug Delivery Reviews. 2019 Mar 15;143:68–96. [CrossRef]
- Lotke R, Petersen M, Sauter D. Restriction of Viral Glycoprotein Maturation by Cellular Protease Inhibitors. Viruses. 2024 Feb 22;16(3):332. [CrossRef]
- Stein P, Chothia C. Serpin tertiary structure transformation. Journal of Molecular Biology. 1991 Sep 20;221(2):615–21. [CrossRef]
- Nichols DB, De Martini W, Cottrell J. Poxviruses Utilize Multiple Strategies to Inhibit Apoptosis. Viruses. 2017 Aug;9(8):215. [CrossRef]
- Tarighi P, Eftekhari S, Chizari M, Sabernavaei M, Jafari D, Mirzabeigi P. A review of potential suggested drugs for coronavirus disease (COVID-19) treatment. Eur J Pharmacol. 2021 Mar 15;895:173890. [CrossRef]
- Wyde PR, Chetty SN, Jewell AM, Boivin G, Piedra PA. Comparison of the inhibition of human metapneumovirus and respiratory syncytial virus by ribavirin and immune serum globulin in vitro. Antiviral Res. 2003 Sep;60(1):51–9. [CrossRef]
- Thomas G, Couture F, Kwiatkowska A. The Path to Therapeutic Furin Inhibitors: From Yeast Pheromones to SARS-CoV-2. International Journal of Molecular Sciences. 2022 Jan;23(7):3435.
- Essalmani R, Jain J, Susan-Resiga D, Andréo U, Evagelidis A, Derbali RM, et al. Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity. J Virol. 96(8):e00128-22.
- 125. Someya A, Tanaka N, Okuyama A. Inhibition of influenza virus AWSN replication by a trypsin inhibitor, 6-amidino-2-naphthyl p-guanidinobenzoate. Biochemical and Biophysical Research Communications. 1990 May 31;169(1):148–52. [CrossRef]
- Hosoya M, Matsuyama S, Baba M, Suzuki H, Shigeta S. Effects of protease inhibitors on replication of various myxoviruses. Antimicrob Agents Chemother. 1992 Jul;36(7):1432–6. [CrossRef]
- Hosoya M, Shigeta S, Ishii T, Suzuki H, Clercq ED. Comparative Inhibitory Effects of Various Nucleoside and Nonnucleoside Analogues on Replication of Influenza Virus Types A and B In Vitro and In Ovo. The Journal of Infectious Diseases. 1993 Sep 1;168(3):641–6. [CrossRef]
- Gunst JD, Staerke NB, Pahus MH, Kristensen LH, Bodilsen J, Lohse N, et al. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with Covid-19-a double-blind randomized controlled trial. EClinicalMedicine. 2021 May;35:100849. [CrossRef]
- Zhirnov OP, Klenk HD, Wright PF. Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res. 2011 Oct;92(1):27–36. [CrossRef]
- Grocott HP, Sheng H, Miura Y, Sarraf-Yazdi S, Mackensen GB, Pearlstein RD, et al. The effects of aprotinin on outcome from cerebral ischemia in the rat. Anesth Analg. 1999 Jan;88(1):1–7.
- Mangano DT, Tudor IC, Dietzel C, Multicenter Study of Perioperative Ischemia Research Group, Ischemia Research and Education Foundation. The risk associated with aprotinin in cardiac surgery. N Engl J Med. 2006 Jan 26;354(4):353–65.
- Hamilton BS, Chung C, Cyphers SY, Rinaldi VD, Marcano VC, Whittaker GR. Inhibition of influenza virus infection and hemagglutinin cleavage by the protease inhibitor HAI-2. Biochem Biophys Res Commun. 2014 Jul 25;450(2):1070–5. [CrossRef]
- Straus MR, Kinder JT, Segall M, Dutch RE, Whittaker GR. SPINT2 inhibits proteases involved in activation of both influenza viruses and metapneumoviruses. Virology. 2020 Apr;543:43–53. [CrossRef]
- Shapira T, Monreal IA, Dion SP, Buchholz DW, Imbiakha B, Olmstead AD, et al. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature. 2022;605(7909):340–8.
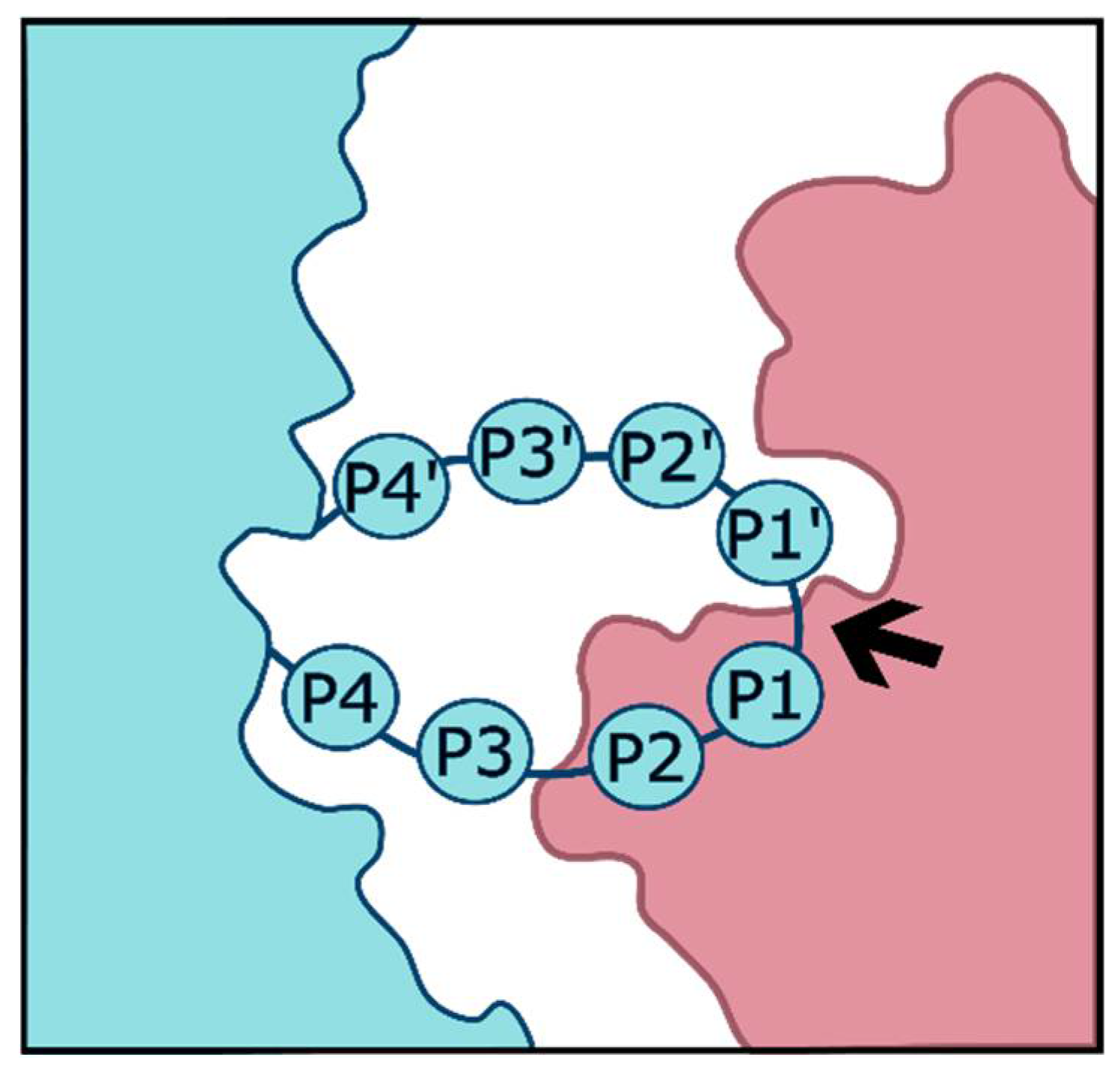
| Serine Proteases | Cleavage Preferences | Exploiting Respiratory Viruses |
|---|---|---|
| Chymase | Aromatic amino acids at P1, aliphatic amino acids from P2-P4, S at P1’, E/D at P2’, A/V/G at P3’ [5] | Mammalian Orthoreovirus [6] |
| Factor Xa | Preference for PXG/AR↓XXD [7] | SARS-CoV-1 [8], SARS-CoV-2 [9] |
| Furin | R-X-X-R↓S [10] | Influenza virus [11], SARS-CoV-2 [12], MERS-CoV[13] , RSV [14], HPIV [15], Measles virus (MeV) [16], Mumps virus [17] |
| kallikrein-related peptidase 13 (KLK13) | V/Y-R/L/F/M-R↓ [18] | HKU-1 [19] |
| KLK1 | R/Y↓S/R [20] | Influenza virus [21] |
| KLK5 | X(aliphatic/aromatic)-R/K -X(polar/aliphatic)- R↓[18] | Influenza virus [21] |
| Neutrophil Elastase | A/V/I/T↓ [22] | Mammalian Orthoreovirus [23], SARS-CoV-1 [24,25] |
| PC5/6 | R-X-R/K-R↓ [26] | Influenza virus [27], RSV [27] |
| Plasmin | R/L↓ [28], preference for aromatic hydrophobic residue at P2 [29] | Influenza virus [30], RSV[31], SARS-CoV-1 [32], SARS-CoV-2 [33] |
| Thrombin | L-X-P-R↓S/A/G/T-X(aromatic)-R [34] | RSV [31], SARS-CoV-2 [9] |
| Trypsin | K/R↓[22] | SARS-CoV-1 [35,36], HMPV [37], RSV [31], HPIV [38] |
| Cathepsin G | Preference for F, Y, W, or L at P1 [39], S at P6, negatively charged amino acid in P2’ position | Mammalian Orthoreovirus [6], RSV [40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





