Submitted:
13 May 2024
Posted:
14 May 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Synthetic Microcosms
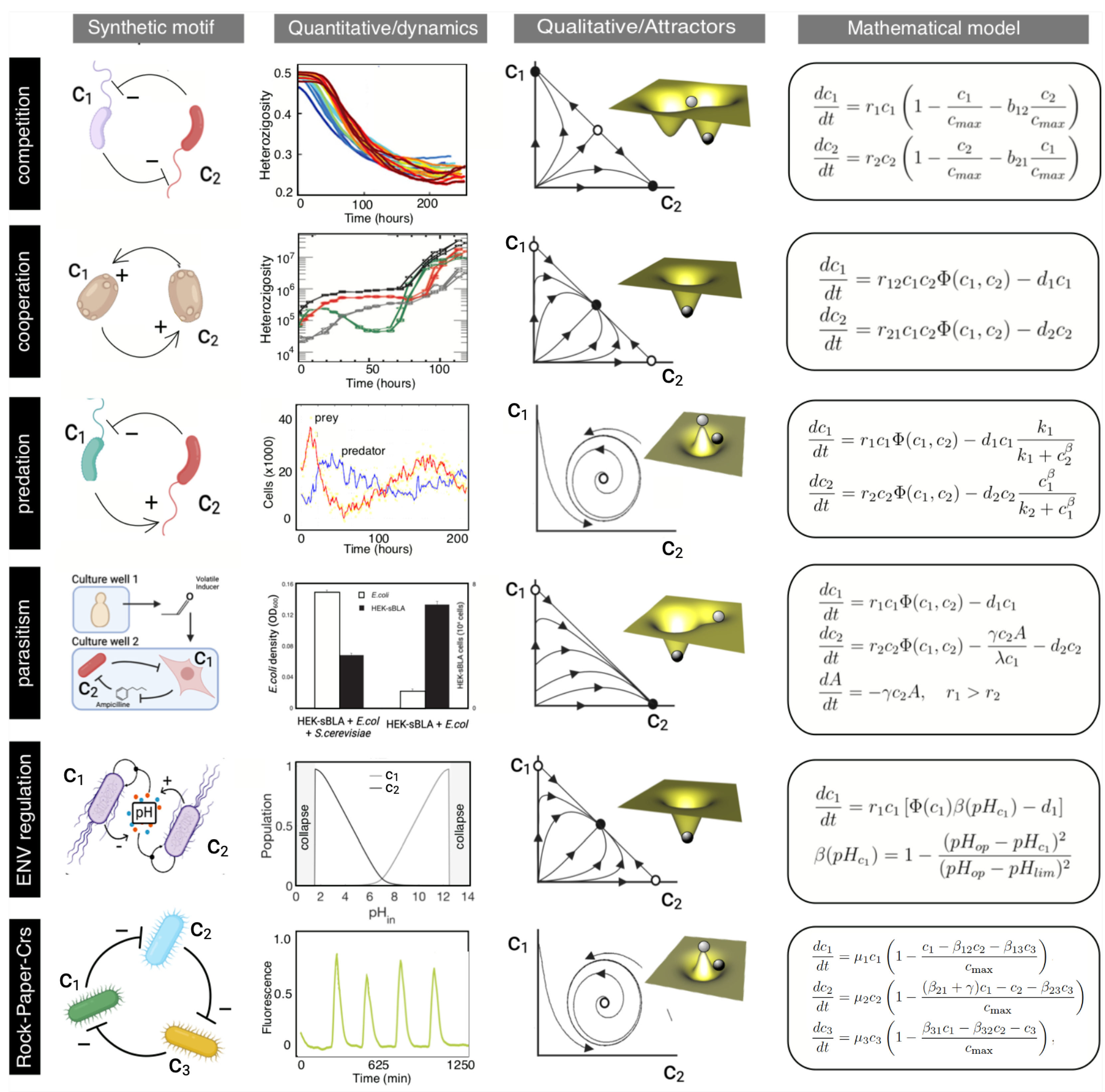
2.1. Competition
2.2. Cooperation
2.3. Parasitism and Predator-Prey Interactions
2.4. Environmental Self-Regulation
2.5. Non-Transitive Interactions
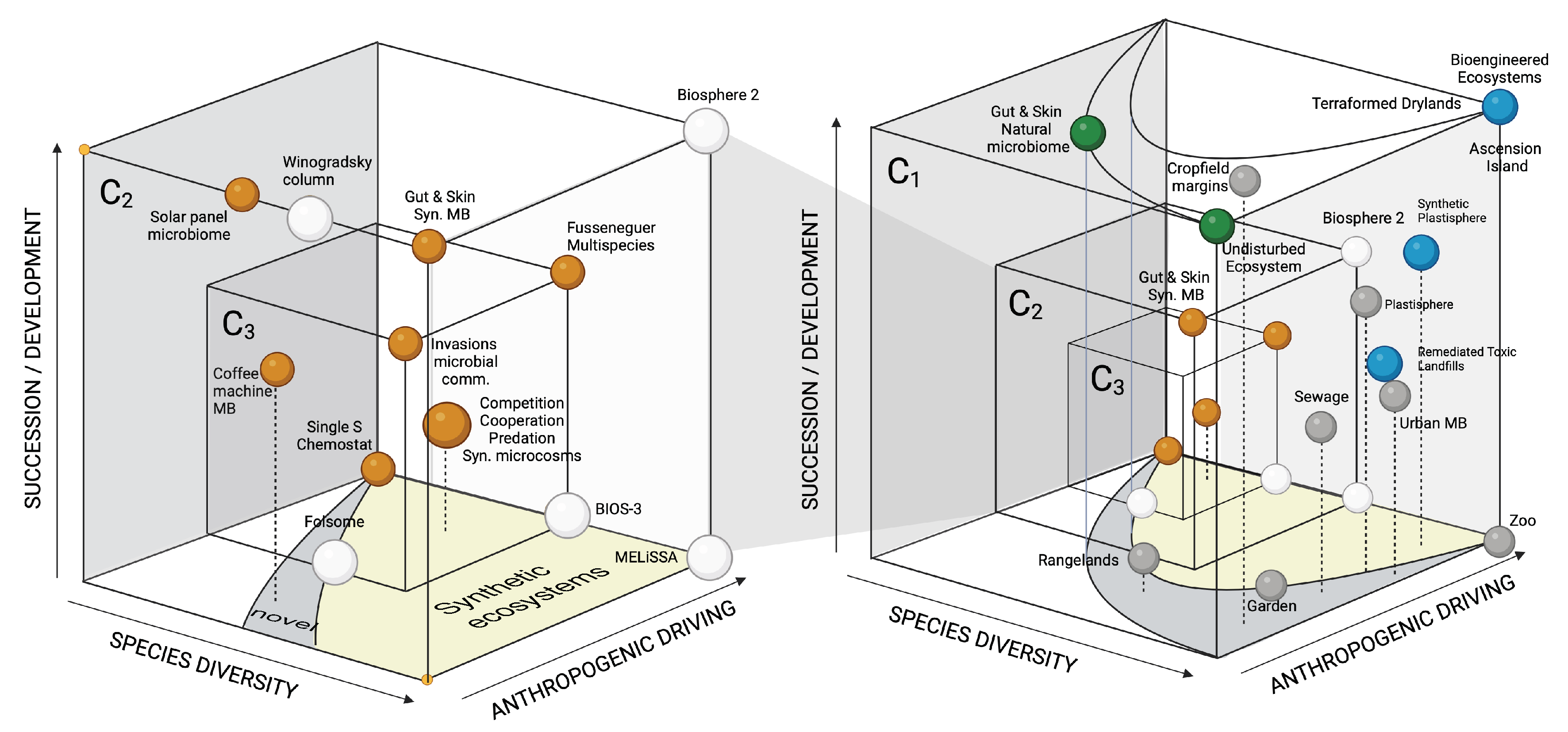
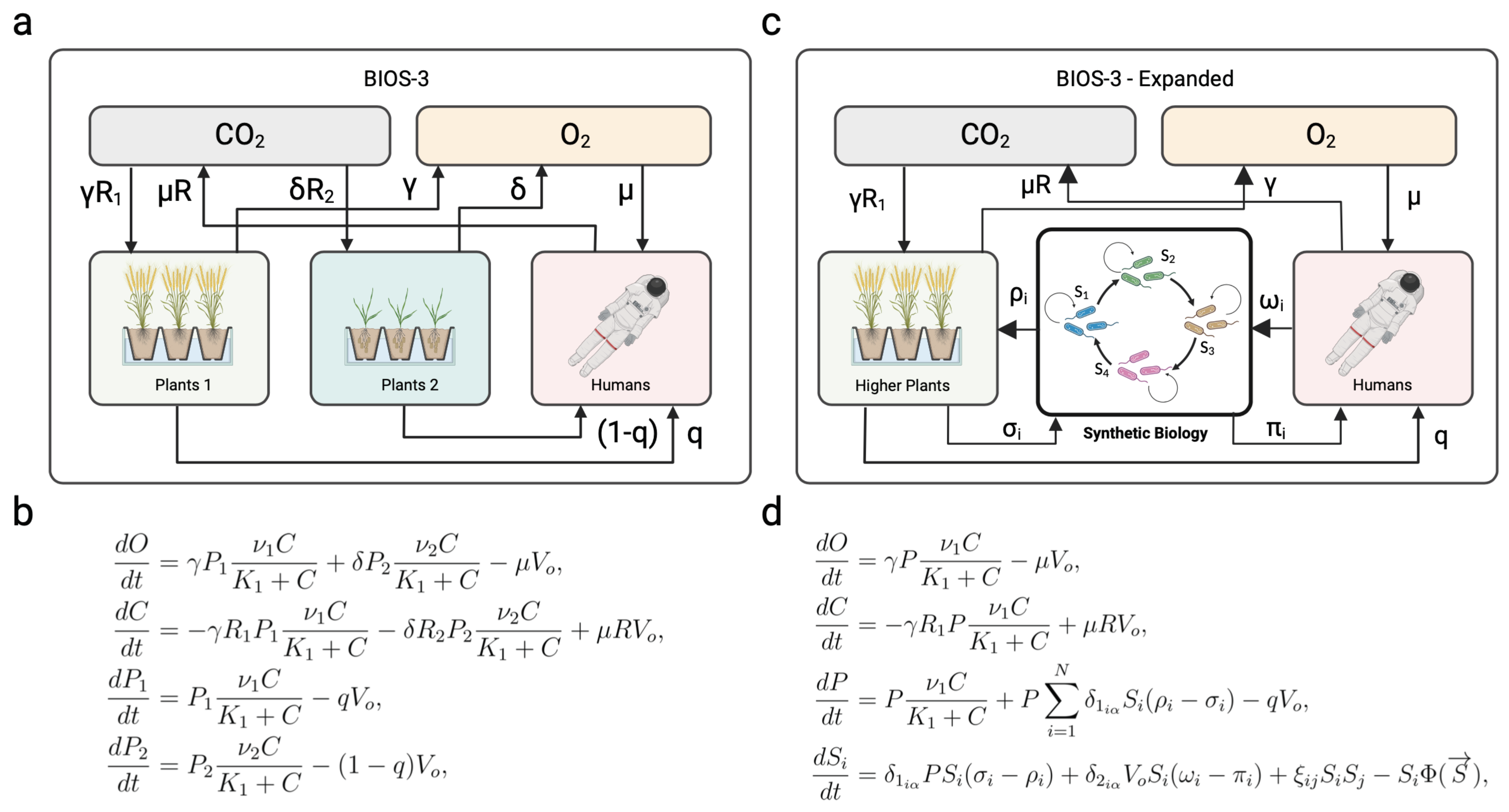
3. Closed Ecospheres and Space-Life Support Systems
|
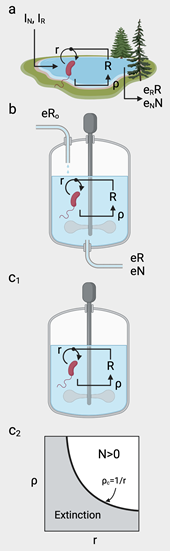
BOX 1. Critical thresholds for synthetic closed ecosystems The simplest model approach to closed ecosystems includes a population of size N of a given species that exploits some limited (inorganic) resource R. In the open scenario (Figure a), the equations read: Mathematically, a closed ecosystem is a special case of the previous model, where the dilution rate is nonexistent (). Now (Figure c1), the system only depends on the initial condition, the intrinsic growth dynamics of the species and the recycling rate of resource generation. Additionally, closure imposes a maximum carrying capacity, defined by logistic growth1. The equations read: |
 |
4. Synthetic Biospheres
| BOX 2. Emergent bioengineering: systems biology of ecosystem terraformation |
| What can be expected if we inoculate a given synthetic strain in a community? Mathematical models help predict the outcome of this intervention and the role played by biodiversity. One general scenario involves a set of species , namely where , and a set of resources , where . The growth rates for each resource and species are represented by and , respectively [138]. One is a strain () engineered from a resident species, thus sharing all the ecological interactions with the wild type. This synthetic strain can reduce the loss of a given, shared resource. The equations read: |
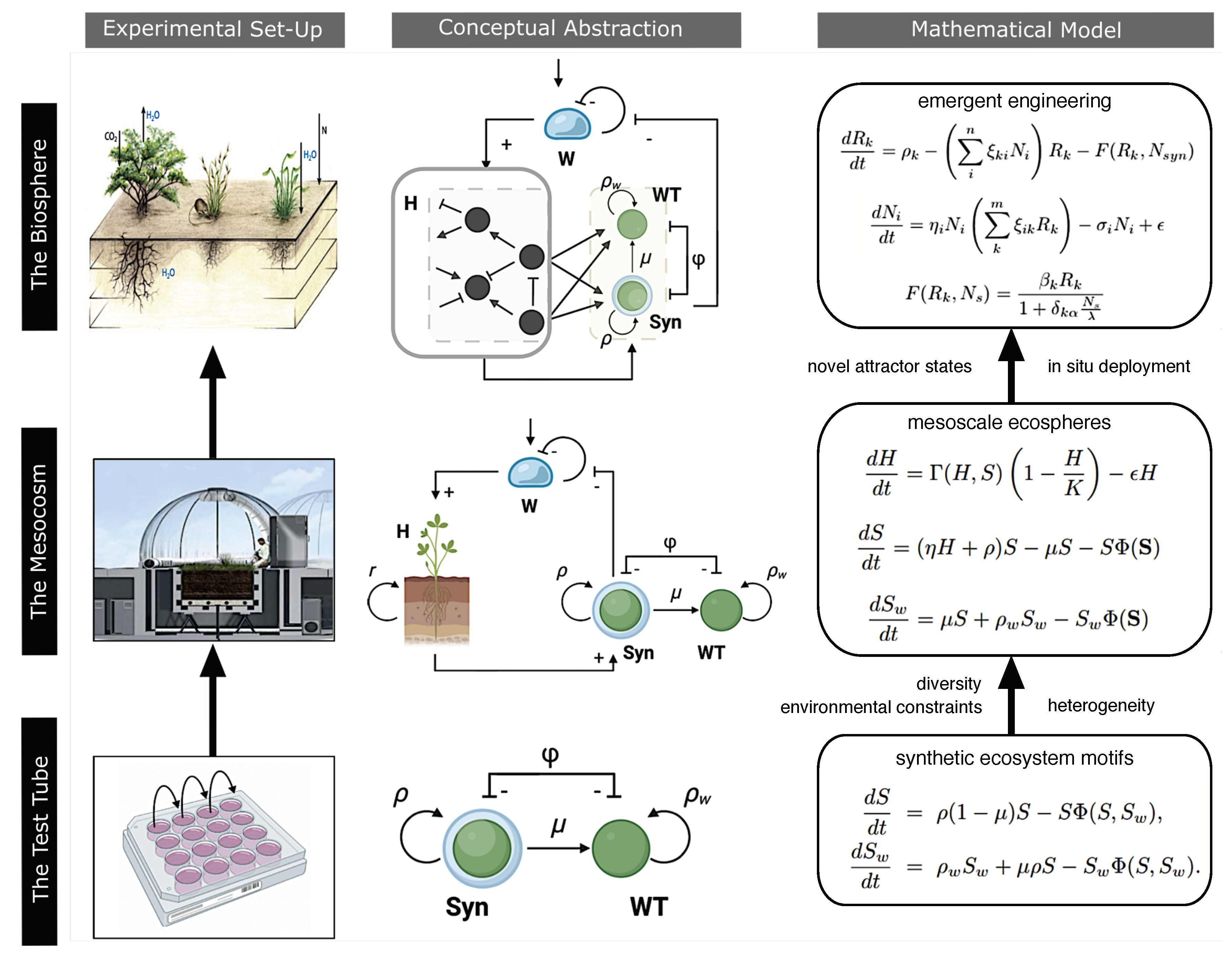
5. Discussion
- Although the microbial consortia described above follow the logic of ecological interactions in nature, this constraint does not necessarily limit the potential design principles, particularly when engineering computations with living cells [154]. In particular, it has been shown that complex functionalities can be built on engineered consortia that do not follow either biological or standard engineering designs [155]. What are the corresponding motifs and the alternative designs for ecological engineering?
- Who needs to be targeted when modifying ecospheres or whole communities in the wild? If we keep the single-species scenario described above, ecosystem engineers are a clear choice. However, considering single-species changes is one particular scenario within engineering complex communities, which can be obtained using either synthetic biology [87,156] or directed evolution [157]. Importantly, despite the success of microbiome transplants (which would be the non-engineered counterpart of our proposal) little is known in terms of theoretical models about how these transplants actually work. Developing synthetic models to address this problem would benefit both biomedical research and new ecosystem bioengineering strategies.
-
Detailed quantitative models of ecological communities require estimates about the sign and weight of species-species interactions. This is a challenging problem that has received much attention and mathematical models involving several (n) species can be easily defined using generalised Lotka-Volterra equations, namely:[158,159] This is the standard model used in most quantitative analyses of microbiome dynamics, particularly the gut microbiome [160,161,162,163,164]. The challenge here is to deal with appropriate estimates of the matrix that captures the strength of competitive interactions [165]. Of particular importance if to determine the resilience of these communities and their responses to engineering them. Recent work on community transitions [166] provides a useful estimate of these properties (based on an entropy measure) that could be used as a quantitative approach to determine the likelihood of microbiome shifts.
- Engineering efforts affecting extant communities, such as drylands, present an important challenge in terms of the strategies to be followed given the interactions between scales. The microbiome affects soil carbon content, which influences plant cover and the quality of the plant community influences back soil properties. On the other hand, we are considering changes that can percolate across whole landscapes, which can display several sources of heterogeneity. The simple scheme summarized in table 2 might require an extended approach that considers all these sources of variability. In this context, future efforts might benefit from machine learning techniques that can provide a system-level integration of data-driven information about responses across scales. Future models of terraformation should address this multiscale picture.
- In our previous examples, we assumed that synthetic biology has microbial candidates as potential chassis for engineered designs, and the population dynamics occur at the bacterial-resource scale. Viruses, on the other hand, and despite their tiny contribution to biomass, are acknowledged as crucial players in the dynamics of microbiomes, and they are largely understudied [167]. The impact of viruses and their potential role as vectors for ecosystem engineering should be explored using mathematical models that introduce the virome in an explicit manner [168].
- Under an evolutionary perspective, the nature of innovations (or major evolutionary transitions, see [169]) is one particularly relevant one. These transitions include for example the origins of the first cells and early forms of multicellularity. It has been proposed that an approach to these questions can be grounded in considering "Synthetic transitions" [170] where potential paths to innovation could be recreated using synthetic biology, artificial life or evolutionary robotics. Along with the standard list of MET, it has been suggested that synthetic ecosystems can shed light into a variety of open problems regarding the evolution of complexity beyond the species level [171]. Here too, ongoing efforts to build and augment in vitro synthetic interaction networks [172,173,174], along with new population dynamics models could help define scenarios of ecosystem transitions and the role played by contingency versus robust network properties on the tempo and mode of microbiome evolution.
Acknowledgements
References
- Lotka, A.J. Elements of mathematical biology; Dover Publications: Mineola, NY, 1956. [Google Scholar]
- Bacaër, N. Histoires de mathématiques et de populations; Cassini, 2008. [Google Scholar]
- Vandermeer, J.H.; Goldberg, D.E. Population Ecology, 2 ed.; Princeton University Press: Princeton, NJ, 2013. [Google Scholar]
- Weitz, J.S. Quantitative Biosciences: Dynamics Across Cells, Organisms, and Populations; Princeton University Press, 2024. [Google Scholar]
- Sole, R.V.; Bascompte, J. Self-organization in complex ecosystems.; Monographs in Population Biology, Princeton University Press: Princeton, NJ, 2006. [Google Scholar]
- Botsford, L.W.; Wilson White, J.; Hastings, A. Population dynamics for conservation; Oxford University Press: London, England, 2019. [Google Scholar]
- Anderson, R.M.; May, R.M. Infectious diseases of humans; Oxford science publications, Oxford University Press: London, England, 1991. [Google Scholar]
- Lockwood, J.L.; Hoopes, M.F.; Marchetti, M.P. Invasion Ecology, 2 ed.; Wiley-Blackwell: Chichester, England, 2013. [Google Scholar]
- Odum, E.P.; Barrett, G.W.; others. Fundamentals of ecology, Vol. 3, Saunders Philadelphia, 1971.
- Pimm, S.L. The balance of nature?: ecological issues in the conservation of species and communities; University of Chicago Press, 1991. [Google Scholar]
- May, R.M. Stability and complexity in model ecosystems; Princeton Landmarks in Biology, Princeton University Press: Princeton, NJ, 2001. [Google Scholar]
- Beyers, R.J.; Odum, H.T. Ecological Microcosms, 1993 ed.; Springer Advanced Texts in Life Sciences, Springer: New York, NY, 1993. [Google Scholar]
- De Roy, K.; Marzorati, M.; Van den Abbeele, P.; Van de Wiele, T.; Boon, N. Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities. Environmental microbiology 2014, 16, 1472–1481. [Google Scholar] [CrossRef]
- Friedman, J.; Gore, J. Ecological systems biology: The dynamics of interacting populations. Current Opinion in Systems Biology 2017, 1, 114–121. [Google Scholar] [CrossRef]
- Goldford, J.E.; Lu, N.; Bajić, D.; Estrela, S.; Tikhonov, M.; Sanchez-Gorostiaga, A.; Segrè, D.; Mehta, P.; Sanchez, A. Emergent simplicity in microbial community assembly. Science 2018, 361, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Amor, D.R.; Barbier, M.; Bunin, G.; Gore, J. Emergent phases of ecological diversity and dynamics mapped in microcosms. Science 2022, 378, 85–89. [Google Scholar] [CrossRef] [PubMed]
- Skwara, A.; Gowda, K.; Yousef, M.; Diaz-Colunga, J.; Raman, A.S.; Sanchez, A.; Tikhonov, M.; Kuehn, S. Statistically learning the functional landscape of microbial communities. Nature Ecology & Evolution 2023, 7, 1823–1833. [Google Scholar]
- Arya, S.; George, A.B.; O’Dwyer, J.P. Sparsity of higher-order landscape interactions enables learning and prediction for microbiomes. Proceedings of the National Academy of Sciences 2023, 120, e2307313120. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, W.R.; Grilli, J. Investigating macroecological patterns in coarse-grained microbial communities using the stochastic logistic model of growth. bioRxiv 2023, 2023–03. [Google Scholar]
- Odum, E.P. The Mesocosm. BioScience 1984, 34, 558–562. [Google Scholar] [CrossRef]
- Nelson, M.; Burgess, T.L.; Alling, A.; Alvarez-Romo, N.; Dempster, W.F.; Walford, R.L.; Allen, J.P. Using a closed ecological system to study Earth’s biosphere: Initial results from Biosphere 2. Bioscience 1993, 43, 225–236. [Google Scholar] [CrossRef]
- Cohen, J.E.; Tilman, D. Biosphere 2 and Biodiversity–The Lessons So Far. Science 1996, 274, 1150–1151. [Google Scholar] [CrossRef]
- Levin, S.A. Ecosystems and the Biosphere as Complex Adaptive Systems. Ecosystems 1998, 1, 431–436. [Google Scholar] [CrossRef]
- Levin, S.A. Multiple Scales and the Maintenance of Biodiversity. Ecosystems 2000, 3, 498–506. [Google Scholar] [CrossRef]
- Widder, S.; Allen, R.J.; Pfeiffer, T.; Curtis, T.P.; Wiuf, C.; Sloan, W.T.; Cordero, O.X.; Brown, S.P.; Momeni, B.; Shou, W.; others. Challenges in microbial ecology: building predictive understanding of community function and dynamics. The ISME journal 2016, 10, 2557–2568. [Google Scholar] [CrossRef]
- Wilkinson, D.M. The parable of Green Mountain: Ascension Island, ecosystem construction and ecological fitting, 2004.
- Amaral-Zettler, L.A.; Zettler, E.R.; Mincer, T.J. Ecology of the plastisphere. Nature Reviews Microbiology 2020, 18, 139–151. [Google Scholar] [CrossRef]
- Schwartzkopf, S.H. Design of a controlled ecological life support system. Bioscience 1992, 42, 526–535. [Google Scholar] [CrossRef]
- Escalante, A.E.; Rebolleda-Gómez, M.; Benítez, M.; Travisano, M. Ecological perspectives on synthetic biology: insights from microbial population biology. Frontiers in microbiology 2015, 6, 126260. [Google Scholar] [CrossRef]
- Zomorrodi, A.R.; Segrè, D. Synthetic ecology of microbes: mathematical models and applications. Journal of molecular biology 2016, 428, 837–861. [Google Scholar] [CrossRef]
- Müller, M.; Neugeboren, B.; Nelson, D.; Murray, A. Genetic drift opposes mutualism during spatial population expansion. Proc Natl Acad Sci 2014, 111, 1037–42. [Google Scholar] [CrossRef]
- Amor, D.R.; Montañez, R.; Duran-Nebreda, S.; Solé, R. Spatial dynamics of synthetic microbial mutualists and their parasites. PLOS Computational Biology 2017, 13, e1005689. [Google Scholar] [CrossRef]
- Knödlseder, N.; Fábrega, M.J.; Santos-Moreno, J.; Manils, J.; Toloza, L.; Marín Vilar, M.; Fernández, C.; Broadbent, K.; Maruotti, J.; Lemenager, H.; others. Delivery of a sebum modulator by an engineered skin microbe in mice. Nature Biotechnology 2024, 1–6. [Google Scholar] [CrossRef]
- de Lorenzo, V. Systems biology approaches to bioremediation. Current opinion in biotechnology 2008, 19, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; de Lorenzo, V. Synthetic constructs in/for the environment: managing the interplay between natural and engineered biology. FEBS letters 2012, 586, 2199–2206. [Google Scholar] [CrossRef] [PubMed]
- de Lorenzo, V. Environmental Galenics: large-scale fortification of extant microbiomes with engineered bioremediation agents. Philosophical Transactions of the Royal Society B 2022, 377, 20210395. [Google Scholar] [CrossRef] [PubMed]
- Solé, R. Bioengineering the biosphere? Ecological Complexity 2015, 22, 40–49. [Google Scholar] [CrossRef]
- Solé, R.V.; Montañez, R.; Duran-Nebreda, S. Synthetic circuit designs for earth terraformation. Biology Direct 2015, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- de Lorenzo, V.; Marliere, P.; Sole, R. Bioremediation at a global scale: from the test tube to planet Earth. Microbial biotechnology 2016, 9, 618–625. [Google Scholar] [CrossRef] [PubMed]
- Conde-Pueyo, N.; Vidiella, B.; Sardanyés, J.; Berdugo, M.; Maestre, F.T.; De Lorenzo, V.; Solé, R. Synthetic biology for terraformation lessons from mars, earth, and the microbiome. life 2020, 10, 14. [Google Scholar] [CrossRef] [PubMed]
- Lotka, A.J. The growth of mixed populations: two species competing for a common food supply. Journal of the Washington Academy of Sciences 1932, 22, 461–469. [Google Scholar]
- Volterra, V. Fluctuations in the Abundance of a Species considered Mathematically. Nature 1926, 118, 558–560. [Google Scholar] [CrossRef]
- Gause, G.F. Experimental Studies on the Struggle for Existence: I. Mixed Population of Two Species of Yeast. The Journal of Experimental Biology 1932, 9, 389–402. [Google Scholar] [CrossRef]
- Solé, R.V.; Bascompte, J.; Valls, J. Stability and complexity of spatially extended two-species competition. Journal of theoretical Biology 1992, 159, 469–480. [Google Scholar] [CrossRef]
- Mitri, S.; Clarke, E.; Foster, K.R. Resource limitation drives spatial organization in microbial groups. The ISME journal 2016, 10, 1471–1482. [Google Scholar] [CrossRef] [PubMed]
- Eigen, M.; Schuster, P. The Hypercycle; Springer: Berlin, Germany, 1979. [Google Scholar]
- Sardanyés, J.; Solé, R.V. Bifurcations and phase transitions in spatially extended two-member hypercycles. Journal of Theoretical Biology 2006, 243, 468–482. [Google Scholar] [CrossRef] [PubMed]
- Shou, W.; Ram, S.; Vilar, J.M. Synthetic cooperation in engineered yeast populations. Proceedings of the National Academy of Sciences 2007, 104, 1877–1882. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, K.; Suzuki, S.; Yamauchi, Y.; Shiroguchi, Y.; Kashiwagi, A.; Ono, N.; Mori, K.; Yomo, T. Cooperative Adaptation to Establishment of a Synthetic Bacterial Mutualism. PLoS ONE 2011, 6, e17105. [Google Scholar] [CrossRef] [PubMed]
- Momeni, B.; Waite, A.J.; Shou, W. Spatial self-organization favors heterotypic cooperation over cheating. Elife 2013, 2, e00960. [Google Scholar] [CrossRef] [PubMed]
- Szathmáry, E. The origin of replicators and reproducers. Philosophical Transactions of the Royal Society B: Biological Sciences 2006, 361, 1761–1776. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Hypercycles and the origin of life. Nature 1979, 280, 541–541. [Google Scholar] [CrossRef]
- Boerlijst, M.C.; Hogeweg, P. Spiral wave structure in pre-biotic evolution: hypercycles stable against parasites. Physica D: Nonlinear Phenomena 1991, 48, 17–28. [Google Scholar] [CrossRef]
- Pande, S.; Kaftan, F.; Lang, S.; Svatoš, A.; Germerodt, S.; Kost, C. Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. The ISME Journal 2016, 10, 1413–1423. [Google Scholar] [CrossRef]
- Weber, W.; Daoud-El Baba, M.; Fussenegger, M. Synthetic ecosystems based on airborne inter- and intrakingdom communication. Proceedings of the National Academy of Sciences 2007, 104, 10435–10440. [Google Scholar] [CrossRef] [PubMed]
- Lotka, A.J. Contribution to the Theory of Periodic Reactions. The Journal of Physical Chemistry 1910, 14, 271–274. [Google Scholar] [CrossRef]
- Martin, M.O. Predatory prokaryotes: an emerging research opportunity. Journal of Molecular Microbiology and Biotechnology 2002, 4, 467–478. [Google Scholar] [PubMed]
- Pérez, J.; Moraleda-Muñoz, A.; Marcos-Torres, F.J.; Muõz Dorado, J. Bacterial predation: 75 years and counting! Environmental Microbiology 2016, 18, 766–779. [Google Scholar] [CrossRef] [PubMed]
- Balagaddé, F.K.; Song, H.; Ozaki, J.; Collins, C.H.; Barnet, M.; Arnold, F.H.; Quake, S.R.; You, L. A synthetic Escherichia coli predator-prey ecosystem. Molecular Systems Biology 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, M.; Diggle, S.P.; Greenberg, E.P. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Barabás, G.; Michalska-Smith, M.J.; Allesina, S. Self-regulation and the stability of large ecological networks. Nature Ecology & Evolution 2017, 1, 1870–1875. [Google Scholar] [CrossRef] [PubMed]
- Konopka, A.; Lindemann, S.; Fredrickson, J. Dynamics in microbial communities: unraveling mechanisms to identify principles. The ISME Journal 2014, 9, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Ranheim Sveen, T.; Hannula, S.; Bahram, M. Microbial regulation of feedbacks to ecosystem change. Trends in Microbiology 2024, 32, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.J.; Lovelock, J.E. Biological homeostasis of the global environment: the parable of Daisyworld. Tellus B: Chemical and Physical Meteorology 1983, 35, 284. [Google Scholar] [CrossRef]
- Maull, V.; Pla Mauri, J.; Conde Pueyo, N.; Solé, R. A synthetic microbial Daisyworld: planetary regulation in the test tube. Journal of The Royal Society Interface 2024, 21. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, R.; Hamilton, W.D. The Evolution of Cooperation. Science 1981, 211, 1390–1396. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M.; Price, G.R. The Logic of Animal Conflict. Nature 1973, 246, 15–18. [Google Scholar] [CrossRef]
- Frank, S.A. Models of Parasite Virulence. The Quarterly Review of Biology 1996, 71, 37–78. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, T.; Schuster, S. Game-theoretical approaches to studying the evolution of biochemical systems. Trends in Biochemical Sciences 2005, 30, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Riley, M.A.; Wertz, J.E. Bacteriocins: Evolution, Ecology, and Application. Annual Review of Microbiology 2002, 56, 117–137. [Google Scholar] [CrossRef]
- Granato, E.T.; Meiller-Legrand, T.A.; Foster, K.R. The Evolution and Ecology of Bacterial Warfare. Current Biology 2019, 29, R521–R537. [Google Scholar] [CrossRef] [PubMed]
- Kerr, B.; Riley, M.A.; Feldman, M.W.; Bohannan, B.J.M. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 2002, 418, 171–174. [Google Scholar] [CrossRef] [PubMed]
- Neumann, G.; Schuster, S. Continuous model for the rock-scissors-paper game between bacteriocin producing bacteria. Journal of Mathematical Biology 2007, 54, 815–846. [Google Scholar] [CrossRef]
- van den Berg, N.I.; Machado, D.; Santos, S.; Rocha, I.; Chacón, J.; Harcombe, W.; Mitri, S.; Patil, K.R. Ecological modelling approaches for predicting emergent properties in microbial communities. Nature Ecology & Evolution 2022, 6, 855–865. [Google Scholar] [CrossRef]
- Shetty, S.A.; Smidt, H.; de Vos, W.M. Reconstructing functional networks in the human intestinal tract using synthetic microbiomes. Current Opinion in Biotechnology 2019, 58, 146–154. [Google Scholar] [CrossRef]
- Bartsev, S.; Okhonin, V. Potentialities of theoretical and experimental prediction of life support systems reliability. Advances in Space Research 1999, 24, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Menezes, A.A.; Cumbers, J.; Hogan, J.A.; Arkin, A.P. Towards synthetic biological approaches to resource utilization on space missions. Journal of the Royal Society interface 2015, 12, 20140715. [Google Scholar] [CrossRef] [PubMed]
- Santomartino, R.; Averesch, N.J.; Bhuiyan, M.; Cockell, C.S.; Colangelo, J.; Gumulya, Y.; Lehner, B.; Lopez-Ayala, I.; McMahon, S.; Mohanty, A.; others. Toward sustainable space exploration: a roadmap for harnessing the power of microorganisms. Nature communications 2023, 14, 1391. [Google Scholar] [CrossRef]
- Foster, K.R.; Schluter, J.; Coyte, K.Z.; Rakoff-Nahoum, S. The evolution of the host microbiome as an ecosystem on a leash. Nature 2017, 548, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Folsome, C.E.; Hanson, J.A. The emergence of materially-closed-system ecology. Ecosystem Theory and Application, 1986; 269–299. [Google Scholar]
- Ulanowicz, R.E. Identifying the structure of cycling in ecosystems. Mathematical Biosciences 1983, 65, 219–237. [Google Scholar] [CrossRef]
- Loreau, M. Material cycling and the stability of ecosystems. The American Naturalist 1994, 143, 508–513. [Google Scholar] [CrossRef]
- Rillig, M.C.; Antonovics, J. Microbial biospherics: The experimental study of ecosystem function and evolution. Proceedings of the National Academy of Sciences 2019, 116, 11093–11098. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, W.R.; Jones, S.E.; Muscarella, M.E.; Behringer, M.G.; Lehmkuhl, B.K.; Lennon, J.T. Microbial population dynamics and evolutionary outcomes under extreme energy limitation. Proceedings of the National Academy of Sciences 2021, 118, e2101691118. [Google Scholar] [CrossRef]
- de Jesús Astacio, L.M.; Prabhakara, K.H.; Li, Z.; Mickalide, H.; Kuehn, S. Closed microbial communities self-organize to persistently cycle carbon. Proceedings of the National Academy of Sciences 2021, 118, e2013564118. [Google Scholar] [CrossRef]
- Goyal, A.; Flamholz, A.I.; Petroff, A.P.; Murugan, A. Closed ecosystems extract energy through self-organized nutrient cycles. Proceedings of the National Academy of Sciences 2023, 120, e2309387120. [Google Scholar] [CrossRef] [PubMed]
- McCarty, N.S.; Ledesma-Amaro, R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends in biotechnology 2019, 37, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Bajic, D.; Diaz-Colunga, J.; Skwara, A.; Vila, J.C.; Kuehn, S. The community-function landscape of microbial consortia. Cell Systems 2022, 14, 122–134. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Colunga, J.; Catalan, P.; San Roman, M.; Arrabal, A.; Sanchez, A. Full factorial construction of synthetic microbial communities. bioRxiv 2024, 2024–05. [Google Scholar]
- Ben Said, S.; Or, D. Synthetic microbial ecology: engineering habitats for modular consortia. Frontiers in microbiology 2017, 8, 252185. [Google Scholar] [CrossRef] [PubMed]
- Menezes, A.A.; Montague, M.G.; Cumbers, J.; Hogan, J.A.; Arkin, A.P. Grand challenges in space synthetic biology. Journal of The Royal Society Interface 2015, 12, 20150803. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, I.; Terskov, I.; Kovrov, B.; Lisovskii, G.; Okladnikov, Y.N.; Sid’Ko, F.Y.; Trubachev, I.; Shilenko, M.; Alekseev, S.; Pan’kova, I.; others. Long-term experiments on man’s stay in biological life-support system. Advances in Space Research 1989, 9, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, J.I.; Lisovsky, G.M.; MacElroy, R.D. Man-Made Closed Ecological Systems; Earth space institute book series; CRC Press: London, England, 2002. [Google Scholar]
- Fulget, N.; Poughon, L.; Richalet, J.; Lasseur, C. MELISSA: global control strategy of the artificial ecosystem by using first principles models of the compartments. Advances in Space Research 1999, 24, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Lasseur, C.; Brunet, J.; De Weever, H.; Dixon, M.; Dussap, G.; Godia, F.; Leys, N.; Mergeay, M.; Van Der Straeten, D. MELiSSA: the European project of closed life support system. Gravitational and Space Research 2010, 23. [Google Scholar]
- Ciurans, C.; Bazmohammadi, N.; Poughon, L.; Vasquez, J.C.; Dussap, C.G.; Gòdia, F.; Guerrero, J.M. Hierarchically controlled ecological life support systems. Computers & Chemical Engineering 2022, 157, 107625. [Google Scholar]
- Bartsev, S. Stoichiometric constraints and complete closure of long-term life support systems. Advances in Space Research 2004, 34, 1509–1516. [Google Scholar] [CrossRef]
- Eigen, M.; Schuster, P. The hypercycle: a principle of natural self-organization, Springer Science & Business Media, 2012.
- Conde-Pueyo, N.; Vidiella, B.; Sardanyés, J.; Berdugo, M.; Maestre, F.T.; De Lorenzo, V.; Solé, R. Synthetic biology for terraformation lessons from mars, earth, and the microbiome. life 2020, 10, 14. [Google Scholar] [CrossRef]
- Mee, M.T.; Collins, J.J.; Church, G.M.; Wang, H.H. Syntrophic exchange in synthetic microbial communities. Proceedings of the National Academy of Sciences 2014, 111, E2149–E2156. [Google Scholar] [CrossRef] [PubMed]
- Mapstone, L.J.; Leite, M.N.; Purton, S.; Crawford, I.A.; Dartnell, L. Cyanobacteria and microalgae in supporting human habitation on Mars. Biotechnology Advances 2022, 59, 107946. [Google Scholar] [CrossRef] [PubMed]
- Billi, D.; Baqué, M.; Smith, H.; McKay, C.; others. Cyanobacteria from extreme deserts to space. Advances in Microbiology 2013, 3, 80–86. [Google Scholar] [CrossRef]
- Billi, D. Desert cyanobacteria under space and planetary simulations: a tool for searching for life beyond Earth and supporting human space exploration. International Journal of Astrobiology 2019, 18, 483–489. [Google Scholar] [CrossRef]
- Verseux, C.; Baqué, M.; Lehto, K.; de Vera, J.P.P.; Rothschild, L.J.; Billi, D. Sustainable life support on Mars–the potential roles of cyanobacteria. International Journal of Astrobiology 2016, 15, 65–92. [Google Scholar] [CrossRef]
- Holling, C.S. Resilience and stability of ecological systems. Annual review of ecology and systematics 1973, 4, 1–23. [Google Scholar] [CrossRef]
- May, R.M. Thresholds and breakpoints in ecosystems with a multiplicity of stable states. Nature 1977, 269, 471–477. [Google Scholar] [CrossRef]
- Scheffer, M.; Carpenter, S.; Foley, J.A.; Folke, C.; Walker, B. Catastrophic shifts in ecosystems. Nature 2001, 413, 591–596. [Google Scholar] [CrossRef]
- Scheffer, M. Critical transitions in nature and society; Vol. 16, Princeton University Press, 2020.
- Rockström, J.; Steffen, W.; Noone, K.; Persson, Å.; Chapin III, F.S.; Lambin, E.; Lenton, T.M.; Scheffer, M.; Folke, C.; Schellnhuber, H.J.; others. Planetary boundaries: exploring the safe operating space for humanity. Ecology and society 2009, 14. [Google Scholar] [CrossRef]
- Turner, M.G.; Calder, W.J.; Cumming, G.S.; Hughes, T.P.; Jentsch, A.; LaDeau, S.L.; Lenton, T.M.; Shuman, B.N.; Turetsky, M.R.; Ratajczak, Z.; others. Climate change, ecosystems and abrupt change: science priorities. Philosophical Transactions of the Royal Society B 2020, 375, 20190105. [Google Scholar] [CrossRef] [PubMed]
- Solé, R.; Levin, S. Ecological complexity and the biosphere: the next 30 years, 2022.
- Jansson, J.K.; McClure, R.; Egbert, R.G. Soil microbiome engineering for sustainability in a changing environment. Nature Biotechnology 2023, 41, 1716–1728. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Ripple, W.J.; Timmis, K.N.; Azam, F.; Bakken, L.R.; Baylis, M.; Behrenfeld, M.J.; Boetius, A.; Boyd, P.W.; Classen, A.T.; others. Scientists’ warning to humanity: microorganisms and climate change. Nature Reviews Microbiology 2019, 17, 569–586. [Google Scholar] [CrossRef] [PubMed]
- Maestre, F.T.; Sole, R.; Singh, B.K. Microbial biotechnology as a tool to restore degraded drylands. Microbial biotechnology 2017, 10, 1250–1253. [Google Scholar] [CrossRef] [PubMed]
- Vidiella, B.; Sardanyés, J.; Solé, R.V. Synthetic soil crusts against green-desert transitions: a spatial model. Royal Society open science 2020, 7, 200161. [Google Scholar] [CrossRef] [PubMed]
- DeLisi, C. The role of synthetic biology in climate change mitigation. Biology Direct 2019, 14, 1–5. [Google Scholar]
- Goodchild-Michelman, I.M.; Church, G.M.; Schubert, M.G.; Tang, T.C. Light and carbon: Synthetic biology toward new cyanobacteria-based living biomaterials. Materials Today Bio 2023, 19, 100583. [Google Scholar] [CrossRef] [PubMed]
- Piaggio, A.J.; Segelbacher, G.; Seddon, P.J.; Alphey, L.; Bennett, E.L.; Carlson, R.H.; Friedman, R.M.; Kanavy, D.; Phelan, R.; Redford, K.H.; others. Is it time for synthetic biodiversity conservation? Trends in ecology & evolution 2017, 32, 97–107. [Google Scholar]
- Maestre, F.T.; Eldridge, D.J.; Soliveres, S.; Kéfi, S.; Delgado-Baquerizo, M.; Bowker, M.A.; García-Palacios, P.; Gaitán, J.; Gallardo, A.; Lázaro, R.; others. Structure and functioning of dryland ecosystems in a changing world. Annual review of ecology, evolution, and systematics 2016, 47, 215–237. [Google Scholar] [CrossRef]
- Berdugo, M.; Delgado-Baquerizo, M.; Soliveres, S.; Hernández-Clemente, R.; Zhao, Y.; Gaitán, J.J.; Gross, N.; Saiz, H.; Maire, V.; Lehmann, A.; others. Global ecosystem thresholds driven by aridity. Science 2020, 367, 787–790. [Google Scholar] [CrossRef]
- Berdugo, M.; Vidiella, B.; Solé, R.V.; Maestre, F.T. Ecological mechanisms underlying aridity thresholds in global drylands. Functional Ecology 2022, 36, 4–23. [Google Scholar] [CrossRef]
- Pointing, S.B.; Belnap, J. Microbial colonization and controls in dryland systems. Nature Reviews Microbiology 2012, 10, 551–562. [Google Scholar] [CrossRef] [PubMed]
- Maestre, F.T.; Delgado-Baquerizo, M.; Jeffries, T.C.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Quero, J.L.; García-Gómez, M.; Gallardo, A.; Ulrich, W.; others. Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proceedings of the National Academy of Sciences 2015, 112, 15684–15689. [Google Scholar] [CrossRef] [PubMed]
- Coleine, C.; Delgado-Baquerizo, M.; DiRuggiero, J.; Guirado, E.; Harfouche, A.L.; Perez-Fernandez, C.; Singh, B.K.; Selbmann, L.; Egidi, E. Dryland microbiomes reveal community adaptations to desertification and climate change. The ISME Journal 2024, 18, wrae056. [Google Scholar] [CrossRef] [PubMed]
- Maestre, F.T.; Martín, N.; Díez, B.; Lopez-Poma, R.; Santos, F.; Luque, I.; Cortina, J. Watering, fertilization, and slurry inoculation promote recovery of biological crust function in degraded soils. Microbial Ecology 2006, 52, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Rojas, M.; Román, J.; Roncero-Ramos, B.; Erickson, T.; Merritt, D.; Aguila-Carricondo, P.; Cantón, Y. Cyanobacteria inoculation enhances carbon sequestration in soil substrates used in dryland restoration. Science of the Total Environment 2018, 636, 1149–1154. [Google Scholar] [CrossRef] [PubMed]
- Rossi, F.; Li, H.; Liu, Y.; De Philippis, R. Cyanobacterial inoculation (cyanobacterisation): Perspectives for the development of a standardized multifunctional technology for soil fertilization and desertification reversal. Earth-Science Reviews 2017, 171, 28–43. [Google Scholar] [CrossRef]
- Chamizo, S.; Mugnai, G.; Rossi, F.; Certini, G.; De Philippis, R. Cyanobacteria inoculation improves soil stability and fertility on different textured soils: gaining insights for applicability in soil restoration. Frontiers in Environmental Science 2018, 6, 49. [Google Scholar] [CrossRef]
- Kéfi, S.; Rietkerk, M.; van Baalen, M.; Loreau, M. Local facilitation, bistability and transitions in arid ecosystems. Theoretical population biology 2007, 71, 367–379. [Google Scholar] [CrossRef]
- Borgogno, F.; D’Odorico, P.; Laio, F.; Ridolfi, L. Mathematical models of vegetation pattern formation in ecohydrology. Reviews of geophysics 2009, 47. [Google Scholar] [CrossRef]
- Brooker, R.W.; Maestre, F.T.; Callaway, R.M.; Lortie, C.L.; Cavieres, L.A.; Kunstler, G.; Liancourt, P.; Tielbörger, K.; Travis, J.M.; Anthelme, F.; others. Facilitation in plant communities: the past, the present, and the future. Journal of ecology 2008, 18–34. [Google Scholar]
- Wright, O.; Stan, G.B.; Ellis, T. Building-in biosafety for synthetic biology. Microbiology 2013, 159, 1221–1235. [Google Scholar] [CrossRef] [PubMed]
- Lensch, A.; Lindfors, H.A.; Duwenig, E.; Fleischmann, T.; Hjort, C.; Kärenlampi, S.O.; McMurtry, L.; Melton, E.D.; Andersen, M.R.; Skinner, R.; others. Safety aspects of microorganisms deliberately released into the environment. EFB Bioeconomy Journal 2023, 100061. [Google Scholar]
- Maull, V.; Solé, R. Network-level containment of single-species bioengineering. Philosophical Transactions of the Royal Society B 2022, 377, 20210396. [Google Scholar] [CrossRef] [PubMed]
- Samson, F.B.; Knopf, F.L.; Jones, C.G.; Lawton, J.H.; Shachak, M. Organisms as ecosystem engineers. Ecosystem management: selected readings, 1996; 130–147. [Google Scholar]
- Jones, C.G.; Lawton, J.H.; Shachak, M. Positive and negative effects of organisms as physical ecosystem engineers. Ecology 1997, 78, 1946–1957. [Google Scholar] [CrossRef]
- Byers, J.E.; Cuddington, K.; Jones, C.G.; Talley, T.S.; Hastings, A.; Lambrinos, J.G.; Crooks, J.A.; Wilson, W.G. Using ecosystem engineers to restore ecological systems. Trends in ecology & evolution 2006, 21, 493–500. [Google Scholar]
- Maull, V.; Solé, R. Biodiversity as a Firewall to Engineered Microbiomes for Restoration and Conservation. Royal Society open science 2024. ?, ?. [Google Scholar]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial diversity drives multifunctionality in terrestrial ecosystems. Nature communications 2016, 7, 10541. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Singh, B.K.; Maestre, F.T. Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecology letters 2017, 20, 1295–1305. [Google Scholar] [CrossRef]
- Stirling, F.; Bitzan, L.; O’Keefe, S.; Redfield, E.; Oliver, J.W.; Way, J.; Silver, P.A. Rational design of evolutionarily stable microbial kill switches. Molecular cell 2017, 68, 686–697. [Google Scholar] [CrossRef]
- Rottinghaus, A.G.; Ferreiro, A.; Fishbein, S.R.; Dantas, G.; Moon, T.S. Genetically stable CRISPR-based kill switches for engineered microbes. Nature communications 2022, 13, 672. [Google Scholar] [CrossRef]
- Case, T.J. Invasion resistance arises in strongly interacting species-rich model competition communities. Proceedings of the National Academy of Sciences 1990, 87, 9610–9614. [Google Scholar] [CrossRef]
- van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottovā, D.; Krištůfek, V.; Salles, J.F.a. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proceedings of the National Academy of Sciences 2012, 109, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, R.J.; Higgs, E.; Harris, J.A. Novel ecosystems: implications for conservation and restoration. Trends in Ecology & Evolution 2009, 24, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Tamaru, Y.; Takani, Y.; Yoshida, T.; Sakamoto, T. Crucial role of extracellular polysaccharides in desiccation and freezing tolerance in the terrestrial cyanobacterium Nostoc commune. Applied and environmental microbiology 2005, 71, 7327–7333. [Google Scholar] [CrossRef] [PubMed]
- Mager, D. Extracellular polysaccharides from cyanobacterial soil crusts and their role in dryland surface processes. PhD thesis, University Manchester Metropolitan, 2008.
- Adessi, A.; de Carvalho, R.C.; De Philippis, R.; Branquinho, C.; da Silva, J.M. Microbial extracellular polymeric substances improve water retention in dryland biological soil crusts. Soil Biology and Biochemistry 2018, 116, 67–69. [Google Scholar] [CrossRef]
- Ollé-Vila, A.; Duran-Nebreda, S.; Conde-Pueyo, N.; Montañez, R.; Solé, R. A morphospace for synthetic organs and organoids: the possible and the actual. Integrative Biology 2016, 8, 485–503. [Google Scholar] [CrossRef]
- Avena-Koenigsberger, A.; Goñi, J.; Solé, R.; Sporns, O. Network morphospace. Journal of the Royal Society Interface 2015, 12, 20140881. [Google Scholar] [CrossRef] [PubMed]
- Solé, R.; Ollé-Vila, A.; Vidiella, B.; Duran-Nebreda, S.; Conde-Pueyo, N. The road to synthetic multicellularity. Current Opinion in Systems Biology 2018, 7, 60–67. [Google Scholar] [CrossRef]
- Hobbs, R.J.; Higgs, E.S.; Hall, C. Novel Ecosystems: Intervening in the New Ecological World Order; Wiley-Blackwell: Hoboken, NJ, 2013. [Google Scholar]
- Morse, N.B.; Pellissier, P.A.; Cianciola, E.N.; Brereton, R.L.; Sullivan, M.M.; Shonka, N.K.; Wheeler, T.B.; McDowell, W.H. Novel ecosystems in the Anthropocene: a revision of the novel ecosystem concept for pragmatic applications. Ecology and Society 2014, 19. [Google Scholar] [CrossRef]
- Goñi-Moreno, Á. Biocomputation: Moving Beyond Turing with Living Cellular Computers. Communications of the ACM 2024. [Google Scholar] [CrossRef]
- Solé, R.V.; Macia, J. Expanding the landscape of biological computation with synthetic multicellular consortia. Natural Computing 2013, 12, 485–497. [Google Scholar] [CrossRef]
- Johns, N.I.; Blazejewski, T.; Gomes, A.L.; Wang, H.H. Principles for designing synthetic microbial communities. Current Opinion in Microbiology 2016, 31, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.Y.; Vila, J.C.; Bender, M.; Li, R.; Mankowski, M.C.; Bassette, M.; Borden, J.; Golfier, S.; Sanchez, P.G.L.; Waymack, R.; others. Engineering complex communities by directed evolution. Nature ecology & evolution 2021, 5, 1011–1023. [Google Scholar]
- Saavedra, S.; Rohr, R.P.; Bascompte, J.; Godoy, O.; Kraft, N.J.; Levine, J.M. A structural approach for understanding multispecies coexistence. Ecological Monographs 2017, 87, 470–486. [Google Scholar] [CrossRef]
- Saavedra, S.; Medeiros, L.P.; AlAdwani, M. Structural forecasting of species persistence under changing environments. Ecology Letters 2020, 23, 1511–1521. [Google Scholar] [CrossRef] [PubMed]
- Marino, S.; Baxter, N.T.; Huffnagle, G.B.; Petrosino, J.F.; Schloss, P.D. Mathematical modeling of primary succession of murine intestinal microbiota. Proceedings of the National Academy of Sciences 2013, 111, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.R.; Bucci, V.; Toussaint, N.C.; Buffie, C.G.; Rätsch, G.; Pamer, E.G.; Sander, C.; Xavier, J.a.B. Ecological Modeling from Time-Series Inference: Insight into Dynamics and Stability of Intestinal Microbiota. PLoS Computational Biology 2013, 9, e1003388. [Google Scholar] [CrossRef] [PubMed]
- Buffie, C.G.; Bucci, V.; Stein, R.R.; McKenney, P.T.; Ling, L.; Gobourne, A.; No, D.; Liu, H.; Kinnebrew, M.; Viale, A.; Littmann, E.; van den Brink, M.R.M.; Jenq, R.R.; Taur, Y.; Sander, C.; Cross, J.R.; Toussaint, N.C.; Xavier, J.B.; Pamer, E.G. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2014, 517, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Coyte, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef] [PubMed]
- Joseph, T.A.; Shenhav, L.; Xavier, J.B.; Halperin, E.; Pe’er, I. Compositional Lotka-Volterra describes microbial dynamics in the simplex. PLOS Computational Biology 2020, 16, e1007917. [Google Scholar] [CrossRef]
- Remien, C.H.; Eckwright, M.J.; Ridenhour, B.J. Structural identifiability of the generalized Lotka-Volterra model for microbiome studies. Royal Society Open Science 2021, 8, 201378. [Google Scholar] [CrossRef]
- Long, C.; Deng, J.; Nguyen, J.; Liu, Y.Y.; Alm, E.J.; Solé, R.; Saavedra, S. Structured community transitions explain the switching capacity of microbial systems. Proceedings of the National Academy of Sciences 2024, 121, e2312521121. [Google Scholar] [CrossRef] [PubMed]
- Jansson, J.K. Soil viruses: Understudied agents of soil ecology. Environmental Microbiology 2023, 25, 143. [Google Scholar] [CrossRef] [PubMed]
- Weitz, J.S. Quantitative viral ecology: dynamics of viruses and their microbial hosts; Princeton University Press, 2016. [Google Scholar]
- Smith, J.M.; Szathmary, E. The major transitions in evolution; OUP Oxford, 1997. [Google Scholar]
- Solé, R. The major synthetic evolutionary transitions, 2016.
- Sole, R. Synthetic transitions: towards a new synthesis. Philosophical Transactions of the Royal Society B: Biological Sciences 2016, 371, 20150438. [Google Scholar] [CrossRef] [PubMed]
- Venturelli, O.S.; Carr, A.V.; Fisher, G.; Hsu, R.H.; Lau, R.; Bowen, B.P.; Hromada, S.; Northen, T.; Arkin, A.P. Deciphering microbial interactions in synthetic human gut microbiome communities. Molecular systems biology 2018, 14, e8157. [Google Scholar] [CrossRef] [PubMed]
- Weiss, A.S.; Burrichter, A.G.; Durai Raj, A.C.; von Strempel, A.; Meng, C.; Kleigrewe, K.; Münch, P.C.; Rössler, L.; Huber, C.; Eisenreich, W.; others. In vitro interaction network of a synthetic gut bacterial community. The ISME journal 2022, 16, 1095–1109. [Google Scholar] [CrossRef]
- Cheng, A.G.; Ho, P.Y.; Aranda-Díaz, A.; Jain, S.; Feiqiao, B.Y.; Meng, X.; Wang, M.; Iakiviak, M.; Nagashima, K.; Zhao, A.; others. Design, construction, and in vivo augmentation of a complex gut microbiome. Cell 2022, 185, 3617–3636. [Google Scholar] [CrossRef]
| a | A closed microbial system confined in a container would easily impose a carrying capacity due to space constraints, i.e. proximity-dependent inhibition. Conversely, an unbounded scenario would present a much more restrictive solution, being the only condition for long-term . |
| 1 | This is a simple mathematical model of the Allee effect, where a population will thrive, provided that minimal population size is achieved. This is obtained here by first finding the fixed points of the dynamics, i.e., the , such that . In this case, one solution is the extinction (desert) state , whereas two extra states are found by solving the quadratic equation . The solutions of this equation exist only if , which gives the critical condition. |

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




